Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Retrieval and Identification of TPSs
2.2. Multiple Sequence Alignment and Phylogenetic Analyses
2.3. Characterization of TPSs from M. longifolia
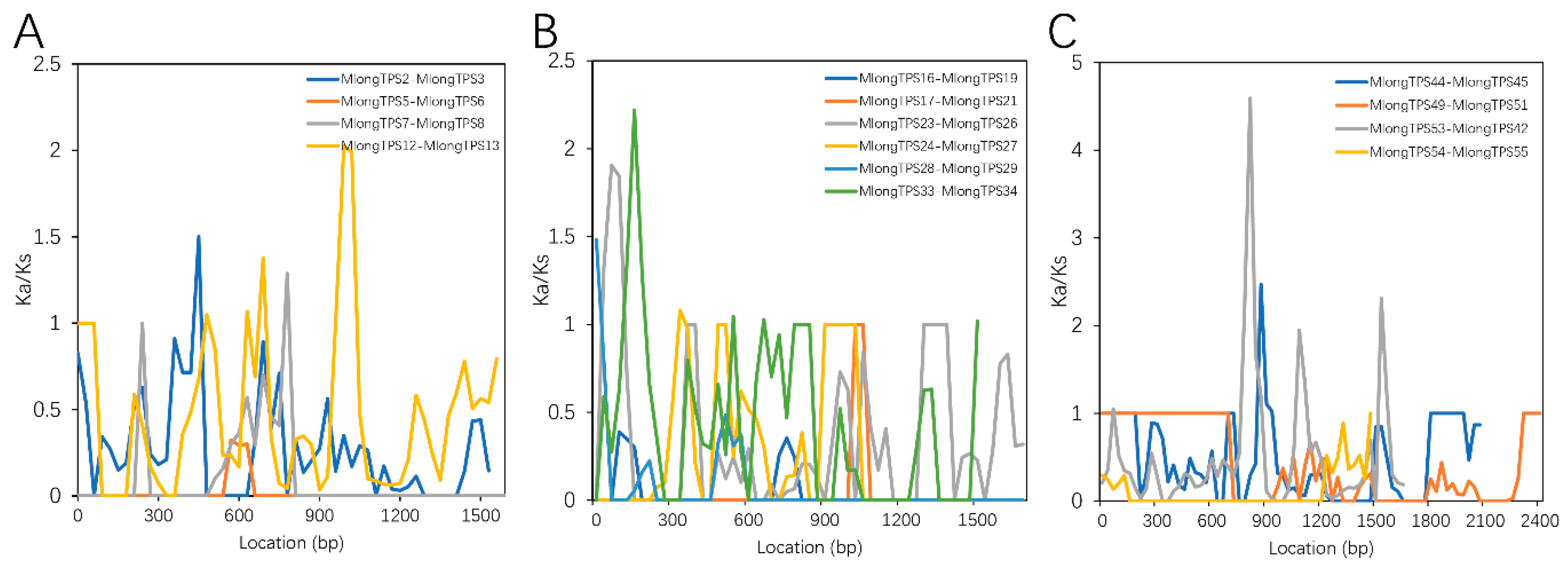
2.4. Adaptive Evolution Analysis of M. longifolia TPSs
2.5. RNA Isolation and MlongTPS29 Cloning
2.6. Expression of Recombinant MlongTPS29 in Escherichia coli and Enzyme Assays
3. Results
3.1. Identification of TPS Genes in M. longifolia Genome Sequence
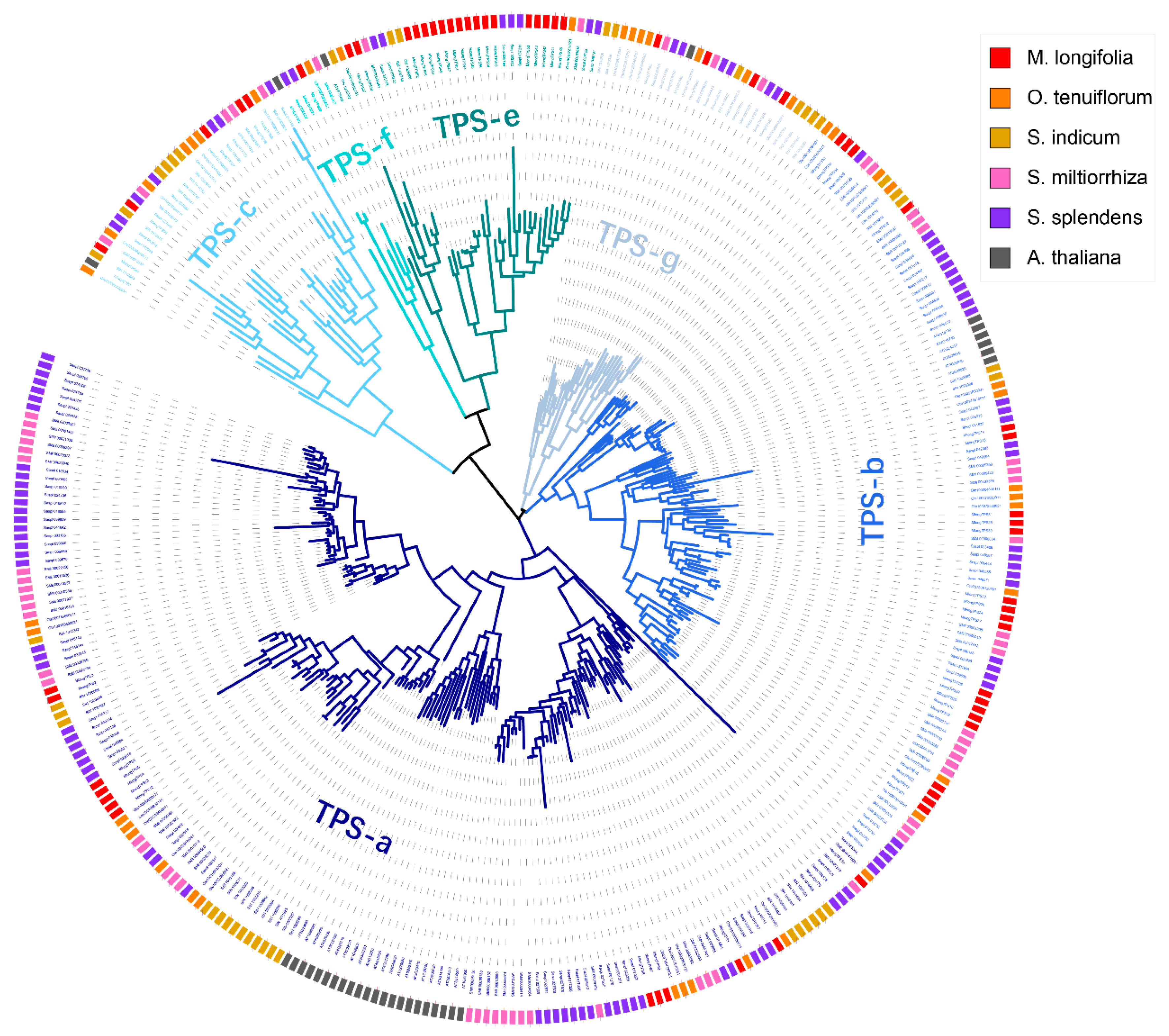
3.2. Phylogenetic Analyses of TPSs from M. longifolia and Other Lamiaceae Plants
3.3. Classification of M. longifolia TPSs Based on the Phylogenetic Tree
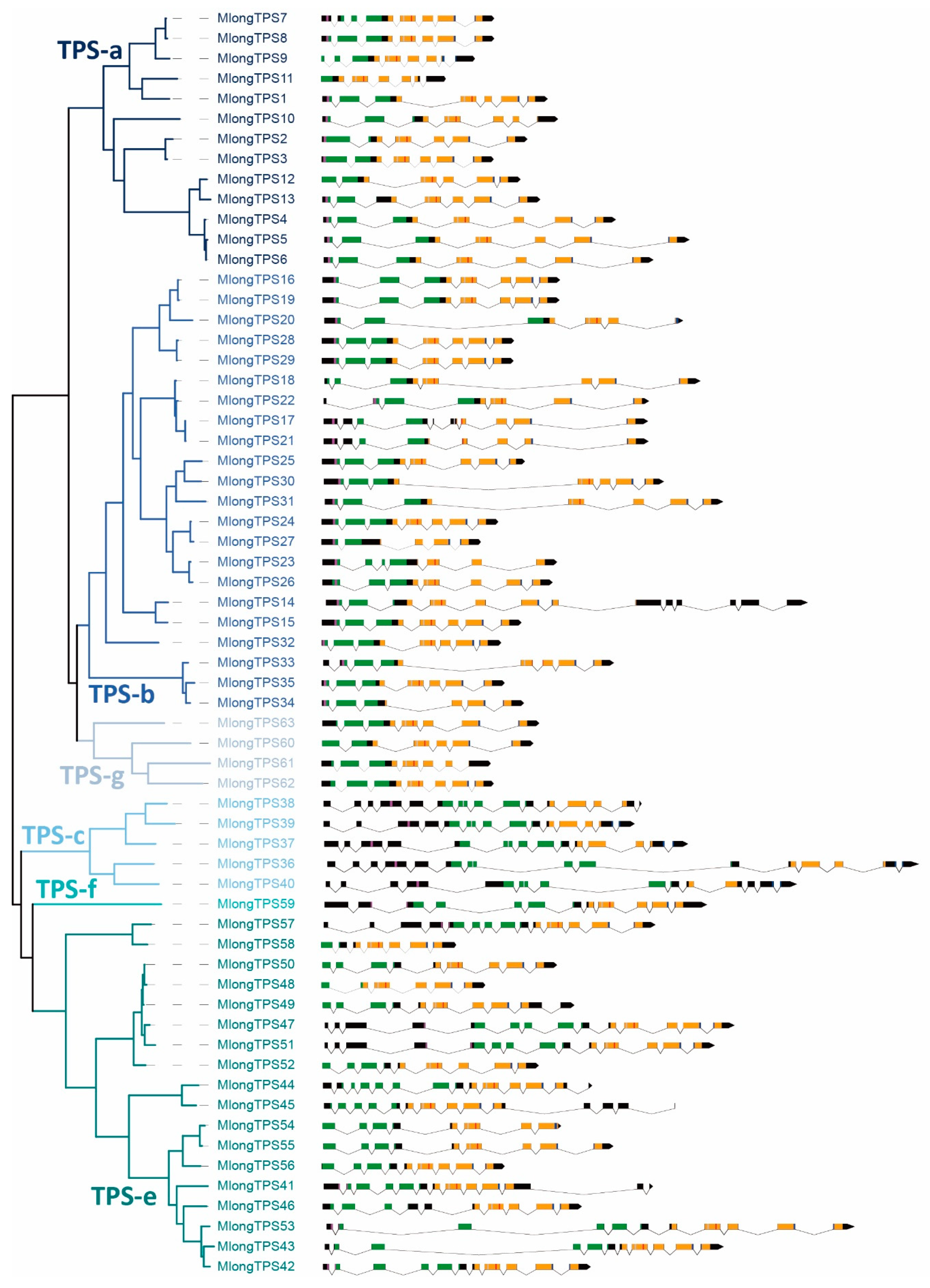
3.4. Exon-Intron Stucture of M. longifolia TPS Genes
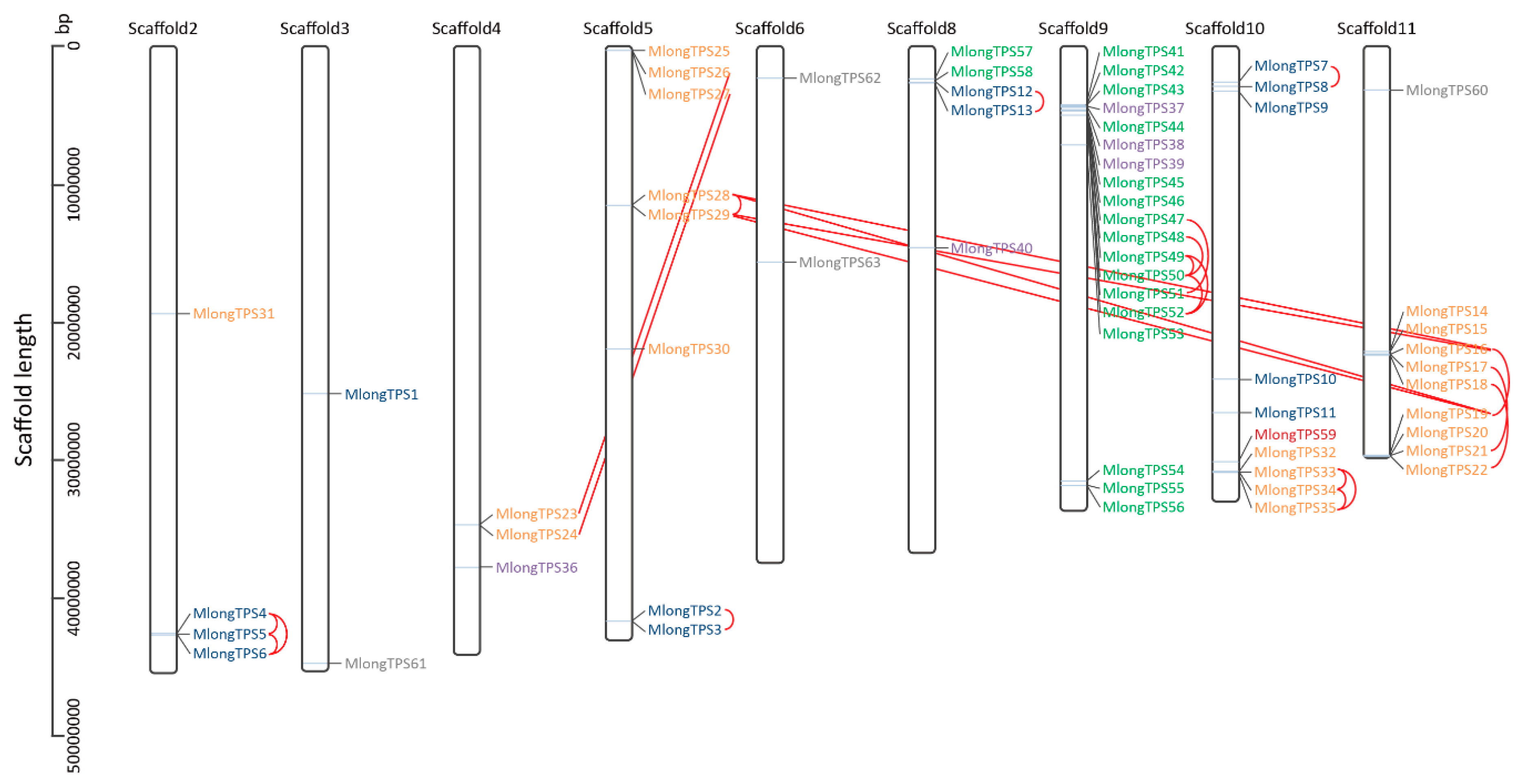
3.5. Genomic Distribution of M. longifolia TPS Genes
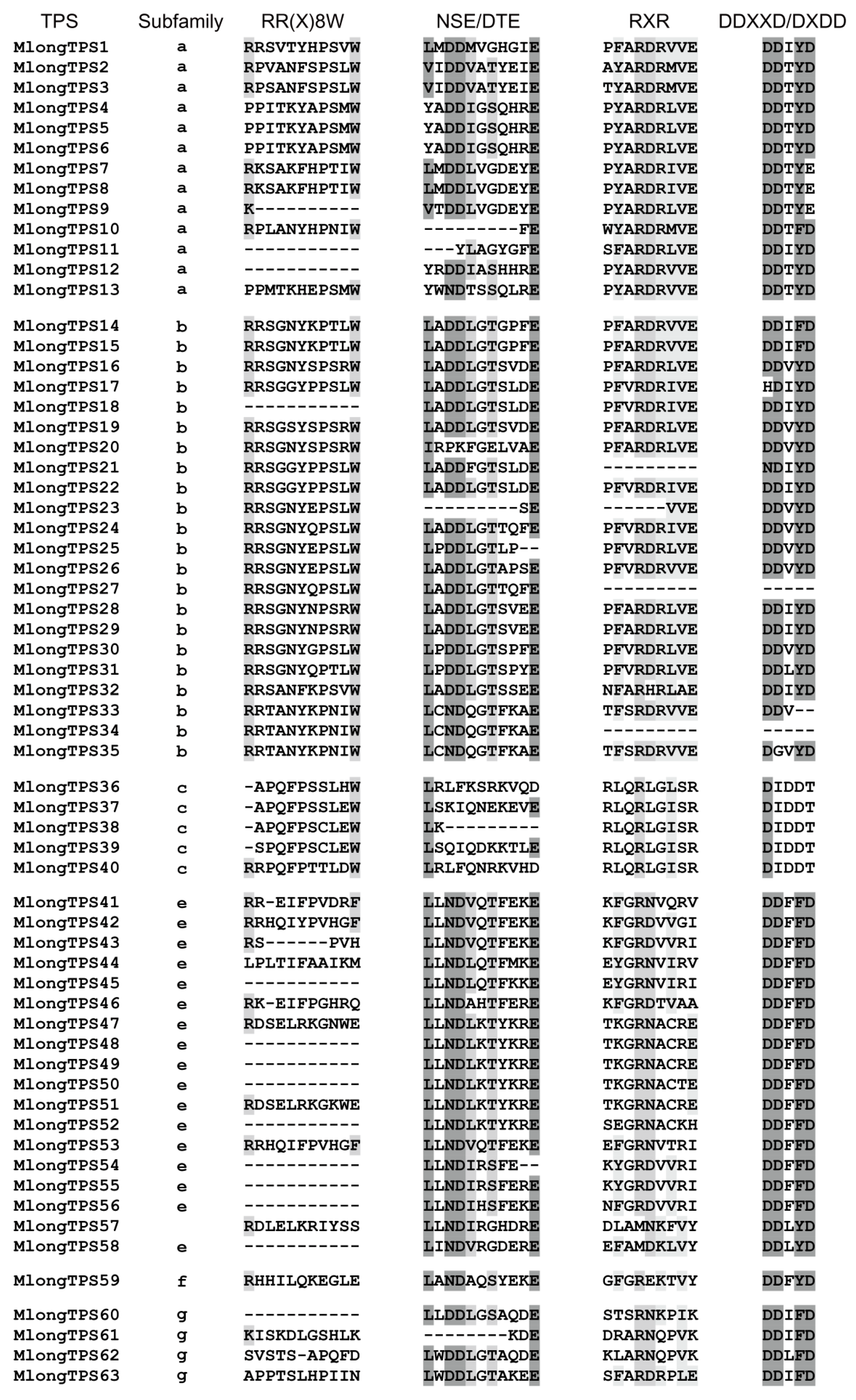
3.6. Conserved Motif Analyses of M. longifolia TPSs
3.7. Adaptive Evolution Analysis of M. longifolia TPSs
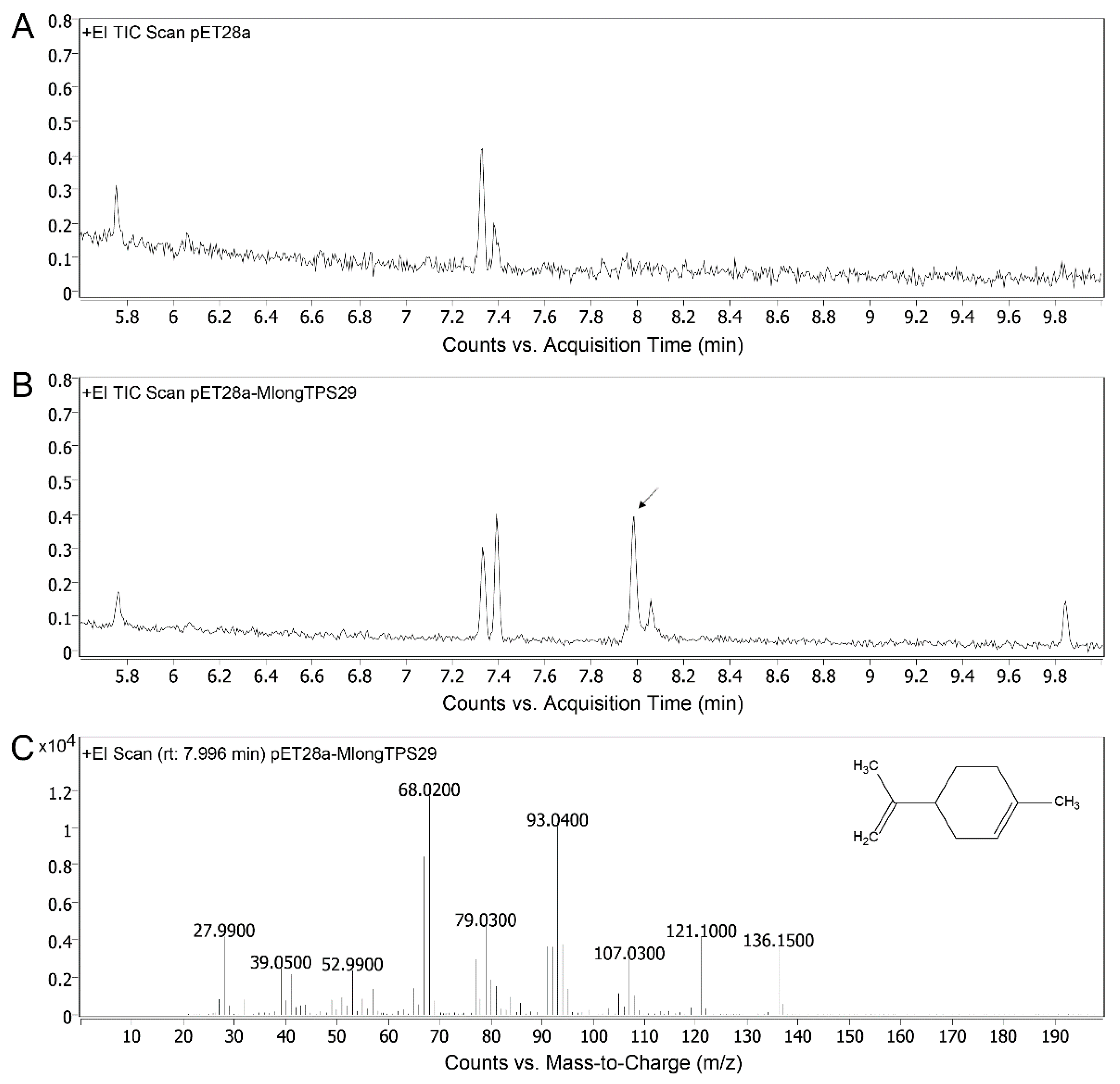
3.8. Enzyme Activity Assays of MlongTPS29
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biot. 2015, 148, 63–106. [Google Scholar]
- Yamada, Y.; Kuzuyama, T.; Komatsu, M.; Shin-Ya, K.; Omura, S.; Cane, D.-E.; Ikeda, H. Terpene synthases are widely distributed in bacteria. Proc. Natl. Acad. Sci. USA 2015, 112, 857–862. [Google Scholar] [CrossRef]
- Christianson, D.W. Structural and Chemical Biology of Terpenoid Cyclases. Chem. Rev. 2017, 117, 11570–11648. [Google Scholar] [CrossRef]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar] [CrossRef]
- Loreto, F.; Dicke, M.; Schnitzler, J.-P.; Turlings, T.C.J. Plant volatiles and the environment. Plant Cell Environ. 2014, 37, 1905–1908. [Google Scholar] [CrossRef]
- Zulak, K.G.; Bohlmann, J. Terpenoid Biosynthesis and Specialized Vascular Cells of Conifer Defense. J. Integr. Plant Biol. 2010, 52, 86–97. [Google Scholar] [CrossRef]
- Van Geldre, E.; Vergauwe, A.; Eeckhout, E.V.D. State of the art of the production of the antimalarial compound artemisinin in plants. Plant Mol. Biol. 1997, 33, 199–209. [Google Scholar] [CrossRef]
- Newman, J.D.; Chappell, J. Isoprenoid Biosynthesis in Plants: Carbon Partitioning Within the Cytoplasmic Pathway. Crit. Rev. Biochem. Mol. Biol. 1999, 34, 95–106. [Google Scholar] [CrossRef]
