Identification of a Splice Variant (c.5074+3A>C) of BRCA1 by RNA Sequencing and TOPO Cloning
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Sequencing
2.2. Bioinformatics
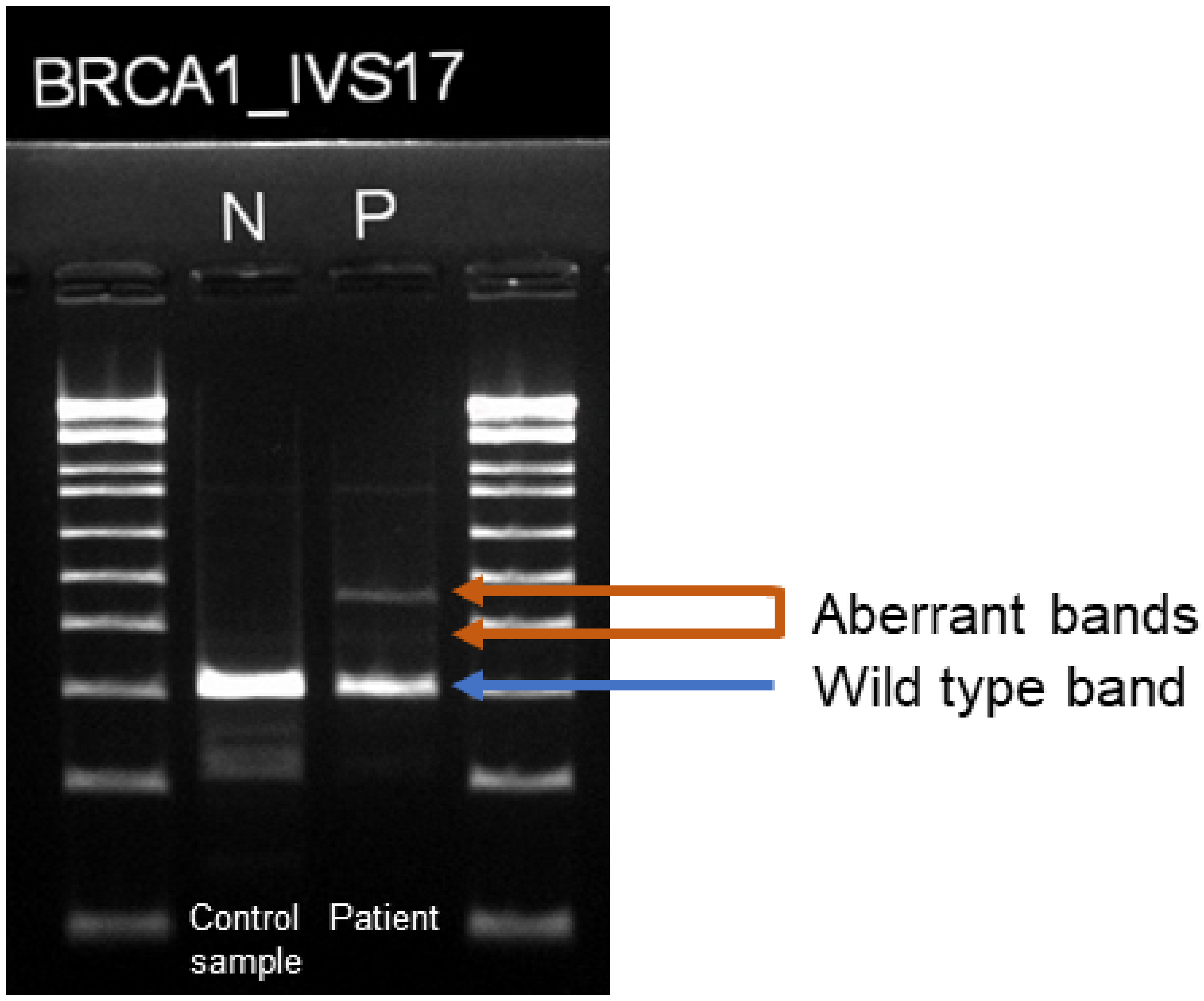
2.3. RNA Analysis
2.4. TOPO Cloning and Sequencing of the BRCA1 c.5074+3A>C Variant
3. Case Description
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66. [Google Scholar] [CrossRef]
- Wooster, R.; Neuhausen, S.L.; Mangion, J.; Quirk, Y.; Ford, D.; Collins, N.; Nguyen, K.; Seal, S.; Tran, T.; Averill, D.; et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science 1994, 265, 2088. [Google Scholar] [CrossRef]
- Gudmundsdottir, K.; Ashworth, A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene 2006, 25, 5864–5874. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. 2007, 25, 1329–1333. [Google Scholar] [CrossRef]
- Pal, T.; Permuth-Wey, J.; Betts, J.A.; Krischer, J.P.; Fiorica, J.; Arango, H.; LaPolla, J.; Hoffman, M.; Martino, M.A.; Wakeley, K.; et al. BRCA1 and BRCA2 mutations account for a large proportion of ovarian carcinoma cases. Cancer 2005, 104, 2807–2816. [Google Scholar] [CrossRef] [PubMed]
- Paluch-Shimon, S.; Cardoso, F.; Sessa, C.; Balmana, J.; Cardoso, M.J.; Gilbert, F.; Senkus, E. Prevention and screening in BRCA mutation carriers and other breast/ovarian hereditary cancer syndromes: ESMO Clinical Practice Guidelines for cancer prevention and screening. Ann. Oncol. 2016, 27, v103–v110. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.L.; Gordon, O.K. Positive Results: Making the Best Decisions When You’re at High Risk for Breast or Ovarian Cancer; Prometheus Books: Amherst, NY, USA, 2010. [Google Scholar]
- Forbes, C.; Fayter, D.; de Kock, S.; Quek, R.G. A systematic review of international guidelines and recommendations for the genetic screening, diagnosis, genetic counseling, and treatment of BRCA-mutated breast cancer. Cancer Manag. Res. 2019, 11, 2321–2337. [Google Scholar] [CrossRef]
- González-Santiago, S.; Ramón y Cajal, T.; Aguirre, E.; Alés-Martínez, J.E.; Andrés, R.; Balmaña, J.; Graña, B.; Herrero, A.; Llort, G.; González-del-Alba, A.; et al. SEOM clinical guidelines in hereditary breast and ovarian cancer (2019). Clin. Transl. Oncol. 2020, 22, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Béroud, C.; Letovsky, S.I.; Braastad, C.D.; Caputo, S.M.; Beaudoux, O.; Bignon, Y.J.; Bressac-De Paillerets, B.; Bronner, M.; Buell, C.M.; Collod-Béroud, G.; et al. BRCA Share: A Collection of Clinical BRCA Gene Variants. Hum Mutat. 2016, 37, 1318–1328. [Google Scholar] [CrossRef]
