Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries
Abstract
1. Introduction
1.1. Body Fat Distribution in Metabolic Diseases
1.2. Measures of Body Fat Distribution
1.3. Genetic and Non-Genetic Determinants of Body Fat Distribution
1.4. The Role of Ancestry in Genetic Studies
2. GWAS for Fat Distribution in European Populations
2.1. Loci Associated with WHR
2.2. Loci Associated with WC and Hip
3. GWAS for Fat Distribution in Asian Populations
3.1. Loci Associated with WHR
3.2. Loci Associated with WC
4. GWAS for Fat Distribution in African Populations
Loci Associated with WHR and WC
5. Sexual Dimorphism of Fat Distribution Loci in Different Ethnicities
5.1. Sexual Dimorphism of FD Loci in Europeans
5.2. Sexual Dimorphism of FD Loci in Asian
5.3. Sexual Dimorphism of FD Loci in Africans
6. Potential Regulatory Genes for Ectopic Fat Deposition
7. Molecular Mechanisms Underlying Trans-Population Differences in Fat Distribution
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Blüher, M. Obesity: Global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 2019, 15, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Karelis, A.D.; Faraj, M.; Bastard, J.-P.; St-Pierre, D.H.; Brochu, M.; Prud’homme, D.; Rabasa-Lhoret, R. The metabolically healthy but obese individual presents a favorable inflammation profile. J. Clin. Endocrinol. Metab. 2005, 90, 4145–4150. [Google Scholar] [CrossRef]
- Karelis, A.D.; St-Pierre, D.H.; Conus, F.; Rabasa-Lhoret, R.; Poehlman, E.T. Metabolic and body composition factors in subgroups of obesity: What do we know? J. Clin. Endocrinol. Metab. 2004, 89, 2569–2575. [Google Scholar] [CrossRef] [PubMed]
- Sims, E.A. Are there persons who are obese, but metabolically healthy? Metab. Clin. Exp. 2001, 50, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Kramer, C.K.; Zinman, B.; Retnakaran, R. Are metabolically healthy overweight and obesity benign conditions? A systematic review and meta-analysis. Ann. Intern. Med. 2013, 159, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.A.; Kivimaki, M.; Hamer, M. Metabolically healthy obesity and risk of incident type 2 diabetes: A meta-analysis of prospective cohort studies. Obes. Rev. 2014, 15, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Eckel, N.; Meidtner, K.; Kalle-Uhlmann, T.; Stefan, N.; Schulze, M.B. Metabolically healthy obesity and cardiovascular events: A systematic review and meta-analysis. Eur. J. Prev. Cardiol. 2016, 23, 956–966. [Google Scholar] [CrossRef]
- Lin, H.; Zhang, L.; Zheng, R.; Zheng, Y. The prevalence, metabolic risk and effects of lifestyle intervention for metabolically healthy obesity: A systematic review and meta-analysis: A PRISMA-compliant article. Medicine 2017, 96, e8838. [Google Scholar] [CrossRef]
- Fingeret, M.; Marques-Vidal, P.; Vollenweider, P. Incidence of type 2 diabetes, hypertension, and dyslipidemia in metabolically healthy obese and non-obese. Nutr. Metab. Cardiovasc. Dis. 2018, 28, 1036–1044. [Google Scholar] [CrossRef]
- Hwang, Y.-C.; Hayashi, T.; Fujimoto, W.Y.; Kahn, S.E.; Leonetti, D.L.; McNeely, M.J.; Boyko, E.J. Visceral abdominal fat accumulation predicts the conversion of metabolically healthy obese subjects to an unhealthy phenotype. Int. J. Obes. 2015, 39, 1365–1370. [Google Scholar] [CrossRef]
- Krotkiewski, M.; Björntorp, P.; Sjöström, L.; Smith, U. Impact of obesity on metabolism in men and women. Importance of regional adipose tissue distribution. J. Clin. Investig. 1983, 72, 1150–1162. [Google Scholar] [CrossRef]
- Björntorp, P. Metabolic implications of body fat distribution. Diabetes Care 1991, 14, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Rimm, E.B.; Stampfer, M.J.; Willett, W.C.; Hu, F.B. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am. J. Clin. Nutr. 2005, 81, 555–563. [Google Scholar] [CrossRef]
- Liu, J.; Fox, C.S.; Hickson, D.A.; May, W.D.; Hairston, K.G.; Carr, J.J.; Taylor, H.A. Impact of abdominal visceral and subcutaneous adipose tissue on cardiometabolic risk factors: The Jackson Heart Study. J. Clin. Endocrinol. Metab. 2010, 95, 5419–5426. [Google Scholar] [CrossRef]
- Lee, S.W.; Son, J.Y.; Kim, J.M.; Hwang, S.-S.; Han, J.S.; Heo, N.J. Body fat distribution is more predictive of all-cause mortality than overall adiposity. Diabetes Obes. Metab. 2018, 20, 141–147. [Google Scholar] [CrossRef]
- Fox, C.S.; Massaro, J.M.; Hoffmann, U.; Pou, K.M.; Maurovich-Horvat, P.; Liu, C.-Y.; Vasan, R.S.; Murabito, J.M.; Meigs, J.B.; Cupples, L.A.; et al. Abdominal visceral and subcutaneous adipose tissue compartments: Association with metabolic risk factors in the Framingham Heart Study. Circulation 2007, 116, 39–48. [Google Scholar] [CrossRef]
- Klein, S.; Allison, D.B.; Heymsfield, S.B.; Kelley, D.E.; Leibel, R.L.; Nonas, C.; Kahn, R. Waist circumference and cardiometabolic risk: A consensus statement from Shaping America’s Health: Association for Weight Management and Obesity Prevention; NAASO, The Obesity Society; the American Society for Nutrition; and the American Diabetes Association. Am. J. Clin. Nutr. 2007, 85, 1197–1202. [Google Scholar] [CrossRef] [PubMed]
- Stefan, N.; Häring, H.-U.; Hu, F.B.; Schulze, M.B. Metabolically healthy obesity: Epidemiology, mechanisms, and clinical implications. Lancet Diabetes Endocrinol. 2013, 1, 152–162. [Google Scholar] [CrossRef]