- Vranová, E.; Coman, D.; Gruissem, W. Network Analysis of the MVA and MEP Pathways for Isoprenoid Synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar] [CrossRef]
- McGarvey, D.J.; Croteau, R. Terpenoid Metabolism. Plant Cell 1995, 7, 1015. [Google Scholar] [CrossRef]
- Tholl, D. Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr. Opin. Plant Biol. 2006, 9, 297–304. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Jiang, S.-Y.; Jin, J.; Sarojam, R.; Ramachandran, S. A Comprehensive Survey on the Terpene Synthase Gene Family Provides New Insight into Its Evolutionary Patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef]
- Dudareva, N.; Martin, D.; Kish, C.M.; Kolosova, N.; Gorenstein, N.; Fäldt, J.; Miller, B.; Bohlmann, J. (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 2003, 15, 1227–1241. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef]
- Trapp, S.C.; Croteau, R.B. Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 2001, 158, 811–832. [Google Scholar]
- Lange, B.M.; Ahkami, A. Metabolic engineering of plant monoterpenes, sesquiterpenes and diterpenes-current status and future opportunities. Plant Biotechnol. J. 2012, 11, 169–196. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.M. Biosynthesis and biotechnology of high-value p-menthane monoterpenes, including menthol, carvone, and limonene. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 319–353. [Google Scholar]
- Ahkami, A.; Johnson, S.R.; Srividya, N.; Lange, B.M. Multiple levels of regulation determine monoterpenoid essential oil compositional variation in the mint family. Mol. Plant 2015, 8, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Turner, G.W.; Croteau, R. Organization of Monoterpene Biosynthesis in Mentha. Immunocytochemical Localizations of Geranyl Diphosphate Synthase, Limonene-6-Hydroxylase, Isopiperitenol Dehydrogenase, and Pulegone Reductase. Plant Physiol. 2004, 136, 4215–4227. [Google Scholar] [CrossRef] [PubMed]
- Croteau, R.B.; Davis, E.M.; Ringer, K.L.; Wildung, M.R. (−)-Menthol biosynthesis and molecular genetics. Naturwissenschaften 2005, 92, 562–577. [Google Scholar] [CrossRef] [PubMed]
- Vining, K.; Zhang, Q.; Tucker, A.; Smith, C.; Davis, T. Mentha longifolia (L.) L.: A Model Species for Mint Genetic Research. HortScience 2005, 40, 1225–1229. [Google Scholar] [CrossRef]