- Cline, M.S.; Liao, R.G.; Parsons, M.T.; Paten, B.; Alquaddoomi, F.; Antoniou, A.; Baxter, S.; Brody, L.; Cook-Deegan, R.; Coffin, A.; et al. BRCA Challenge: BRCA Exchange as a global resource for variants in BRCA1 and BRCA2. PLoS Genet. 2018, 14, e1007752. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Liu, X.; Wu, C.; Li, C.; Boerwinkle, E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016, 37, 235–241. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Abecasis, G.R.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Culp, M.; Johnson, K.; Michailides, G. ada: An R Package for Stochastic Boosting. J. Stat. Softw. 2006, 17, 9. [Google Scholar] [CrossRef][Green Version]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2001, 23, 18–22. [Google Scholar]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Menéndez, M.; Castellsagué, J.; Mirete, M.; Pros, E.; Feliubadaló, L.; Osorio, A.; Calaf, M.; Tornero, E.; del Valle, J.; Fernández-Rodríguez, J.; et al. Assessing the RNA effect of 26 DNA variants in the BRCA1 and BRCA2 genes. Breast Cancer Res. Treat. 2012, 132, 979–992. [Google Scholar] [CrossRef]
- Findlay, G.M.; Daza, R.M.; Martin, B.; Zhang, M.D.; Leith, A.P.; Gasperini, M.; Janizek, J.D.; Huang, X.; Starita, L.M.; Shendure, J. Accurate classification of BRCA1 variants with saturation genome editing. Nature 2018, 562, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Boerwinkle, E.; Liu, X. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014, 42, 13534–13544. [Google Scholar] [CrossRef]
- Desmet, F.-O.; Hamroun, D.; Lalande, M.; Collod-Béroud, G.; Claustres, M.; Béroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed]
- Yeo, G.; Burge, C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004, 11, 377–394. [Google Scholar] [CrossRef]
- Shapiro, M.B.; Senapathy, P. RNA splice junctions of different classes of eukaryotes: Sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987, 15, 7155–7174. [Google Scholar] [CrossRef] [PubMed]