- Heid, I.M.; Jackson, A.U.; Randall, J.C.; Winkler, T.W.; Qi, L.; Steinthorsdottir, V.; Thorleifsson, G.; Zillikens, M.C.; Speliotes, E.K.; Mägi, R.; et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010, 42, 949–960. [Google Scholar] [CrossRef]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Winkler, T.W.; Justice, A.E.; Graff, M.; Barata, L.; Feitosa, M.F.; Chu, S.; Czajkowski, J.; Esko, T.; Fall, T.; Kilpeläinen, T.O.; et al. The Influence of Age and Sex on Genetic Associations with Adult Body Size and Shape: A Large-Scale Genome-Wide Interaction Study. PLoS Genet. 2015, 11, e1005378. [Google Scholar] [CrossRef] [PubMed]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Lotta, L.A.; Wittemans, L.B.L.; Zuber, V.; Stewart, I.D.; Sharp, S.J.; Luan, J.; Day, F.R.; Li, C.; Bowker, N.; Cai, L.; et al. Association of Genetic Variants Related to Gluteofemoral vs Abdominal Fat Distribution With Type 2 Diabetes, Coronary Disease, and Cardiovascular Risk Factors. JAMA 2018, 320, 2553–2563. [Google Scholar] [CrossRef] [PubMed]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef]
- Wen, W.; Cho, Y.-S.; Zheng, W.; Dorajoo, R.; Kato, N.; Qi, L.; Chen, C.-H.; Delahanty, R.J.; Okada, Y.; Tabara, Y.; et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat. Genet. 2012, 44, 307–311. [Google Scholar] [CrossRef]
- Wen, W.; Kato, N.; Hwang, J.-Y.; Guo, X.; Tabara, Y.; Li, H.; Dorajoo, R.; Yang, X.; Tsai, F.-J.; Li, S.; et al. Genome-wide association studies in East Asians identify new loci for waist-hip ratio and waist circumference. Sci. Rep. 2016, 6, 17958. [Google Scholar] [CrossRef]
- Cho, Y.S.; Go, M.J.; Kim, Y.J.; Heo, J.Y.; Oh, J.H.; Ban, H.-J.; Yoon, D.; Lee, M.H.; Kim, D.-J.; Park, M.; et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009, 41, 527–534. [Google Scholar] [CrossRef]
- Scott, W.R.; Zhang, W.; Loh, M.; Tan, S.-T.; Lehne, B.; Afzal, U.; Peralta, J.; Saxena, R.; Ralhan, S.; Wander, G.S.; et al. Investigation of Genetic Variation Underlying Central Obesity amongst South Asians. PLoS ONE 2016, 11, e0155478. [Google Scholar] [CrossRef]
- Liu, C.-T.; Buchkovich, M.L.; Winkler, T.W.; Heid, I.M.; Borecki, I.B.; Fox, C.S.; Mohlke, K.L.; North, K.E.; Adrienne Cupples, L. Multi-ethnic fine-mapping of 14 central adiposity loci. Hum. Mol. Genet. 2014, 23, 4738–4744. [Google Scholar] [CrossRef]
- Yoneyama, S.; Yao, J.; Guo, X.; Fernandez-Rhodes, L.; Lim, U.; Boston, J.; Buzková, P.; Carlson, C.S.; Cheng, I.; Cochran, B.; et al. Generalization and fine mapping of European ancestry-based central adiposity variants in African ancestry populations. Int. J. Obes. 2017, 41, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Wajchenberg, B.L. Subcutaneous and visceral adipose tissue: Their relation to the metabolic syndrome. Endocr. Rev. 2000, 21, 697–738. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.C.K. Sexual dimorphism of body composition. Best Pract. Res. Clin. Endocrinol. Metab. 2007, 21, 415–430. [Google Scholar] [CrossRef]
- Selby, J.V.; Newman, B.; Quesenberry, C.P.; Fabsitz, R.R.; Carmelli, D.; Meaney, F.J.; Slemenda, C. Genetic and behavioral influences on body fat distribution. Int. J. Obes. 1990, 14, 593–602. [Google Scholar]
- Rose, K.M.; Newman, B.; Mayer-Davis, E.J.; Selby, J.V. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 1998, 6, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Rask-Andersen, M.; Karlsson, T.; Ek, W.E.; Johansson, Å. Genome-wide association study of body fat distribution identifies adiposity loci and sex-specific genetic effects. Nat. Commun. 2019, 10, 339. [Google Scholar] [CrossRef]
- Justice, A.E.; Karaderi, T.; Highland, H.M.; Young, K.L.; Graff, M.; Lu, Y.; Turcot, V.; Auer, P.L.; Fine, R.S.; Guo, X.; et al. Protein-coding variants implicate novel genes related to lipid homeostasis contributing to body-fat distribution. Nat. Genet. 2019, 51, 452–469. [Google Scholar] [CrossRef] [PubMed]
- Pinnick, K.E.; Nicholson, G.; Manolopoulos, K.N.; McQuaid, S.E.; Valet, P.; Frayn, K.N.; Denton, N.; Min, J.L.; Zondervan, K.T.; Fleckner, J.; et al. Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes 2014, 63, 3785–3797. [Google Scholar] [CrossRef] [PubMed]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Després, J.-P.; Hu, F.B. Sugar-sweetened beverages, obesity, type 2 diabetes mellitus, and cardiovascular disease risk. Circulation 2010, 121, 1356–1364. [Google Scholar] [CrossRef]
- Schleinitz, D.; Böttcher, Y.; Blüher, M.; Kovacs, P. The genetics of fat distribution. Diabetologia 2014, 57, 1276–1286. [Google Scholar] [CrossRef]
- Pérusse, L.; Després, J.P.; Lemieux, S.; Rice, T.; Rao, D.C.; Bouchard, C. Familial aggregation of abdominal visceral fat level: Results from the Quebec family study. Metab. Clin. Exp. 1996, 45, 378–382. [Google Scholar] [CrossRef]
- Malis, C.; Rasmussen, E.L.; Poulsen, P.; Petersen, I.; Christensen, K.; Beck-Nielsen, H.; Astrup, A.; Vaag, A.A. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes. Res. 2005, 13, 2139–2145. [Google Scholar] [CrossRef]
- Visscher, P.M.; Hill, W.G.; Wray, N.R. Heritability in the genomics era—Concepts and misconceptions. Nat. Rev. Genet. 2008, 9, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Thornton, J.C.; Russell, M.; Burastero, S.; Heymsfield, S.; Pierson, R.N. Asians have lower body mass index (BMI) but higher percent body fat than do whites: Comparisons of anthropometric measurements. Am. J. Clin. Nutr. 1994, 60, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Deurenberg, P.; Yap, M.; van Staveren, W.A. Body mass index and percent body fat: A meta analysis among different ethnic groups. Int. J. Obes. Relat. Metab. Disord. 1998, 22, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Deurenberg-Yap, M.; Schmidt, G.; van Staveren, W.A.; Deurenberg, P. The paradox of low body mass index and high body fat percentage among Chinese, Malays and Indians in Singapore. Int. J. Obes. Relat. Metab. Disord. 2000, 24, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Lear, S.A.; Humphries, K.H.; Kohli, S.; Chockalingam, A.; Frohlich, J.J.; Birmingham, C.L. Visceral adipose tissue accumulation differs according to ethnic background: Results of the Multicultural Community Health Assessment Trial (M-CHAT). Am. J. Clin. Nutr. 2007, 86, 353–359. [Google Scholar] [CrossRef]
