Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Development of the Micro-Tom EMS Population
2.2. Powdery Mildew Disease Assays and Quantification of Relative Fungal Biomass
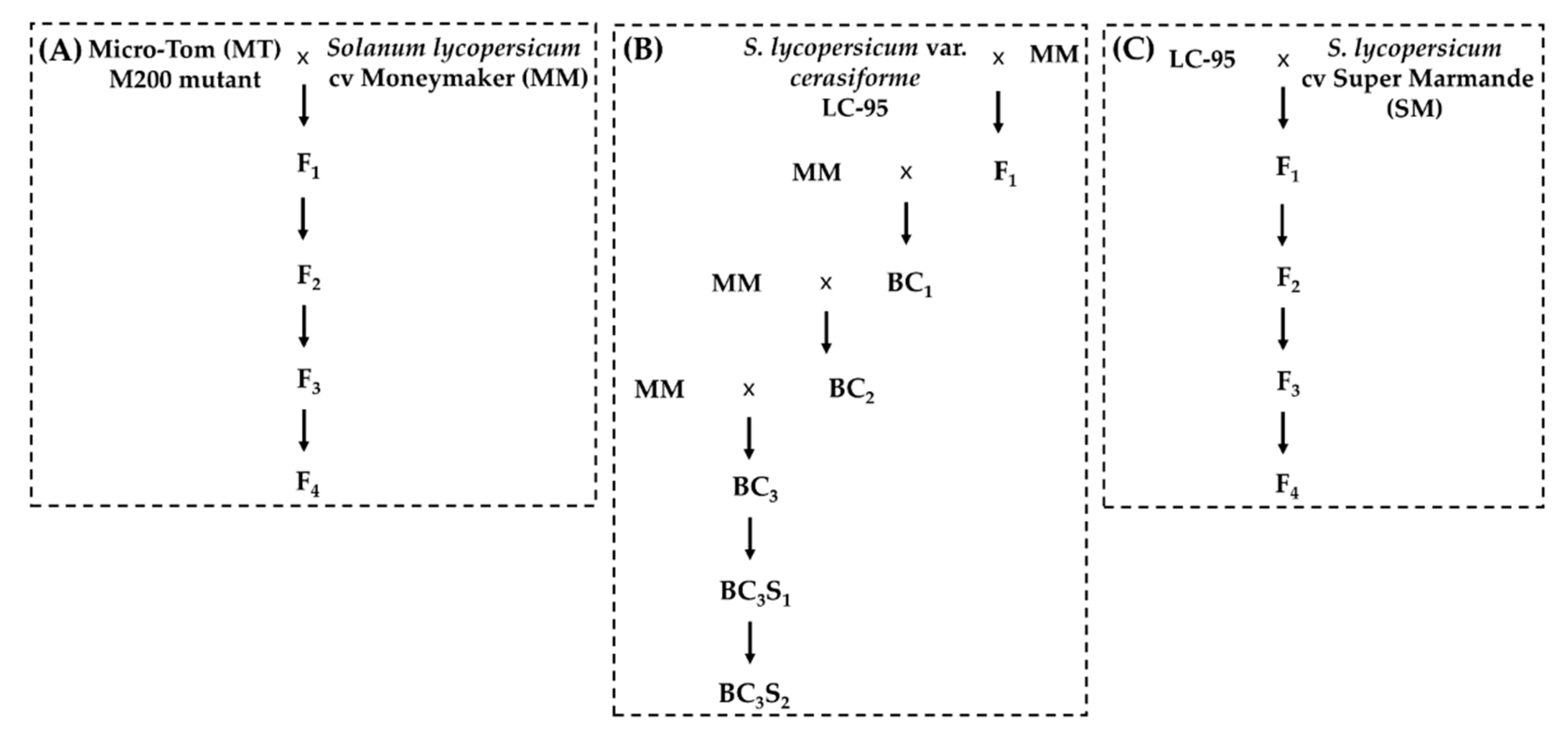
2.3. Plant Materials
2.4. Cloning of the SlMLO1 Coding Sequence from the Mutagenized Resistant Micro-Tom Plant M200
2.5. Development of a HRM Marker for Detection of the Mutation in the SlMLO1 Gene
2.6. Histological Analysis
3. Results
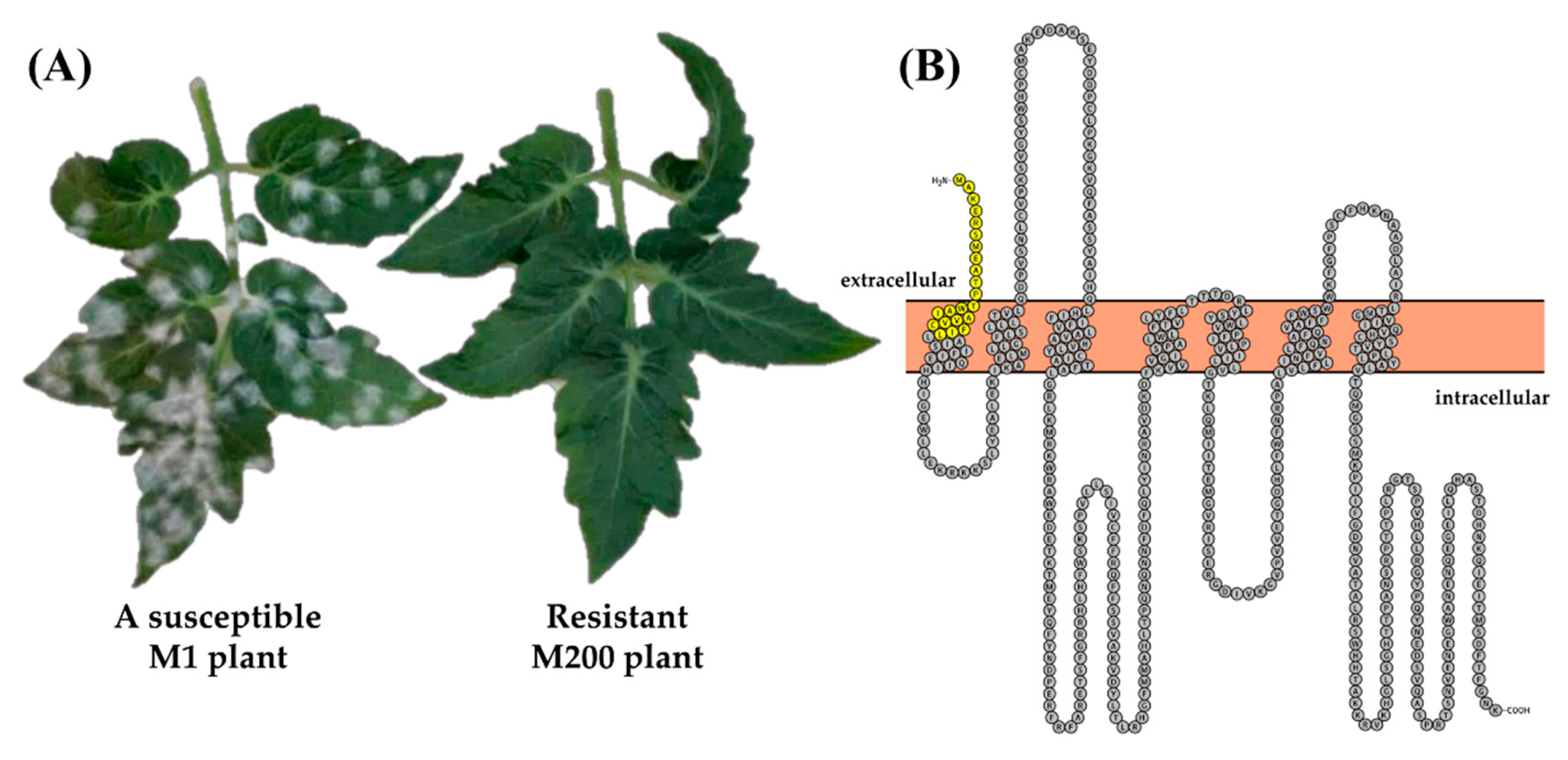
3.1. A Novel EMS mlo Mutant (M200) Shows Resistance to Powdery Mildew
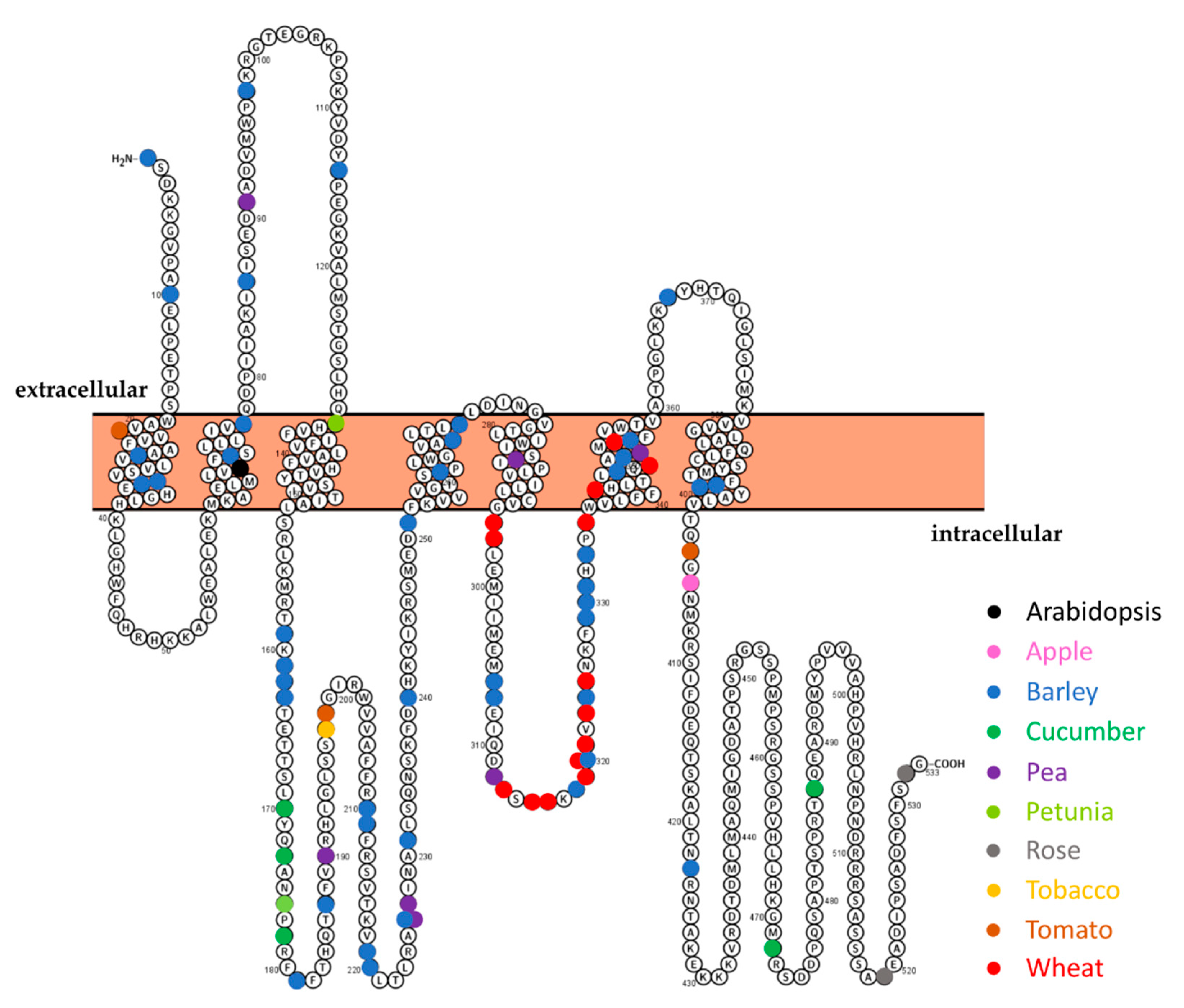
3.2. The m200 Allele Is a Unique SlMlo1 Loss-of-Function Allele
3.3. The Resistant Phenotype Fully Cosegregates with the Novel m200 Allele
3.4. Full Resistance Provided by the m200 Allele
3.5. Papilla Formation Is Associated with Resistance in the m200 Mutant
4. Discussion
4.1. Is the m200 Mutation a Real Product of the EMS Mutagenesis?
4.2. Is the Resistance Level of Slmlo Mutants Dependent on Papilla Formation?
4.3. Is the Level of mlo-Based Resistance Influenced by the Position of the Mutation?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nat. Cell Biol. 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Moco, S.; Bino, R.J.; Vorst, O.; Verhoeven, H.A.; De Groot, J.; Van Beek, T.A.; Vervoort, J.; De Vos, C.H.R. A Liquid Chromatography-Mass Spectrometry-Based Metabolome Database for Tomato. Plant. Physiol. 2006, 141, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.-S.; Yao, Q.-H.; Peng, R.-H.; Li, X.; Han, P.-L.; Fan, H.-Q. Different effects on ACC oxidase gene silencing triggered by RNA interference in transgenic tomato. Plant. Cell Rep. 2004, 23, 639–646. [Google Scholar] [CrossRef]
- Schijlen, E.G.; de Vos, C.R.; Martens, S.; Jonker, H.H.; Rosin, F.M.; Molthoff, J.W.; Tikunov, Y.M.; Angenent, G.C.; van Tunen, A.J.; Bovy, A.G. RNA Interference Silencing of Chalcone Synthase, the First Step in the Flavonoid Biosynthesis Pathway, Leads to Parthenocarpic Tomato Fruits. Plant. Physiol. 2007, 144, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.; Wolters-Arts, M.; Feron, R.; Mariani, C.; Vriezen, W.H. The Solanum lycopersicumauxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant. J. 2009, 57, 160–170. [Google Scholar] [CrossRef] [PubMed]
- Lor, V.S.; Starker, C.G.; Voytas, D.F.; Weiss, D.; Olszewski, N.E. Targeted Mutagenesis of the Tomato PROCERA Gene Using Transcription Activator-Like Effector Nucleases. Plant. Physiol. 2014, 166, 1288–1291. [Google Scholar] [CrossRef]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient Gene Editing in Tomato in the First Generation Using the Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated9 System. Plant. Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.; Zhu, H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef]