- Vining, K.J.; Johnson, S.R.; Ahkami, A.; Lange, I.; Parrish, A.N.; Trapp, S.C.; Croteau, R.B.; Straub, S.C.; Pandelova, I.; Lange, B.M. Draft Genome Sequence of Mentha longifolia and Development of Resources for Mint Cultivar Improvement. Mol. Plant 2017, 10, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Song, J.; Luo, H.; Zhang, Y.; Li, Q.; Zhu, Y.; Xu, J.; Li, Y.; Song, C.; Wang, B.; et al. Analysis of the Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. Mol. Plant 2016, 9, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.K.; Chacko, A.R.; Gandhimathi, A.; Ghosh, P.; Harini, K.; Joseph, A.P.; Joshi, A.G.; Karpe, S.D.; Kaushik, S.; Kuravadi, N.; et al. Genome sequencing of herb Tulsi (Ocimum tenuiflorum) unravels key genes behind its strong medicinal properties. BMC Plant Biol. 2015, 15, 212. [Google Scholar] [CrossRef]
- Wang, L.; Xia, Q.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Ni, X.; Gao, Y.; Xiang, H.; Wei, X.; et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Dong, A.-X.; Xin, H.-B.; Li, Z.-J.; Liu, H.; Sun, Y.-Q.; Nie, S.; Zhao, Z.-N.; Cui, R.-F.; Zhang, R.-G.; Yun, Q.-Z.; et al. High-quality assembly of the reference genome for scarlet sage, Salvia splendens, an economically important ornamental plant. GigaScience 2018, 7, 7. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic anditerative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT-The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef]
- Burge, C.; Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997, 268, 78–94. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Delcher, A.L.; Wortman, J.R.; Salzberg, S.L. DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics 2004, 20, 3643–3646. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-Y.; Christoffels, A.; Ramamoorthy, R.; Ramachandran, S. Expansion Mechanisms and Functional Annotations of Hypothetical Genes in the Rice Genome. Plant Physiol. 2009, 150, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y.W. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef]
- Nielsen, R.; Yang, Z. Likelihood Models for Detecting Positively Selected Amino Acid Sites and Applications to the HIV-1 Envelope Gene. Genetics 1998, 148, 929–936. [Google Scholar] [CrossRef]
- Wong, W.S.W.; Yang, Z.; Goldman, N.; Nielsen, R. Accuracy and Power of Statistical Methods for Detecting Adaptive Evolution in Protein Coding Sequences and for Identifying Positively Selected Sites. Genetics 2004, 168, 1041–1051. [Google Scholar] [CrossRef]