- Goedecke, J.H.; Tootla, M.; Keswell, D. Ethnic differences in regional adipose tissue oestrogen receptor gene expression. Endocr. Connect. 2019, 8, 32–38. [Google Scholar] [CrossRef]
- Rønn, P.F.; Andersen, G.S.; Lauritzen, T.; Christensen, D.L.; Aadahl, M.; Carstensen, B.; Jørgensen, M.E. Ethnic differences in anthropometric measures and abdominal fat distribution: A cross-sectional pooled study in Inuit, Africans and Europeans. J. Epidemiol. Community Health 2017, 71, 536–543. [Google Scholar] [CrossRef]
- Lee, S.; Kuk, J.L. Visceral fat is associated with the racial differences in liver fat between black and white adolescent boys with obesity. Pediatr. Diabetes 2017, 18, 660–663. [Google Scholar] [CrossRef]
- Yoon, K.-H.; Lee, J.-H.; Kim, J.-W.; Cho, J.H.; Choi, Y.-H.; Ko, S.-H.; Zimmet, P.; Son, H.-Y. Epidemic obesity and type 2 diabetes in Asia. Lancet 2006, 368, 1681–1688. [Google Scholar] [CrossRef]
- Wong, R.J.; Chou, C.; Sinha, S.R.; Kamal, A.; Ahmed, A. Ethnic disparities in the association of body mass index with the risk of hypertension and diabetes. J. Community Health 2014, 39, 437–445. [Google Scholar] [CrossRef]
- Gray, S.L.; Vidal-Puig, A.J. Adipose tissue expandability in the maintenance of metabolic homeostasis. Nutr. Rev. 2007, 65, S7–S12. [Google Scholar] [CrossRef]
- Sohail, M.; Maier, R.M.; Ganna, A.; Bloemendal, A.; Martin, A.R.; Turchin, M.C.; Chiang, C.W.; Hirschhorn, J.; Daly, M.J.; Patterson, N.; et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. Elife 2019, 8. [Google Scholar] [CrossRef]
- Duncan, L.; Shen, H.; Gelaye, B.; Meijsen, J.; Ressler, K.; Feldman, M.; Peterson, R.; Domingue, B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019, 10, 3328. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Kovacs, P.; Guiu-Jurado, E. Genetics of Obesity in East Asians. Front. Genet. 2020, 11, 575049. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.J.; Harpak, A.; Sinnott-Armstrong, N.; Joergensen, A.M.; Mostafavi, H.; Field, Y.; Boyle, E.A.; Zhang, X.; Racimo, F.; Pritchard, J.K.; et al. Reduced signal for polygenic adaptation of height in UK Biobank. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Bamshad, M.; Wooding, S.; Salisbury, B.A.; Stephens, J.C. Deconstructing the relationship between genetics and race. Nat. Rev. Genet. 2004, 5, 598–609. [Google Scholar] [CrossRef]
- Rito, T.; Vieira, D.; Silva, M.; Conde-Sousa, E.; Pereira, L.; Mellars, P.; Richards, M.B.; Soares, P. A dispersal of Homo sapiens from southern to eastern Africa immediately preceded the out-of-Africa migration. Sci. Rep. 2019, 9, 4728. [Google Scholar] [CrossRef]
- Nielsen, R.; Akey, J.M.; Jakobsson, M.; Pritchard, J.K.; Tishkoff, S.; Willerslev, E. Tracing the peopling of the world through genomics. Nature 2017, 541, 302–310. [Google Scholar] [CrossRef]
- Mathieson, I.; Lazaridis, I.; Rohland, N.; Mallick, S.; Patterson, N.; Roodenberg, S.A.; Harney, E.; Stewardson, K.; Fernandes, D.; Novak, M.; et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 2015, 528, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, P.d.B.; Marchi, N.; Rasmussen, S.; Peyrot, M.; Renaud, G.; Korneliussen, T.; Moreno-Mayar, J.V.; Pedersen, M.W.; Goldberg, A.; Usmanova, E.; et al. 137 ancient human genomes from across the Eurasian steppes. Nature 2018, 557, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, V.M.; Patterson, N.; Moorjani, P.; Rohland, N.; Bernardos, R.; Mallick, S.; Lazaridis, I.; Nakatsuka, N.; Olalde, I.; Lipson, M.; et al. The formation of human populations in South and Central Asia. Science 2019, 365. [Google Scholar] [CrossRef]
- McColl, H.; Racimo, F.; Vinner, L.; Demeter, F.; Gakuhari, T.; Moreno-Mayar, J.V.; van Driem, G.; Gram Wilken, U.; Seguin-Orlando, A.; de La Fuente Castro, C.; et al. The prehistoric peopling of Southeast Asia. Science 2018, 361, 88–92. [Google Scholar] [CrossRef]
- Novembre, J.; Johnson, T.; Bryc, K.; Kutalik, Z.; Boyko, A.R.; Auton, A.; Indap, A.; King, K.S.; Bergmann, S.; Nelson, M.R.; et al. Genes mirror geography within Europe. Nature 2008, 456, 98–101. [Google Scholar] [CrossRef]
- Karakachoff, M.; Duforet-Frebourg, N.; Simonet, F.; Le Scouarnec, S.; Pellen, N.; Lecointe, S.; Charpentier, E.; Gros, F.; Cauchi, S.; Froguel, P.; et al. Fine-scale human genetic structure in Western France. Eur. J. Hum. Genet. 2015, 23, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-Y.; Novembre, J.; Eskin, E.; Halperin, E. A model-based approach for analysis of spatial structure in genetic data. Nat. Genet. 2012, 44, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, C.M.; Heid, I.M.; Randall, J.C.; Lamina, C.; Steinthorsdottir, V.; Qi, L.; Speliotes, E.K.; Thorleifsson, G.; Willer, C.J.; Herrera, B.M.; et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009, 5, e1000508. [Google Scholar] [CrossRef]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Süveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef] [PubMed]
- Kraja, A.T.; Chasman, D.I.; North, K.E.; Reiner, A.P.; Yanek, L.R.; Kilpeläinen, T.O.; Smith, J.A.; Dehghan, A.; Dupuis, J.; Johnson, A.D.; et al. Pleiotropic genes for metabolic syndrome and inflammation. Mol. Genet. Metab. 2014, 112, 317–338. [Google Scholar] [CrossRef]
- Cha, S.; Park, A.Y.; Kang, C. A Genome-Wide Association Study Uncovers a Genetic Locus Associated with Thoracic-to-Hip Ratio in Koreans. PLoS ONE 2015, 10, e0145220. [Google Scholar] [CrossRef] [PubMed]