- Rodríguez, G.R.; Muños, S.; Anderson, C.; Sim, S.-C.; Michel, A.; Causse, M.; Gardener, B.B.M.; Francis, D.; van der Knaap, E. Distribution of SUN, OVATE, LC, and FAS in the Tomato Germplasm and the Relationship to Fruit Shape Diversity. Plant. Physiol. 2011, 156, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Kissoudis, C.; Van De Wiel, C.; Visser, R.G.; Van Der Linden, G. Future-proof crops: Challenges and strategies for climate resilience improvement. Curr. Opin. Plant. Biol. 2016, 30, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.-Y.; Forster, B.P.; Nakagawa, H. Principles and applications of plant mutation breeding. In Plant Mutation Breeding and Biotechnology; Shu, Q.-Y., Forster, B.P., Nakagawa, H., Eds.; CABI: Wallingford, UK, 2012; pp. 301–325. [Google Scholar] [CrossRef]
- Greene, E.A.; Codomo, C.A.; Taylor, N.E.; Henikoff, J.G.; Till, B.J.; Reynolds, S.H.; Enns, L.C.; Burtner, C.; Johnson, J.E.; Odden, A.R.; et al. Spectrum of Chemically Induced Mutations from a Large-Scale Reverse-Genetic Screen in Arabidopsis. Genetics 2003, 164, 731–740. [Google Scholar] [CrossRef]
- Till, B.J.; Reynolds, S.H.; Weil, C.; Springer, N.; Burtner, C.; Young, K.; Bowers, E.; Codomo, C.A.; Enns, L.C.; Odden, A.R.; et al. Discovery of induced point mutations in maize genes by TILLING. BMC Plant. Biol. 2004, 4, 12. [Google Scholar] [CrossRef]
- Till, B.J.; Cooper, J.; Tai, T.H.; Colowit, P.; Greene, E.A.; Henikoff, S.; Comai, L. Discovery of chemically induced mutations in rice by TILLING. BMC Plant. Biol. 2007, 7, 19. [Google Scholar] [CrossRef]
- Meissner, R.; Jacobson, Y.; Melamed, S.; Levyatuv, S.; Shalev, G.; Ashri, A.; Elkind, Y.; Levy, A. A new model system for tomato genetics. Plant. J. 1997, 12, 1465–1472. [Google Scholar] [CrossRef]
- Menda, N.; Semel, Y.; Peled, D.; Eshed, Y.; Zamir, D. In silico screening of a saturated mutation library of tomato. Plant J. 2004, 38, 861–872. [Google Scholar] [CrossRef]
- Gady, A.L.; Hermans, F.W.; Van De Wal, M.H.; Van Loo, E.N.; Visser, R.G.; Bachem, C.W. Implementation of two high through-put techniques in a novel application: Detecting point mutations in large EMS mutated plant populations. Plant. Methods 2009, 5, 13. [Google Scholar] [CrossRef] [PubMed]
- Saito, T.; Asamizu, E.; Mizoguchi, T.; Fukuda, N.; Matsukura, C.; Ezura, H. Mutant Resources for the Miniature Tomato (Solanum lycopersicum L.) ‘Micro-Tom’. J. Jpn. Soc. Hortic. Sci. 2009, 78, 6–13. [Google Scholar] [CrossRef][Green Version]
- Minoia, S.; Petrozza, A.; D’Onofrio, O.; Piron, F.; Mosca, G.; Sozio, G.; Cellini, F.; Bendahmane, A.; Carriero, F. A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 2010, 3, 69. [Google Scholar] [CrossRef]