- Ding, G.; Zhang, S.; Ma, B.; Liang, J.; Li, H.; Luo, Y.; He, N. Origin and functional differentiation of (E)-β-ocimene synthases reflect the expansion of monoterpenes in angiosperms. J. Exp. Bot. 2020, 71, 6571–6586. [Google Scholar] [CrossRef]
- Zhou, K.; Peters, R.J. Investigating the conservation pattern of a putative second terpene synthase divalent metal binding motif in plants. Phytochemistry 2009, 70, 366–369. [Google Scholar] [CrossRef]
- Williams, D.C.; McGarvey, D.J.; Katahira, E.J.; Croteau, R. Truncation of Limonene Synthase Preprotein Provides a Fully Active ‘Pseudomature’ Form of This Monoterpene Cyclase and Reveals the Function of the Amino-Terminal Arginine Pair†. Biochemistry 1998, 37, 12213–12220. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef] [PubMed]
- Starks, C.M.; Back, K.; Chappell, J.; Noel, J.P. Structural Basis for Cyclic Terpene Biosynthesis by Tobacco 5-Epi-Aristolochene Synthase. Science 1997, 277, 1815–1820. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Wise, M.L.; Urbansky, M.; Coates, R.M.; Croteau, R.B.; Christianson, D.W. Nonlinear partial differential equations and applications: Bornyl diphosphate synthase: Structure and strategy for carbocation manipulation by a terpenoid cyclase. Proc. Natl. Acad. Sci. USA 2002, 99, 15375–15380. [Google Scholar] [CrossRef] [PubMed]
- Prisic, S.; Xu, J.; Coates, R.M.; Peters, R.J. Probing the Role of the DXDD Motif in Class II Diterpene Cyclases. ChemBioChem 2007, 8, 869–874. [Google Scholar] [CrossRef] [PubMed]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Mint Evolutionary Genomics Consortium. Phylogenomic mining of the mints reveals multiple mechanisms contributing to the evolution of chemical diversity in Lamiaceae. Mol. Plant 2018, 11, 1084–1096. [Google Scholar] [CrossRef]
- Hyatt, D.C.; Youn, B.; Zhao, Y.; Santhamma, B.; Coates, R.M.; Croteau, R.B.; Kang, C. Structure of limonene synthase, a simple model for terpenoid cyclase catalysis. Proc. Natl. Acad. Sci. USA 2007, 104, 5360–5365. [Google Scholar] [CrossRef] [PubMed]
| Gene ID | Scaffold | Start | End | Strand | Gene Length (bp) | CDS (bp) | Amino Acid | Exon Number | pI | Mw (kDa) | Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MlongTPS1 | scaffold3 | 25207839 | 25211071 | − | 3233 | 1635 | 544 | 7 | 5.08 | 62.93 | Chloroplast a/Cytoplasm b |