- Patlaka, C.; Mai, H.A.; Lång, P.; Andersson, G. The growth factor-like adipokine tartrate-resistant acid phosphatase 5a interacts with the rod G3 domain of adipocyte-produced nidogen-2. Biochem. Biophys. Res. Commun. 2014, 454, 446–452. [Google Scholar] [CrossRef]
- Zhao, L.P.; Alshiekh, S.; Zhao, M.; Carlsson, A.; Larsson, H.E.; Forsander, G.; Ivarsson, S.A.; Ludvigsson, J.; Kockum, I.; Marcus, C.; et al. Next-Generation Sequencing Reveals That HLA-DRB3, -DRB4, and -DRB5 May Be Associated With Islet Autoantibodies and Risk for Childhood Type 1 Diabetes. Diabetes 2016, 65, 710–718. [Google Scholar] [CrossRef]
- Wu, Y.; Duan, H.; Tian, X.; Xu, C.; Wang, W.; Jiang, W.; Pang, Z.; Zhang, D.; Tan, Q. Genetics of Obesity Traits: A Bivariate Genome-Wide Association Analysis. Front. Genet. 2018, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Chambers, J.C.; Elliott, P.; Zabaneh, D.; Zhang, W.; Li, Y.; Froguel, P.; Balding, D.; Scott, J.; Kooner, J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008, 40, 716–718. [Google Scholar] [CrossRef]
- Berndt, S.I.; Gustafsson, S.; Mägi, R.; Ganna, A.; Wheeler, E.; Feitosa, M.F.; Justice, A.E.; Monda, K.L.; Croteau-Chonka, D.C.; Day, F.R.; et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013, 45, 501–512. [Google Scholar] [CrossRef]
- Lango Allen, H.; Estrada, K.; Lettre, G.; Berndt, S.I.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature 2010, 467, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-T.; Monda, K.L.; Taylor, K.C.; Lange, L.; Demerath, E.W.; Palmas, W.; Wojczynski, M.K.; Ellis, J.C.; Vitolins, M.Z.; Liu, S.; et al. Genome-wide association of body fat distribution in African ancestry populations suggests new loci. PLoS Genet. 2013, 9, e1003681. [Google Scholar] [CrossRef]
- Morgan, M.D.; Pairo-Castineira, E.; Rawlik, K.; Canela-Xandri, O.; Rees, J.; Sims, D.; Tenesa, A.; Jackson, I.J. Genome-wide study of hair colour in UK Biobank explains most of the SNP heritability. Nat. Commun. 2018, 9, 5271. [Google Scholar] [CrossRef]
- Garaulet, M.; Pérex-Llamas, F.; Fuente, T.; Zamora, S.; Tebar, F.J. Anthropometric, computed tomography and fat cell data in an obese population: Relationship with insulin, leptin, tumor necrosis factor-alpha, sex hormone-binding globulin and sex hormones. Eur. J. Endocrinol. 2000, 143, 657–666. [Google Scholar] [CrossRef]
- Zillikens, M.C.; Yazdanpanah, M.; Pardo, L.M.; Rivadeneira, F.; Aulchenko, Y.S.; Oostra, B.A.; Uitterlinden, A.G.; Pols, H.A.P.; van Duijn, C.M. Sex-specific genetic effects influence variation in body composition. Diabetologia 2008, 51, 2233–2241. [Google Scholar] [CrossRef]
- Trouwborst, I.; Bowser, S.M.; Goossens, G.H.; Blaak, E.E. Ectopic Fat Accumulation in Distinct Insulin Resistant Phenotypes; Targets for Personalized Nutritional Interventions. Front. Nutr. 2018, 5, 77. [Google Scholar] [CrossRef]
- Shulman, G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N. Engl. J. Med. 2014, 371, 2237–2238. [Google Scholar] [CrossRef]
- Rosito, G.A.; Massaro, J.M.; Hoffmann, U.; Ruberg, F.L.; Mahabadi, A.A.; Vasan, R.S.; O’Donnell, C.J.; Fox, C.S. Pericardial fat, visceral abdominal fat, cardiovascular disease risk factors, and vascular calcification in a community-based sample: The Framingham Heart Study. Circulation 2008, 117, 605–613. [Google Scholar] [CrossRef]
- Ding, J.; Hsu, F.-C.; Harris, T.B.; Liu, Y.; Kritchevsky, S.B.; Szklo, M.; Ouyang, P.; Espeland, M.A.; Lohman, K.K.; Criqui, M.H.; et al. The association of pericardial fat with incident coronary heart disease: The Multi-Ethnic Study of Atherosclerosis (MESA). Am. J. Clin. Nutr. 2009, 90, 499–504. [Google Scholar] [CrossRef]
- Fox, C.S.; Liu, Y.; White, C.C.; Feitosa, M.; Smith, A.V.; Heard-Costa, N.; Lohman, K.; Johnson, A.D.; Foster, M.C.; Greenawalt, D.M.; et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012, 8, e1002695. [Google Scholar] [CrossRef] [PubMed]
- Chu, A.Y.; Deng, X.; Fisher, V.A.; Drong, A.; Zhang, Y.; Feitosa, M.F.; Liu, C.-T.; Weeks, O.; Choh, A.C.; Duan, Q.; et al. Multiethnic genome-wide meta-analysis of ectopic fat depots identifies loci associated with adipocyte development and differentiation. Nat. Genet. 2017, 49, 125–130. [Google Scholar] [CrossRef]
- Foster, M.C.; Yang, Q.; Hwang, S.-J.; Hoffmann, U.; Fox, C.S. Heritability and genome-wide association analysis of renal sinus fat accumulation in the Framingham Heart Study. BMC Med. Genet. 2011, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.S.; White, C.C.; Lohman, K.; Heard-Costa, N.; Cohen, P.; Zhang, Y.; Johnson, A.D.; Emilsson, V.; Liu, C.-T.; Chen, Y.-D.I.; et al. Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS Genet. 2012, 8, e1002705. [Google Scholar] [CrossRef]
- Mahajan, A.; Go, M.J.; Zhang, W.; Below, J.E.; Gaulton, K.J.; Ferreira, T.; Horikoshi, M.; Johnson, A.D.; Ng, M.C.Y.; Prokopenko, I.; et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat. Genet. 2014, 46, 234–244. [Google Scholar] [CrossRef] [PubMed]
- Gruzdeva, O.; Borodkina, D.; Uchasova, E.; Dyleva, Y.; Barbarash, O. Localization of fat depots and cardiovascular risk. Lipids Health Dis. 2018, 17, 218. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Shin, J.-Y.; Yee, J.; Park, T.; Park, M. Structural equation modeling for hypertension and type 2 diabetes based on multiple SNPs and multiple phenotypes. PLoS ONE 2019, 14, e0217189. [Google Scholar] [CrossRef]
- Akiyama, M.; Okada, Y.; Kanai, M.; Takahashi, A.; Momozawa, Y.; Ikeda, M.; Iwata, N.; Ikegawa, S.; Hirata, M.; Matsuda, K.; et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat. Genet. 2017, 49, 1458–1467. [Google Scholar] [CrossRef]
- Division of Cancer Epidemiology & Genetics. LDlink. Available online: https://ldlink.nci.nih.gov/?tab=home (accessed on 1 August 2020).