- Tautz, D.; Renz, M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal. Biochem. 1983, 132, 14–19. [Google Scholar] [CrossRef]
- Brnich, S.E.; Abou Tayoun, A.N.; Couch, F.J.; Cutting, G.R.; Greenblatt, M.S.; Heinen, C.D.; Kanavy, D.M.; Luo, X.; McNulty, S.M.; Starita, L.M.; et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019, 12, 3. [Google Scholar] [CrossRef]
- Roest, P.A.M.; Roberts, R.G.; Sugino, S.; van Ommen, G.-J.B.; den Dunnen, J.T. Protein truncation test (PTT) for rapid detection of translation-terminating mutations. Hum. Mol. Genet. 1993, 2, 1719–1721. [Google Scholar] [CrossRef]
- Guidugli, L.; Carreira, A.; Caputo, S.M.; Ehlen, A.; Galli, A.; Monteiro, A.N.A.; Neuhausen, S.L.; Hansen, T.V.O.; Couch, F.J.; Vreeswijk, M.P.G.; et al. Functional Assays for Analysis of Variants of Uncertain Significance in BRCA2. Hum Mutat. 2014, 35, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Lindeboom, R.G.H.; Supek, F.; Lehner, B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016, 48, 1112–1118. [Google Scholar] [CrossRef] [PubMed]
- Perrin-Vidoz, L.; Sinilnikova, O.M.; Stoppa-Lyonnet, D.; Lenoir, G.M.; Mazoyer, S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum. Mol. Genet. 2002, 11, 2805–2814. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.; Kim, J.H.; Ahn, S.H.; Gu, H.; Chang, S.; Lee, W.; Kim, D.-Y.; Chun, S.; Min, W.-K. Identification of a Splice Variant (c.5074+3A>C) of BRCA1 by RNA Sequencing and TOPO Cloning. Genes 2021, 12, 810. https://doi.org/10.3390/genes12060810
Hong J, Kim JH, Ahn SH, Gu H, Chang S, Lee W, Kim D-Y, Chun S, Min W-K. Identification of a Splice Variant (c.5074+3A>C) of BRCA1 by RNA Sequencing and TOPO Cloning. Genes. 2021; 12(6):810. https://doi.org/10.3390/genes12060810
Chicago/Turabian StyleHong, Jinyoung, Ji Hyun Kim, Se Hee Ahn, Hyunjung Gu, Suhwan Chang, Woochang Lee, Dae-Yeon Kim, Sail Chun, and Won-Ki Min. 2021. "Identification of a Splice Variant (c.5074+3A>C) of BRCA1 by RNA Sequencing and TOPO Cloning" Genes 12, no. 6: 810. https://doi.org/10.3390/genes12060810
APA StyleHong, J., Kim, J. H., Ahn, S. H., Gu, H., Chang, S., Lee, W., Kim, D.-Y., Chun, S., & Min, W.-K. (2021). Identification of a Splice Variant (c.5074+3A>C) of BRCA1 by RNA Sequencing and TOPO Cloning. Genes, 12(6), 810. https://doi.org/10.3390/genes12060810