| MlongTPS2 | scaffold5 | 41734433 | 41737382 | − | 2950 | 1488 | 495 | 6 | 5.28 | 57.36 | Chloroplast a/Cytoplasm b |
| MlongTPS3 | scaffold5 | 41781767 | 41784235 | − | 2469 | 1638 | 545 | 6 | 4.99 | 63.01 | Chloroplast a/Cytoplasm b |
| MlongTPS4 | scaffold2 | 42600236 | 42604433 | + | 4198 | 1626 | 541 | 7 | 5.63 | 63.19 | Chloroplast a/Cytoplasm b |
| MlongTPS5 | scaffold2 | 42646914 | 42652153 | + | 5240 | 1626 | 541 | 7 | 5.56 | 63.06 | Chloroplast a/Cytoplasm b |
| MlongTPS6 | scaffold2 | 42808876 | 42813607 | + | 4732 | 1641 | 546 | 7 | 5.70 | 63.65 | Chloroplast a/Cytoplasmb |
| MlongTPS7 | scaffold10 | 2519038 | 2521515 | + | 2478 | 1572 | 523 | 8 | 5.01 | 60.86 | Chloroplast a/Cytoplasmb |
| MlongTPS8 | scaffold10 | 2869515 | 2871994 | + | 2480 | 1674 | 557 | 7 | 5.11 | 65.04 | Chloroplast a/Cytoplasm b |
| MlongTPS9 | scaffold10 | 3245887 | 3248093 | + | 2207 | 1311 | 436 | 8 | 5.82 | 51.00 | Chloroplast a,b |
| MlongTPS10 | scaffold10 | 24101862 | 24105239 | + | 3378 | 1341 | 446 | 7 | 5.94 | 52.67 | Chloroplast a/Cytoplasm b |
| MlongTPS11 | scaffold10 | 26605063 | 26606857 | − | 1795 | 1155 | 384 | 6 | 6.97 | 44.60 | Chloroplast a/Cytoplasm b |
| MlongTPS12 | scaffold8 | 2619187 | 2622034 | − | 2848 | 1482 | 493 | 6 | 5.44 | 57.37 | Chloroplast a/Cytoplasm b |
| MlongTPS13 | scaffold8 | 2629991 | 2633116 | − | 3126 | 1563 | 520 | 7 | 5.59 | 60.59 | Chloroplast a/Cytoplasm b |
| MlongTPS14 | scaffold11 | 22094766 | 22101682 | + | 6917 | 2589 | 862 | 13 | 5.30 | 100.10 | Chloroplast a,b |
| MlongTPS15 | scaffold11 | 22132562 | 22135423 | + | 2862 | 1791 | 596 | 7 | 5.26 | 69.43 | Chloroplast a,b |
| MlongTPS16 | scaffold11 | 22353164 | 22356569 | + | 3406 | 1782 | 593 | 7 | 5.73 | 68.84 | Chloroplast a,b |
| MlongTPS17 | scaffold11 | 22376541 | 22381192 | − | 4652 | 1560 | 519 | 10 | 5.78 | 60.82 | Chloroplast a,b |
| MlongTPS18 | scaffold11 | 22424761 | 22430157 | − | 5397 | 1449 | 482 | 7 | 5.46 | 56.62 | Chloroplast a,b |
| MlongTPS19 | scaffold11 | 29807062 | 29810465 | + | 3404 | 1782 | 593 | 7 | 5.65 | 68.76 | Chloroplast a,b |
| MlongTPS20 | scaffold11 | 29816966 | 29822114 | + | 5149 | 1362 | 453 | 6 | 7.12 | 52.57 | Chloroplast a,b |
| MlongTPS21 | scaffold11 | 29845320 | 29849984 | − | 4665 | 1320 | 439 | 8 | 5.79 | 51.21 | Chloroplast a,b |