- Kohfeldt, E.; Sasaki, T.; Göhring, W.; Timpl, R. Nidogen-2: A new basement membrane protein with diverse binding properties. J. Mol. Biol. 1998, 282, 99–109. [Google Scholar] [CrossRef]
- Miosge, N.; Holzhausen, S.; Zelent, C.; Sprysch, P.; Herken, R. Nidogen-1 and nidogen-2 are found in basement membranes during human embryonic development. Histochem. J. 2001, 33, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Miosge, N.; Sasaki, T.; Timpl, R. Evidence of nidogen-2 compensation for nidogen-1 deficiency in transgenic mice. Matrix Biol. 2002, 21, 611–621. [Google Scholar] [CrossRef]
- Spracklen, C.N.; Horikoshi, M.; Kim, Y.J.; Lin, K.; Bragg, F.; Moon, S.; Suzuki, K.; Tam, C.H.T.; Tabara, Y.; Kwak, S.-H.; et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 2020, 582, 240–245. [Google Scholar] [CrossRef]
- Zhang, H.; Mo, X.-B.; Xu, T.; Bu, X.-Q.; Lei, S.-F.; Zhang, Y.-H. Novel Genes Affecting Blood Pressure Detected Via Gene-Based Association Analysis. G3 2015, 5, 1035–1042. [Google Scholar] [CrossRef]
- Kim, J.; Oh, B.; Lim, J.E.; Kim, M.K. No Interaction with Alcohol Consumption, but Independent Effect of C12orf51 (HECTD4) on Type 2 Diabetes Mellitus in Korean Adults Aged 40-69 Years: The KoGES_Ansan and Ansung Study. PLoS ONE 2016, 11, e0149321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Kang, L.; Bond, W.; Zhang, N. Interaction between syntaxin 8 and HECTd3, a HECT domain ligase. Cell. Mol. Neurobiol. 2009, 29, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Lancha, A.; López-Garrido, S.; Rodríguez, A.; Catalán, V.; Ramírez, B.; Valentí, V.; Moncada, R.; Silva, C.; Gil, M.J.; Salvador, J.; et al. Expression of syntaxin 8 in visceral adipose tissue is increased in obese patients with type 2 diabetes and related to markers of insulin resistance and inflammation. Arch. Med. Res. 2015, 46, 47–53. [Google Scholar] [CrossRef]
- Pham, D.D.; Ku, B.; Shin, C.; Cho, N.H.; Cha, S.; Kim, J.Y. Thoracic-to-hip circumference ratio as a novel marker of type 2 diabetes, independent of body mass index and waist-to-hip ratio, in Korean adults. Diabetes Res. Clin. Pract. 2014, 104, 273–280. [Google Scholar] [CrossRef]
- Weinstein, L.S.; Xie, T.; Zhang, Q.-H.; Chen, M. Studies of the regulation and function of the Gs alpha gene Gnas using gene targeting technology. Pharmacol. Ther. 2007, 115, 271–291. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Chen, M.; Zhang, Q.-H.; Ma, Z.; Weinstein, L.S. Beta cell-specific deficiency of the stimulatory G protein alpha-subunit Gsalpha leads to reduced beta cell mass and insulin-deficient diabetes. Proc. Natl. Acad. Sci. USA 2007, 104, 19601–19606. [Google Scholar] [CrossRef]
- Taneera, J.; Dhaiban, S.; Mohammed, A.K.; Mukhopadhyay, D.; Aljaibeji, H.; Sulaiman, N.; Fadista, J.; Salehi, A. GNAS gene is an important regulator of insulin secretory capacity in pancreatic β-cells. Gene 2019, 715, 144028. [Google Scholar] [CrossRef]
- Chen, M.; Feng, H.-Z.; Gupta, D.; Kelleher, J.; Dickerson, K.E.; Wang, J.; Hunt, D.; Jou, W.; Gavrilova, O.; Jin, J.-P.; et al. G(s)alpha deficiency in skeletal muscle leads to reduced muscle mass, fiber-type switching, and glucose intolerance without insulin resistance or deficiency. Am. J. Physiol. Cell Physiol. 2009, 296, C930–C940. [Google Scholar] [CrossRef]