- Scott, J.W.; Harbaugh, B.K. Micro-Tom: A Miniature Dwarf Tomato; Agricultural Experiment Station, Institute of Food and Agricultural Sciences, University of Florida: Gainesville, FL, USA, 1989; p. 370. [Google Scholar]
- Saito, T.; Ariizumi, T.; Okabe, Y.; Asamizu, E.; Hiwasa-Tanase, K.; Fukuda, N.; Mizoguchi, T.; Yamazaki, Y.; Aoki, K.; Ezura, H. TOMATOMA: A Novel Tomato Mutant Database Distributing Micro-Tom Mutant Collections. Plant. Cell Physiol. 2011, 52, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant. Biotechnol. J. 2015, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2009, 25, 1–12. [Google Scholar] [CrossRef]
- Van Schie, C.C.; Takken, F.L. Susceptibility Genes 101: How to Be a Good Host. Annu. Rev. Phytopathol. 2014, 52, 551–581. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, J.H. Comparison of Induced Mutant Genes to Spontaneous Genes in Barley Conditioning Resistance to Powdery Mildew; International Atomic Energy Agency: Vienna, Austria, 1971; pp. 117–124. [Google Scholar]
- Jørgensen, I.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Brown, J.K. Durable Resistance of Crops to Disease: A Darwinian Perspective. Annu. Rev. Phytopathol. 2015, 53, 513–539. [Google Scholar] [CrossRef] [PubMed]
- Devoto, A.; Piffanelli, P.; Nilsson, I.; Wallin, E.; Panstruga, R.; von Heijne, G.; Schulze-Lefert, P. Topology, Subcellular Localization, and Sequence Diversity of the Mlo Family in Plants. J. Biol. Chem. 1999, 274, 34993–35004. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Panstruga, R.; Elliott, C.; Müller, J.; Devoto, A.; Yoon, H.W.; Park, H.C.; Cho, M.J.; Schulze-Lefert, P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nat. Cell Biol. 2002, 416, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Kusch, S.; Panstruga, R. mlo-Based Resistance: An Apparently Universal “Weapon” to Defeat Powdery Mildew Disease. Mol. Plant. Microbe Interact. 2017, 30, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Bidzinski, P.; Noir, S.; Shahi, S.; Reinstädler, A.; Gratkowska, D.M.; Panstruga, R. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant. Cell Environ. 2014, 37, 2738–2753. [Google Scholar] [CrossRef]
- Chen, Z.; Noir, S.; Kwaaitaal, M.; Hartmann, H.A.; Wu, M.-J.; Mudgil, Y.; Sukumar, P.; Muday, G.; Panstruga, R.; Jones, A.M. Two Seven-Transmembrane Domain MILDEW RESISTANCE LOCUS O Proteins Cofunction in Arabidopsis Root Thigmomorphogenesis. Plant. Cell 2009, 21, 1972–1991. [Google Scholar] [CrossRef]
- Kessler, S.A.; Shimosato-Asano, H.; Keinath, N.F.; Wuest, S.E.; Ingram, G.; Panstruga, R.; Grossniklaus, U. Conserved Molecular Components for Pollen Tube Reception and Fungal Invasion. Science 2010, 330, 968–971. [Google Scholar] [CrossRef] [PubMed]
- Lorrain, S.; Vailleau, F.; Balagué, C.; Roby, D. Lesion mimic mutants: Keys for deciphering cell death and defense pathways in plants? Trends Plant. Sci. 2003, 8, 263–271. [Google Scholar] [CrossRef]