| MlongTPS22 | scaffold11 | 29920867 | 29925533 | − | 4667 | 1476 | 491 | 7 | 5.61 | 57.69 | Chloroplast a,b |
| MlongTPS23 | scaffold4 | 34738619 | 34741984 | + | 3366 | 1374 | 457 | 7 | 5.74 | 53.33 | Chloroplast a,b |
| MlongTPS24 | scaffold4 | 34742308 | 34744838 | − | 2531 | 1800 | 599 | 7 | 5.41 | 69.98 | Chloroplast a,b |
| MlongTPS25 | scaffold5 | 285351 | 288259 | + | 2909 | 1734 | 577 | 7 | 5.18 | 67.16 | Chloroplast a,b |
| MlongTPS26 | scaffold5 | 291563 | 294867 | + | 3305 | 1737 | 578 | 7 | 5.46 | 67.19 | Chloroplast a,b |
| MlongTPS27 | scaffold5 | 296099 | 298389 | − | 2291 | 1383 | 460 | 5 | 5.78 | 53.55 | Chloroplast a,b |
| MlongTPS28 | scaffold5 | 11506827 | 11509585 | − | 2759 | 1800 | 599 | 7 | 5.32 | 69.92 | Chloroplast a,b |
| MlongTPS29 | scaffold5 | 11621067 | 11623817 | − | 2751 | 1800 | 599 | 7 | 5.43 | 69.91 | Chloroplast a,b |
| MlongTPS30 | scaffold5 | 21893670 | 21898545 | − | 4876 | 1779 | 592 | 7 | 6.23 | 69.34 | Chloroplast a,b |
| MlongTPS31 | scaffold2 | 19325281 | 19331000 | + | 5720 | 1737 | 578 | 7 | 5.36 | 67.30 | Chloroplast a,b |
| MlongTPS32 | scaffold10 | 30749715 | 30752287 | + | 2573 | 1653 | 550 | 7 | 5.55 | 63.29 | Chloroplast a,b |
| MlongTPS33 | scaffold10 | 30761480 | 30765652 | − | 4173 | 1599 | 532 | 8 | 5.55 | 62.05 | Chloroplast a,b |
| MlongTPS34 | scaffold10 | 30776115 | 30779012 | − | 2898 | 1374 | 457 | 6 | 6.07 | 53.11 | Chloroplast a/Cytoplasm b |
| MlongTPS35 | scaffold10 | 30785670 | 30788296 | − | 2627 | 1590 | 529 | 7 | 6.77 | 61.55 | Chloroplast a,b |
| MlongTPS36 | scaffold4 | 37761090 | 37769581 | + | 8492 | 2430 | 809 | 15 | 6.76 | 92.10 | Chloroplast a,b |
| MlongTPS37 | scaffold9 | 4343490 | 4348710 | − | 5221 | 2409 | 802 | 14 | 5.95 | 91.97 | Chloroplast a,b |
| MlongTPS38 | scaffold9 | 4410562 | 4415127 | − | 4566 | 2178 | 725 | 15 | 7.84 | 82.44 | Chloroplast a,b |
| MlongTPS39 | scaffold9 | 4626769 | 4631237 | − | 4469 | 2304 | 767 | 14 | 5.84 | 87.25 | Chloroplast a,b |
| MlongTPS40 | scaffold8 | 14598298 | 14605058 | − | 6761 | 2346 | 781 | 14 | 6.19 | 89.79 | Chloroplast a,b |
| MlongTPS41 | scaffold9 | 4215819 | 4220540 | − | 4722 | 2085 | 694 | 13 | 5.65 | 80.41 | Chloroplast a,b |
| MlongTPS42 | scaffold9 | 4297285 | 4301128 | − | 3844 | 1737 | 578 | 11 | 6.10 | 67.05 | Chloroplast a,b |
| MlongTPS43 | scaffold9 | 4315863 | 4321588 | − | 5726 | 1755 | 584 | 11 | 5.48 | 67.38 | Chloroplast a, b |