- Chen, M.; Chen, H.; Nguyen, A.; Gupta, D.; Wang, J.; Lai, E.W.; Pacak, K.; Gavrilova, O.; Quon, M.J.; Weinstein, L.S. G(s)alpha deficiency in adipose tissue leads to a lean phenotype with divergent effects on cold tolerance and diet-induced thermogenesis. Cell Metab. 2010, 11, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Kashi, Z.; Masoumi, P.; Mahrooz, A.; Hashemi-Soteh, M.B.; Bahar, A.; Alizadeh, A. The variant organic cation transporter 2 (OCT2)-T201M contribute to changes in insulin resistance in patients with type 2 diabetes treated with metformin. Diabetes Res. Clin. Pract. 2015, 108, 78–83. [Google Scholar] [CrossRef]
- Hou, W.; Zhang, D.; Lu, W.; Zheng, T.; Wan, L.; Li, Q.; Bao, Y.; Liu, F.; Jia, W. Polymorphism of organic cation transporter 2 improves glucose-lowering effect of metformin via influencing its pharmacokinetics in Chinese type 2 diabetic patients. Mol. Diagn. Ther. 2015, 19, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Graff, M.; Scott, R.A.; Justice, A.E.; Young, K.L.; Feitosa, M.F.; Barata, L.; Winkler, T.W.; Chu, A.Y.; Mahajan, A.; Hadley, D.; et al. Genome-wide physical activity interactions in adiposity—A meta-analysis of 200,452 adults. PLoS Genet. 2017, 13, e1006528. [Google Scholar] [CrossRef] [PubMed]
- Randall, J.C.; Winkler, T.W.; Kutalik, Z.; Berndt, S.I.; Jackson, A.U.; Monda, K.L.; Kilpeläinen, T.O.; Esko, T.; Mägi, R.; Li, S.; et al. Sex-stratified genome-wide association studies including 270,000 individuals show sexual dimorphism in genetic loci for anthropometric traits. PLoS Genet. 2013, 9, e1003500. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, X.; Peng, D.; Zhang, R.; Sun, X.; Chen, M.; Yan, J.; Wang, S.; Yan, D.; He, Z.; et al. Effects of Obesity Related Genetic Variations on Visceral and Subcutaneous Fat Distribution in a Chinese Population. Sci. Rep. 2016, 6, 20691. [Google Scholar] [CrossRef] [PubMed]
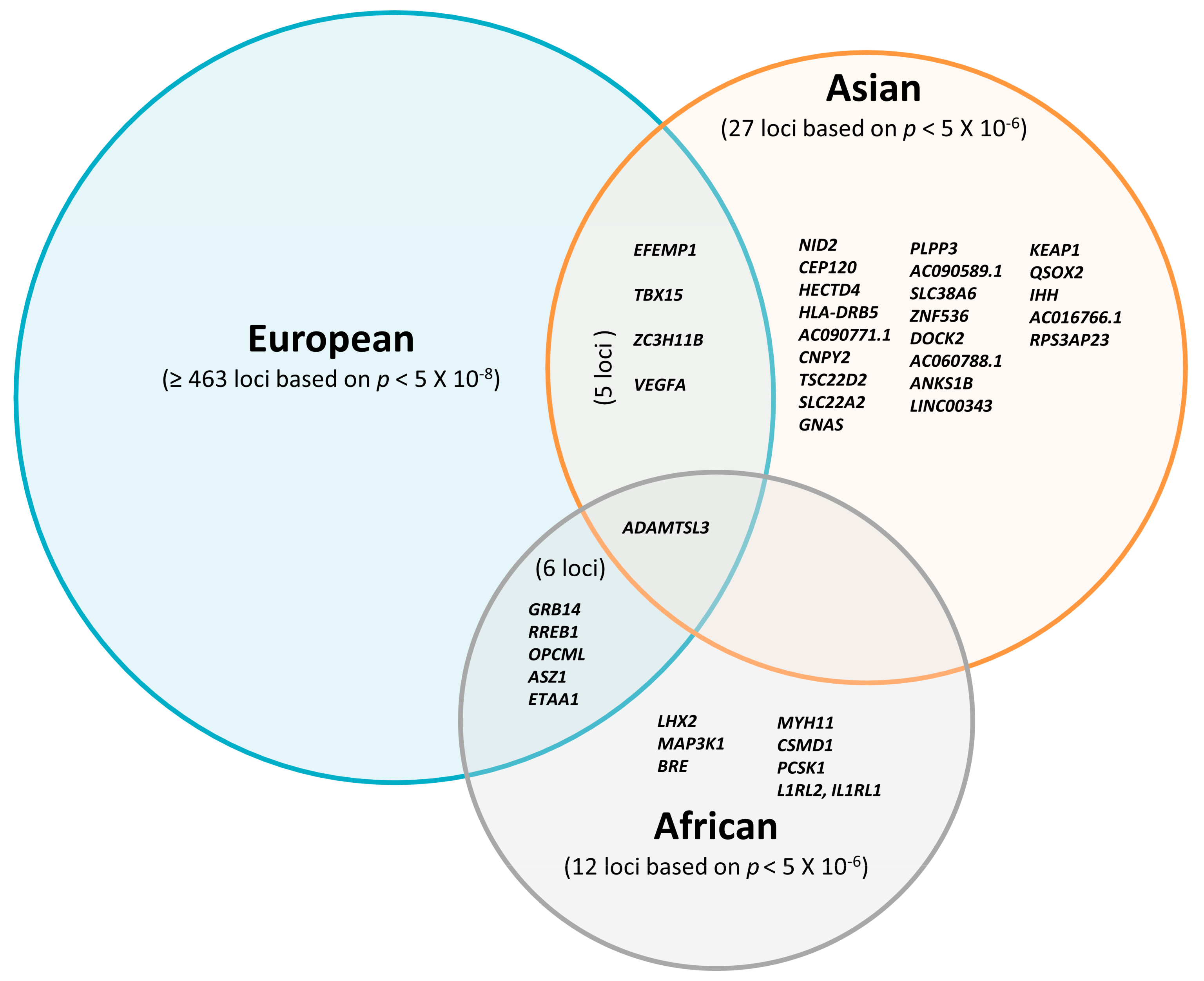
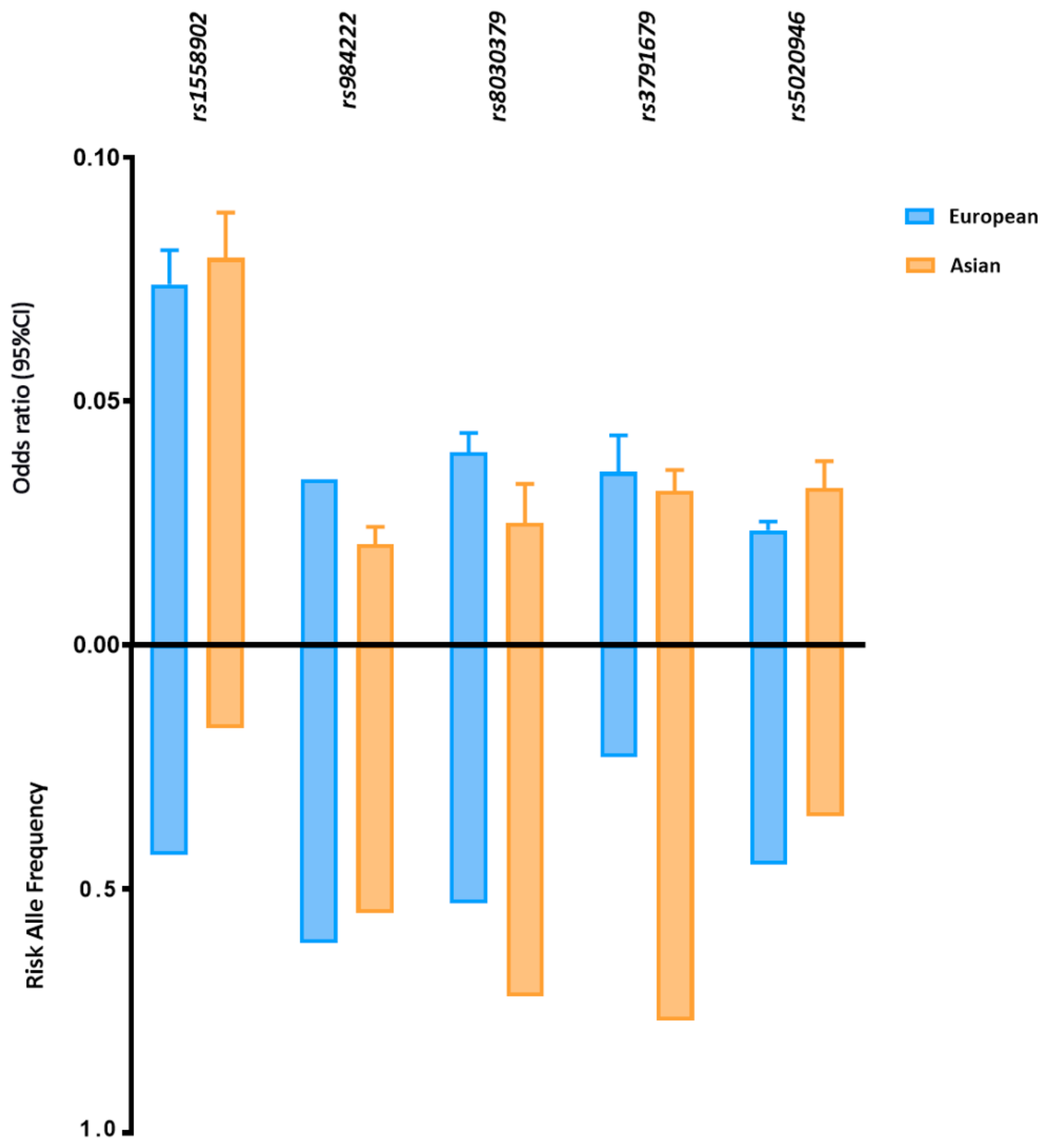
| Study Type | Publication Year | Sample Size | Male/Female | Cohort Age Mean (SD) | Traits | Criteria for Discovery c | N° of Variants d (Discovery Stage) | Criteria for Replication e | N° of Variants f (Replication Stage) | N° of Sexual Dimorphism Variants | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS | 2009 | 140,644 (Dis a: 38,580 European; Rep b: 102,064 European) | NA | 55.73 (9.67) | WC | WC/WHR, p < 1 × 10−5 BMI p > 0.01, Height p > 0.005 | 26 | p < 5 × 10−8 | 2 | NA | [67] |
| WHR | 1 | NA | |||||||||
| Meta-analysis GWAS | 2010 | 190,803 (Dis a: 77,167 European; Rep b: 113,636 European) | 82,483/108,979 | NA | WHR | p < 1.4 × 10−6 | 16 | p < 5 × 10−8 | 14 | 7 | [19] |
| Meta-analysis GWAS | 2015 | 210,088 (Dis a: 142,762 European; Rep b: 67,326 European) | 95,379/114,709 | 50 (10.3) | WHRadjBMI | NA | NA | p < 5 × 10−8 | 49 | 20 | [20] |
| Meta-analysis GWAS | 2015 | 320,485 Europeans | 51,625/60,654 | <=50 | WHRadjBMI | p < 1 × 10−5 | NA | p < 5 × 10−8 | 44 | 44 | [21] |
| 90,988/106,622 | >50 | ||||||||||
| Meta-analysis GWAS | 2018 | 663,598 Europeans | 206,951/245,351 * | 57 (8) * | WHR | p < 5 × 10−8 | 202 | NA | NA | NA | [23] |
| 660,648 Europeans | WHRadjBMI | p < 5 × 10−8 | |||||||||
| Meta-analysis GWAS | 2019 | 694,649 Europeans | NA | NA | WHRadjBMI | p < 5 × 10−9 | 346 | NA | NA | 105 | [24] |
| GWAS | 2017 | 239,856 Europeans (Dia a: 33,811, Rep b: 206,045) | 123,766/116,090 | 51.4 | WHRadjBMI | p < 1 × 10−5 | 85 | p < 5 × 10−8 | 1 | 3 | [69] |