- Wolter, M.; Hollricher, K.; Salamini, F.; Schulze-Lefert, P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Genet. Genom. 1993, 239, 122–128. [Google Scholar] [CrossRef]
- Piffanelli, P.; Zhou, F.; Casais, C.; Orme, J.; Jarosch, B.; Schaffrath, U.; Collins, N.C.; Panstruga, R.; Schulze-Lefert, P. The Barley MLO Modulator of Defense and Cell Death Is Responsive to Biotic and Abiotic Stress Stimuli. Plant. Physiol. 2002, 129, 1076–1085. [Google Scholar] [CrossRef]
- Ruiz-Lozano, J.M.; Gianinazzi, S.; Gianinazzi-Pearson, V. Genes involved in resistance to powdery mildew in barley differentially modulate root colonization by the mycorrhizal fungus Glomus mosseae. Mycorrhiza 1999, 9, 237–240. [Google Scholar] [CrossRef]
- Jansen, C.; von Wettstein, D.; Schäfer, W.; Kogel, K.-H.; Felk, A.; Maier, F.J. Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proc. Natl. Acad. Sci. USA 2005, 102, 16892–16897. [Google Scholar] [CrossRef] [PubMed]
- Jarosch, B.; Kogel, K.-H.; Schaffrath, U. The Ambivalence of the Barley Mlo Locus: Mutations Conferring Resistance Against Powdery Mildew (Blumeria graminis f. sp. hordei) Enhance Susceptibility to the Rice Blast Fungus Magnaporthe grisea. Mol. Plant. Microbe Interact. 1999, 12, 508–514. [Google Scholar] [CrossRef]
- Kumar, J.; Hückelhoven, R.; Beckhove, U.; Nagarajan, S.; Kogel, K.-H. A Compromised Mlo Pathway Affects the Response of Barley to the Necrotrophic Fungus Bipolaris sorokiniana (Teleomorph: Cochliobolus sativus) and Its Toxins. Phytopathology 2001, 91, 127–133. [Google Scholar] [CrossRef] [PubMed]
- McGrann, G.R.D.; Stavrinides, A.; Russell, J.; Corbitt, M.M.; Booth, A.; Chartrain, L.; Thomas, W.T.B.; Brown, J.K.M. A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. J. Exp. Bot. 2014, 65, 1025–1037. [Google Scholar] [CrossRef]
- Kusch, S.; Pesch, L.; Panstruga, R. Comprehensive Phylogenetic Analysis Sheds Light on the Diversity and Origin of the MLO Family of Integral Membrane Proteins. Genome Biol. Evol. 2016, 8, 878–895. [Google Scholar] [CrossRef] [PubMed]
- Feechan, A.; Jermakow, A.M.; Torregrosa, L.; Panstruga, R.; Dry, I.B. Identification of grapevine MLO gene candidates involved in susceptibility to powdery mildew. Funct. Plant. Biol. 2008, 35, 1255–1266. [Google Scholar] [CrossRef] [PubMed]
- Acevedo-Garcia, J.; Kusch, S.; Panstruga, R. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytol. 2014, 204, 273–281. [Google Scholar] [CrossRef]
- Panstruga, R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 2005, 33, 389–392. [Google Scholar] [CrossRef] [PubMed]
- Consonni, C.; Humphry, M.E.; Hartmann, H.A.; Livaja, M.; Durner, J.; Westphal, L.; Vogel, J.; Lipka, V.; Kemmerling, B.; Schulze-Lefert, P.; et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat. Genet. 2006, 38, 716–720. [Google Scholar] [CrossRef] [PubMed]