| MlongTPS44 | scaffold9 | 4400967 | 4404832 | − | 3866 | 1827 | 608 | 14 | 5.90 | 70.06 | Chloroplast a,b |
| MlongTPS45 | scaffold9 | 4663702 | 4668738 | + | 5037 | 1752 | 583 | 14 | 5.43 | 66.94 | Chloroplast a,b |
| MlongTPS46 | scaffold9 | 4696275 | 4699991 | + | 3717 | 1689 | 562 | 10 | 5.58 | 65.28 | Chloroplast a,b |
| MlongTPS47 | scaffold9 | 4746792 | 4752673 | − | 5882 | 2295 | 764 | 14 | 5.88 | 87.58 | Chloroplast a,b |
| MlongTPS48 | scaffold9 | 4791367 | 4793719 | − | 2353 | 1134 | 377 | 6 | 5.31 | 43.28 | Mitochondrion a/Chloroplast b |
| MlongTPS49 | scaffold9 | 4890741 | 4894353 | − | 3613 | 1734 | 577 | 10 | 5.69 | 66.69 | Chloroplast a,b |
| MlongTPS50 | scaffold9 | 4940721 | 4944084 | + | 3364 | 1536 | 511 | 9 | 5.30 | 59.27 | Mitochondrion a/Chloroplast b |
| MlongTPS51 | scaffold9 | 4988299 | 4993896 | + | 5598 | 2292 | 763 | 14 | 5.77 | 87.38 | Chloroplast a,b |
| MlongTPS52 | scaffold9 | 5111972 | 5115082 | + | 3111 | 1515 | 504 | 9 | 5.38 | 58.34 | Mitochondrion a/Chloroplast b |
| MlongTPS53 | scaffold9 | 7132180 | 7139762 | + | 7583 | 1755 | 584 | 11 | 5.38 | 67.56 | Chloroplast a,b |
| MlongTPS54 | scaffold9 | 31439884 | 31443309 | − | 3426 | 1350 | 449 | 8 | 5.03 | 52.24 | Chloroplast a,b |
| MlongTPS55 | scaffold9 | 31907037 | 31911201 | − | 4165 | 1533 | 510 | 9 | 5.09 | 59.61 | Chloroplast a,b |
| MlongTPS56 | scaffold9 | 31917248 | 31919875 | − | 2628 | 1578 | 525 | 9 | 5.53 | 60.86 | Chloroplast a,b |
| MlongTPS57 | scaffold8 | 2453217 | 2457977 | − | 4761 | 2322 | 773 | 14 | 5.62 | 88.21 | Chloroplast a,b |
| MlongTPS58 | scaffold8 | 2469812 | 2471751 | − | 1940 | 1308 | 435 | 7 | 5.22 | 50.43 | Chloroplast a,b |
| MlongTPS59 | scaffold10 | 30078136 | 30083625 | − | 5490 | 2478 | 825 | 12 | 5.99 | 94.00 | Chloroplast a/Cytoplasm b |
| MlongTPS60 | scaffold11 | 3129977 | 3133005 | + | 3029 | 1521 | 506 | 6 | 5.97 | 57.84 | Unknown a/Cytoplasm b |
| MlongTPS61 | scaffold3 | 44742988 | 44745414 | + | 2427 | 1572 | 523 | 7 | 7.04 | 61.62 | Unknown a/Cytoplasm b |
| MlongTPS62 | scaffold6 | 2272054 | 2274523 | + | 2470 | 1728 | 575 | 7 | 5.82 | 66.44 | Unknown a/Cytoplasm b |
| MlongTPS63 | scaffold6 | 15636480 | 15639592 | − | 3113 | 1764 | 587 | 7 | 5.31 | 66.38 | Unknown a/Cytoplasm b |
| Species | Subfamily | Total | |||||
|---|---|---|---|---|---|---|---|