| 54,693 Europeans (Dia a: 34,088, Rep b: 206,053) | 28,276/26,417 | 51.4 | WHR | 80 | 1 | 0 | |||||
| 67,767 Europeans (Dia a: 47,095, Rep b: 206,723) | 34,494/33,273 | 52 | WCadjBMI | 139 | 7 | 3 | |||||
| 68,267 Europeans (Dia a: 47,593, Rep b: 20,6737) | 33,792/34,475 | 50.1 | WC | 118 | 12 | 5 | |||||
| 54,595 Europeans (Dia a: 34,004, Rep b: 205,909) | 28,008/26,587 | 51.4 | HIP | 114 | 7 | 2 |
| Study Type | Publication Year | Sample Size | Male/Female | Cohort Age Mean (SD) | Traits | Criteria for Discovery | N° of Variants d (Discovery Stage) | Criteria for Replication e | N° of Variants f (Replication Stage) | N° of Variants Replicated from g | N° of Variants Successfully Replicated from g | N° of Sexual Dimorphism Variants | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (1) Studies conducted in Asian populations | |||||||||||||
| GWAS | 2009 | 16,703 (Dis a: 8842 East Asian; Rep b: 7861 East Asian) | 7397/9306 | 54.4 (8.4) | WHR | p < 1.0 × 10−5 | 2 | p < 5 × 10−2 | 1 | NA | NA | NA | [27] |
| GWAS | 2016 | 73,596 East Asian (Dis: 48,312, Rep: 25,284) | 31,570/42,026 | 54.4 (9) | WHRadjBMI | p < 1.0 × 10−6 | 33 | p < 5 × 10−8 | 2 | 60 | 11 (1 × 10−3) | 3 (10 from h) | [26] |
| WHRnoBMI | 33 | p < 5 × 10−8 | 0 | 13 (1 × 10−3) | |||||||||
| 2016 | 78,336 East Asian (Dis: 53,052, Rep: 25,284) | 36,310/42,026 | 54.4 (9) | WCadjBMI | p < 1.0 × 10−6 | 33 | p < 5 × 10−8 | 4 | 60 | 7 (1 × 10−3) | |||
| WCnoBMI | 33 | p < 5 × 10−8 | 3 | 60 | 15 (1 × 10−3) | ||||||||
| GWAS | 2016 | 12,240 (Dis a: 10,318 South Asian; Rep b: 1922 South Asian) | 9825/2415 | 50.5 (11.2) | WHR | p < 5 × 10−8 p < 5 × 10−2 | 0 | p < 5 × 10−2 | 0 | 48 | 4 (p < 0.05) | 2 (11from known WHR) | [28] |
| Exome-Wide Association Study | 2016 | 2637 South Asian | 1798/839 | 51.6 (10.1) | WHR | p < 1.5 × 10−6 (single variant) p < 2.5 × 10−6 (gene-based analyses) | 0 | p < 5 × 10−2 | 0 | NA | NA | NA | [28] |
| GWAS | 2018 | 274 East Asian (139 dizygotic twin pairs) | NA | over 30 | BMI-WHR | p < 1.0 × 10−5 | 26 | NA | NA | NA | NA | NA | [74] |
| GWAS | 2008 | 14,639 (Dis a: 2684 South Asian, Rep b: 7394 South Asian and 4561 European) | 9954/4685 | 51.1 (11) | WC | p < 1.0 × 10−5 | 31 | p < 5 × 10−7 | 4 | NA | NA | NA | [75] |
| (2) Studies conducted in African populations | |||||||||||||
| GWAS | 2013 | 33,738 (Dis a: 23,564, Rep b: 19,744 African American or Afro-Caribbean) | 9224/24,514 | 56.1 (9.5) | WC | p < 5 × 10−6 | 25 | p < 5 × 10−8 | 1 | NA | NA | NA | [78] |
| 27,489 (Dis a: 19,744, Rep b: 7745 African American or Afro-Caribbean) | 6446/21,043 | 55.1 (9.7) | WHR | p < 5 × 10−6 | p < 5 × 10−8 | 1 | 14 | 6 | 1 | ||||
| Fine mapping | 2014 | 19,744 African American | 10,318/9426 | 51.4 (9.4) | WHR | NA | NA | p < 5 × 10−8 | NA | 14 | 12 | NA | [29] |
| Fine mapping | 2016 | 15,981 African American | 3884/12,097 | 54.1 (7.8) | WC | NA | NA | p < 9.97 × 10−5 | NA | 17 | 0 | 0 | [30] |
| WHR | NA | NA | p < 9.97 × 10−5 | NA | 17 | 8 | 5 | ||||||
| SNP | Candidate Gene(s) a | Chr b | Allele c ALT/REF d | RAF e, f | ß-Estimates c (SE) e | p-Value | Reported Traits g | Reference | Explained Variance (%) f |
|---|---|---|---|---|---|---|---|---|---|
| rs1982963 | NID2 | 14 | A/G | 0.85 | 0.048 (0.012) | 1.0 × 10−14 | WHRadjBMI | [26] | 0.059 |