- Hückelhoven, R. Powdery mildew susceptibility and biotrophic infection strategies. FEMS Microbiol. Lett. 2005, 245, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Appiano, M.; Pavan, S.; Bracuto, V.; Ricciardi, L.; Visser, R.G.F.; Wolters, A.-M.A.; Bai, Y. Genome-Wide Study of the Tomato SlMLO Gene Family and Its Functional Characterization in Response to the Powdery Mildew Fungus Oidium neolycopersici. Front. Plant. Sci. 2016, 7, 380. [Google Scholar] [CrossRef]
- Berg, J.A.; Appiano, M.; Martínez, M.S.; Hermans, F.W.K.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant. Biol. 2015, 15, 243. [Google Scholar] [CrossRef]
- Cheng, H.; Kong, W.; Lv, J.; Li, J. Analysis of powdery mildew resistance in wild melon MLO mutants. Hortic. Plant. J. 2015, 1, 165–171. [Google Scholar] [CrossRef]
- Humphry, M.; Reinstädler, A.; Ivanov, S.; Bisseling, T.; Panstruga, R. Durable broad-spectrum powdery mildew resistance in pea er1 plants is conferred by natural loss-of-function mutations in PsMLO1. Mol. Plant. Pathol. 2011, 12, 866–878. [Google Scholar] [CrossRef]
- Sun, S.; Fu, H.; Wang, Z.; Duan, C.; Zong, X.; Zhu, Z. Discovery of a Novel er1 Allele Conferring Powdery Mildew Resistance in Chinese Pea (Pisum sativum L.) Landraces. PLoS ONE 2016, 11, e0147624. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Deng, D.; Wang, Z.; Duan, C.; Wu, X.; Wang, X.; Zong, X.; Zhu, Z. A novel er1 allele and the development and validation of its functional marker for breeding pea (Pisum sativum L.) resistance to powdery mildew. Theor. Appl. Genet. 2016, 129, 909–919. [Google Scholar] [CrossRef]
- Kaufmann, H.; Qiu, X.; Wehmeyer, J.; Debener, T. Isolation, Molecular Characterization, and Mapping of Four Rose MLO Orthologs. Front. Plant. Sci. 2012, 3, 244. [Google Scholar] [CrossRef] [PubMed]
- Pessina, S.; Angeli, D.; Martens, S.; Visser, R.G.; Bai, Y.; Salamini, F.; Velasco, R.; Schouten, H.J.; Malnoy, M. The knock-down of the expression of MdMLO19 reduces susceptibility to powdery mildew (Podosphaera leucotricha) in apple (Malus domestica). Plant. Biotechnol. J. 2016, 14, 2033–2044. [Google Scholar] [CrossRef]
- Fujimura, T.; Sato, S.; Tajima, T.; Arai, M. Powdery mildew resistance in the Japanese domestic tobacco cultivar Kokubu is associated with aberrant splicing of MLO orthologues. Plant. Pathol. 2016, 65, 1358–1365. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef]
- Wan, D.-Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.-Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Reinstädler, A.; Müller, J.; Czembor, J.H.; Piffanelli, P.; Panstruga, R. Novel induced mlo mutant alleles in combination with site-directed mutagenesis reveal functionally important domains in the heptahelical barley Mlo protein. BMC Plant. Biol. 2010, 10, 31. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant. Biotechnol. J. 2016, 15, 367–378. [Google Scholar] [CrossRef]