| rs10051787 | CEP120 | 5 | C/T | 0.59 | −0.04 (0.012) | 7.0 × 10−12 | WC | [26] | 0.078 |
| rs2074356 | HECTD4 | 12 | A/G | 0.13 | 0.006 (0.002) | 8.0 × 10−12 | WHR | [27] | 0.001 |
| rs5020946 | HLA-DRB5 | 6 | T/G | 0.35 | 0.031 (0.01) | 1.0 × 10−9 | WHRadjBMI | [26] | 0.044 |
| rs12970134 | AC090771.1 | 18 | A/G | 0.32 | NA | 2.0 × 10−9 | WC | [75] | NA |
| rs3809128 | CNPY2 (AC073896.2) | 12 | T/C | 0.16 | −0.037 (0.012) | 4.0 × 10−9 | WCadjBMI | [26] | 0.037 |
| rs1868673 | TSC22D2 | 3 | C/A | 0.40 | −0.044 (0.016) | 1.0 × 10−8 | WC | [26] | 0.093 |
| rs368123 | SLC22A2 | 6 | G/A | 0.40 | 0.032 (0.0129) | 3.0 × 10−8 | WC | [26] | 0.049 |
| rs2057291 | GNAS | 20 | G/A | 0.77 | 0.025 (0.01) | 4.0 × 10−8 | WCadjBMI | [26] | 0.022 |
| rs4912314 | PLPP3 | 1 | T/C | 0.24 | 0.029 (0.012) | 3.0 × 10−7 | WHRadjBMI | [26] | NA |
| rs2025924 | LINC00343 | 13 | C/T | NA | NA | 4.0 × 10−7 | WHR | [74] | NA |
| rs3100776 | IHH | 2 | C/T | 0.47 | 0.017 (0.011) | 4.0 × 10−7 | WCadjBMI | [26] | NA |
| rs11103390 | QSOX2 | 9 | C/T | 0.25 | 0.017 (0.006) | 5.0 × 10−7 | WCadjBMI | [26] | NA |
| rs1507456 | AC060788.1 | 8 | C/T | NA | NA | 7.0 × 10−7 | WHR | [74] | NA |
| rs17197710 | AC090589.1 | 11 | C/T | 0.05 | −0.06 (0.024) | 8.0 × 10−7 | WHRadjBMI | [26] | NA |
| rs12227147 | ANKS1B | 12 | A/T | NA | NA | 1.0 × 10−6 | WHR | [74] | NA |
| rs139256956 | ZNF536 | 19 | C/A | 0.03 | −0.25 (0.1) | 1.0 × 10−6 | WHRadjBMI | [28] | NA |
| rs57561811 | SLC38A6 | 14 | C/T | 0.25 | −0.07 (0.02) | 2.0 × 10−6 | WHRadjBMI | [28] | NA |
| rs35316183 | DOCK2 | 5 | A/G | NA | NA | 3.0 × 10−6 | WHR | [74] | NA |
| rs17178527 | RPS3AP23 | 6 | A/G | NA | NA | 3.0 × 10−6 | WC | [92] | NA |
| rs79817709 | KEAP1 | 19 | T/G | NA | NA | 5.0 × 10−6 | WHR | [74] | NA |
| rs4667458 | AC016766.1 | 2 | G/A | NA | NA | 5.0 × 10−6 | WC | [92] | NA |
| Identified Fat Distribution Susceptibility Loci from African Population Based on p < 5 × 10−6 | |||||||||
| rs2075064 | LHX2 | 9 | T/C | 0.07 | −0.07 (0.01) | 2.2 × 10−8 | WCadjBMI | [78] | 0.067 |
| rs6867983 | MAP3K1 | 5 | T/C | 0.24 | −0.09 (0.02) | 1.4 × 10−7 | WC_men | [78] | NA |
| rs7601155 | BRE | 2 | T/C | 0.16 | 0.06 (0.05) | 1.7 × 10−7 | WCadjBMI | [78] | NA |
| rs17213965 | MYH11 | 16 | T/C | 0.16 | 0.12 (0.02) | 8.8 × 10−7 | WHRadjBMI_men | [78] | NA |
| rs11777345 | CSMD1 | 8 | G/C | 0.08 | −0.19 (0.04) | 3.2 × 10−6 | WHRadjBMI_men | [78] | NA |
| rs1345301 | IL1RL2; IL1RL1 | 2 | G/A | 0.19 | −0.08 (0.02) | 4.6 × 10−6 | WC_men | [78] | NA |
| rs2570467 | PCSK1 | 5 | G/A | 0.11 | 0.1 (0.02) | 1.2 × 10−6 | WC_men | [78] | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, C.; Kovacs, P.; Guiu-Jurado, E. Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries. Genes 2021, 12, 841. https://doi.org/10.3390/genes12060841
Sun C, Kovacs P, Guiu-Jurado E. Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries. Genes. 2021; 12(6):841. https://doi.org/10.3390/genes12060841
Chicago/Turabian StyleSun, Chang, Peter Kovacs, and Esther Guiu-Jurado. 2021. "Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries" Genes 12, no. 6: 841. https://doi.org/10.3390/genes12060841
APA StyleSun, C., Kovacs, P., & Guiu-Jurado, E. (2021). Genetics of Body Fat Distribution: Comparative Analyses in Populations with European, Asian and African Ancestries. Genes, 12(6), 841. https://doi.org/10.3390/genes12060841