- Pavan, S.; Schiavulli, A.; Appiano, M.; Marcotrigiano, A.R.; Cillo, F.; Visser, R.G.F.; Bai, Y.; Lotti, C.; Ricciardi, L. Pea powdery mildew er1 resistance is associated to loss-of-function mutations at a MLO homologous locus. Theor. Appl. Genet. 2011, 123, 1425–1431. [Google Scholar] [CrossRef]
- Santo, T.; Rashkova, M.; Alabaça, C.; Leitão, J. The ENU-induced powdery mildew resistant mutant pea (Pisum sativum L.) lines S(er1mut1) and F(er1mut2) harbour early stop codons in the PsMLO1 gene. Mol. Breed. 2013, 32, 723–727. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Wilde, H.D. Identification and mutagenesis of disease susceptibility genes of Petunia hybrida. Plant. Cell Tissue Organ. Cult. 2016, 126, 117–125. [Google Scholar] [CrossRef]
- Ciccarese, F.; Amenduni, M.; Schiavone, D.; Cirulli, M. Occurrence and inheritance of resistance to powdery mildew (Oidium lycopersici) in Lycopersicon species. Plant. Pathol. 1998, 47, 417–419. [Google Scholar] [CrossRef]
- De Giovanni, C.; Dell’Orco, P.; Bruno, A.; Ciccarese, F.; Lotti, C.; Ricciardi, L. Identification of PCR-based markers (RAPD, AFLP) linked to a novel powdery mildew resistance gene (ol-2) in tomato. Plant. Sci. 2004, 166, 41–48. [Google Scholar] [CrossRef]
- Pavan, S.; Zheng, Z.; Borisova, M.; Van Den Berg, P.; Lotti, C.; De Giovanni, C.; Lindhout, P.; De Jong, H.; Ricciardi, L.; Visser, R.G.F.; et al. Map- vs. homology-based cloning for the recessive gene ol-2 conferring resistance to tomato powdery mildew. Euphytica 2007, 162, 91–98. [Google Scholar] [CrossRef]
- Bai, Y.; Pavan, S.; Zheng, Z.; Zappel, N.F.; Reinstädler, A.; Lotti, C.; De Giovanni, C.; Ricciardi, L.; Lindhout, P.; Visser, R.; et al. Naturally Occurring Broad-Spectrum Powdery Mildew Resistance in a Central American Tomato Accession Is Caused by Loss of Mlo Function. Mol. Plant. Microbe Interac. 2008, 21, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Van Der Hulst, R.; Bonnema, G.; Marcel, T.C.; Meijer-Dekens, F.; Niks, R.E.; Lindhout, P. Tomato Defense to Oldium neolycopersici: Dominant Ol Genes Confer Isolate-Dependent Resistance Via a Different Mechanism Than Recessive ol-2. Mol. Plant. Microbe Interact. 2005, 18, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Omasits, U.; Ahrens, C.H.; Müller, S.; Wollscheid, B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 2014, 30, 884–886. [Google Scholar] [CrossRef] [PubMed]
- The 100 Tomato Genome Sequencing Consortium; Aflitos, S.A.; Schijlen, E.; De Jong, H.; De Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N.; et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant. J. 2014, 80, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.K. Powdery mildews. In Westcott’s Plant Disease Handbook; Horst, K.R., Ed.; Springer: Dordrecht, The Netherlands, 2013; pp. 285–293. [Google Scholar] [CrossRef]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Sega, G.A. A review of the genetic effects of ethyl methanesulfonate. Mutat. Res. Genet. Toxicol. 1984, 134, 113–142. [Google Scholar] [CrossRef]
- Griffiths, J.F.; Griffiths, A.J.; Miller, J.H.; Suzuki, D.T. An Introduction to Genetic Analysis, 7th ed.; WH Freeman and Company: New York, NY, USA, 2000. [Google Scholar]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; van Daelen, R.; van der Lee, T.; Diergaarde, P.; Groenendijk, J.; et al. The Barley Mlo Gene: A Novel Control Element of Plant Pathogen Resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Kovalchuk, I.; Kovalchuk, O.; Hohn, B. Genome-wide variation of the somatic mutation frequency in transgenic plants. EMBO J. 2000, 19, 4431–4438. [Google Scholar] [CrossRef]
- Bashir, T.; Sailer, C.; Gerber, F.; Loganathan, N.; Bhoopalan, H.; Eichenberger, C.; Grossniklaus, U.; Baskar, R.; Vanholme, B.; Vanholme, R.; et al. Hybridization Alters Spontaneous Mutation Rates in a Parent-of-Origin-Dependent Fashion in Arabidopsis. Plant. Physiol. 2014, 165, 424–437. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledó, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The Rate and Molecular Spectrum of Spontaneous Mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Mondragon-Palomino, M.; Gaut, B.S. Gene Conversion and the Evolution of Three Leucine-Rich Repeat Gene Families in Arabidopsis thaliana. Mol. Biol. Evol. 2005, 22, 2444–2456. [Google Scholar] [CrossRef]
- Feechan, A.; Jermakow, A.M.; Ivancevic, A.; Godfrey, D.; Pak, H.; Panstruga, R.; Dry, I.B. Host Cell Entry of Powdery Mildew Is Correlated with Endosomal Transport of Antagonistically Acting VvPEN1 and VvMLO to the Papilla. Mol. Plant. Microbe Interact. 2013, 26, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, J.; Henderson, M.; Schweizer, P.; Burton, R.A.; Fincher, G.B.; Little, A. Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol. 2014, 204, 650–660. [Google Scholar] [CrossRef] [PubMed]
- Consonni, C.; Bednarek, P.; Humphry, M.; Francocci, F.; Ferrari, S.; Harzen, A.; van Themaat, E.V.L.; Panstruga, R. Tryptophan-Derived Metabolites Are Required for Antifungal Defense in the Arabidopsis mlo2 Mutant. Plant. Physiol. 2010, 152, 1544–1561. [Google Scholar] [CrossRef] [PubMed]
- Molina-Cano, J.L.; Simiand, J.P.; Sopena, A.; Pérez-Vendrell, A.M.; Dorsch, S.; Rubiales, D.; Swanston, J.S.; Jahoor, A. Mildew-resistant mutants induced in North American two- and six-rowed malting barley cultivars. Theor. Appl. Genet. 2003, 107, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Panstruga, R.; Reinstädler, A.; Müller, J.; Molina-Cano, J.L. Molecular characterization of mlo mutants in North American two- and six-rowed malting barley cultivars. Mol. Plant. Pathol. 2005, 6, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Lundqvist, U. Swedish mutation research in barley with plant breeding aspects. A historical review. In Plant Mutation Breeding for Crop Improvement; International Atomic Energy Agency (IAEA): Vienna, Austria, 1991; pp. 135–147. [Google Scholar]
- Müller, J.; Piffanelli, P.; Devoto, A.; Miklis, M.; Elliott, C.; Ortmann, B.; Schulze-Lefert, P.; Panstruga, R. Conserved ERAD-Like Quality Control of a Plant Polytopic Membrane Protein. Plant. Cell 2005, 17, 149–163. [Google Scholar] [CrossRef]
- Elliott, C.; Müller, J.; Miklis, M.; Bhat, R.A.; Schulze-Lefert, P.; Panstruga, R. Conserved extracellular cysteine residues and cytoplasmic loop–loop interplay are required for functionality of the heptahelical MLO protein. Biochem. J. 2005, 385, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Appiano, M.; Catalano, D.; Martínez, M.S.; Lotti, C.; Zheng, Z.; Visser, R.G.F.; Ricciardi, L.; Bai, Y.; Pavan, S. Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant. Biol. 2015, 15, 1–10. [Google Scholar] [CrossRef]
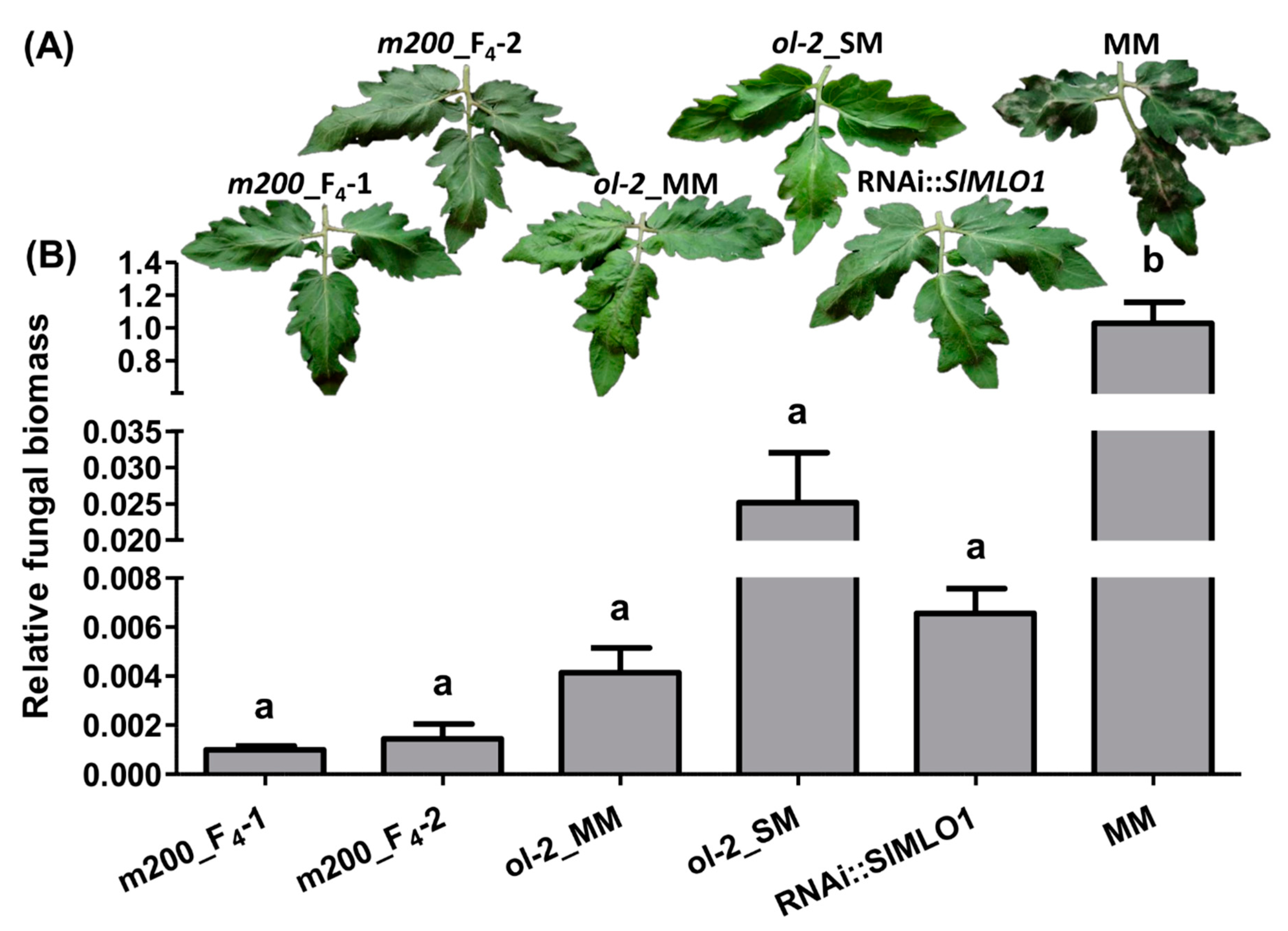

| Genotype | Number of Fungal/Plant Structures Observed | %Papilla/IU | %Haustorium/IU | ||
|---|---|---|---|---|---|
| IU | Papilla | Haustorium | |||
| m200_F4-1 | 97 | 34 | 10 | 35.1 | 10.3 |
| m200_F4-2 | 101 | 33 | 0 | 32.7 | 0 |
| ol-2_MM | 90 | 55 | 4 | 61.1 | 4.4 |
| ol-2_SM | 100 | 51 | 11 | 51 | 11 |
| RNAi::SlMLO1 | 109 | 78 | 6 | 71.6 | 5.5 |
| MM | 102 | 1 | 92 | 0.98 | 90.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Z.; Appiano, M.; van Tuinen, A.; Meijer-Dekens, F.; Schipper, D.; Gao, D.; Huibers, R.; Visser, R.G.F.; Bai, Y.; Wolters, A.-M.A. Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population. Genes 2021, 12, 719. https://doi.org/10.3390/genes12050719
Yan Z, Appiano M, van Tuinen A, Meijer-Dekens F, Schipper D, Gao D, Huibers R, Visser RGF, Bai Y, Wolters A-MA. Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population. Genes. 2021; 12(5):719. https://doi.org/10.3390/genes12050719
Chicago/Turabian StyleYan, Zhe, Michela Appiano, Ageeth van Tuinen, Fien Meijer-Dekens, Danny Schipper, Dongli Gao, Robin Huibers, Richard G. F. Visser, Yuling Bai, and Anne-Marie A. Wolters. 2021. "Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population" Genes 12, no. 5: 719. https://doi.org/10.3390/genes12050719
APA StyleYan, Z., Appiano, M., van Tuinen, A., Meijer-Dekens, F., Schipper, D., Gao, D., Huibers, R., Visser, R. G. F., Bai, Y., & Wolters, A.-M. A. (2021). Discovery and Characterization of a Novel Tomato mlo Mutant from an EMS Mutagenized Micro-Tom Population. Genes, 12(5), 719. https://doi.org/10.3390/genes12050719