| a | b | c | e | f | g | ||
| M. longifolia | 13 | 22 | 5 | 18 | 1 | 4 | 63 |
| O. teruiflorum | 14 | 12 | 7 | 2 | 1 | 7 | 43 |
| S. indicum | 21 | 5 | 6 | 3 | 0 | 7 | 42 |
| S. miltiorrhiza | 32 | 21 | 5 | 2 | 1 | 3 | 64 |
| S. splendens | 52 | 30 | 7 | 7 | 2 | 6 | 104 |
| A. thaliana | 22 | 6 | 1 | 1 | 1 | 1 | 32 |
| TPS SubFamily | Model | np | Ln L | Estimates of Parameters | Model Compared | LRT p-Value | Positive Sites | |||
|---|---|---|---|---|---|---|---|---|---|---|
| TPS-a | M3 | 29 | −6662.29 | p: | 0.300 | 0.605 | 0.095 | M0 vs. M3 | 0.000 | [] |
| ω: | 0.047 | 0.287 | 0.782 | |||||||
| M0 | 25 | −6742.49 | ω0: | 0.225 | Not Allowed | |||||
| M2a | 28 | −6701.40 | p: | 0.819 | 0.044 | 0.138 | M1a vs. M2a | 1.000 | [] | |
| ω: | 0.191 | 1.000 | 1.000 | |||||||
| M1a | 26 | −6701.40 | p: | 0.819 | 0.181 | Not Allowed | ||||
| ω: | 0.191 | 1.000 | ||||||||
| M8 | 28 | −6664.45 | p0 = 0.989 | p = 0.948 | q = 2.701 | M7 vs. M8 | 0.631 | 212 C 0.781 | ||
| p1 = 0.011 | ω = 1.525 | |||||||||
| M7 | 26 | −6664.91 | p= | 0.912 | q= | 2.472 | Not Allowed | |||
| TPS-b | M3 | 47 | −2367.77 | p: | 0.109 | 0.602 | 0.289 | M0 vs. M3 | 0.000 | [] |
| ω: | 0.000 | 0.228 | 0.612 | |||||||
| M0 | 43 | −2393.98 | ω0: | 0.289 | Not Allowed | |||||
| M2a | 46 | −2382.37 | p: | 0.756 | 0.123 | 0.121 | M1a vs. M2a | 1.000 | [] | |
| ω: | 0.230 | 1.000 | 1.000 | |||||||
| M1a | 44 | −2382.37 | p: | 0.756 | 0.244 | Not Allowed | ||||
| ω: | 0.230 | 1.000 | ||||||||
| M8 | 46 | −2374.65 | p0 = 1.000 | p = 1.135 | q = 2.498 | M7 vs. M8 | 1.000 | |||
| p1 = 0.000 | ω = 1.000 | |||||||||
| M7 | 44 | −2374.65 | p= | 1.135 | q= | 2.498 | Not Allowed | |||
| TPS-c | M3 | 13 | −9115.18 | p: | 0.548 | 0.420 | 0.032 | M0 vs. M3 | 0.000 | [] |
| ω: | 0.070 | 0.407 | 8.173 | |||||||
| M0 | 9 | −9231.50 | ω0: | 0.202 | Not Allowed | |||||
| M2a | 12 | −9133.53 | p: | 0.779 | 0.166 | 0.055 | M1a vs. M2a | 1.000 | [] | |
| ω: | 0.129 | 1.000 | 1.000 | |||||||
| M1a | 10 | −9133.53 | p: | 0.779 | 0.221 | Not Allowed | ||||
| ω: | 0.129 | 1.000 | ||||||||
| M8 | 12 | −9115.20 | p0 = 0.968 | p = 0.772 | q = 2.595 | M7 vs. M8 | 0.000 | 8 F 0.567,16 A 0.551,19 L 0.515,28 Y 0.916,32 I 0.748,33 K 0.649,41 E 0.627,212 L 0.711,591 L 0.828,636 E 0.875,637 Q 0.838,639 M 0.851,640 A 0.712,641 A 0.611,643 V 0.944,647 D 0.627,654 K 0.738 | ||
| p1 = 0.032 | ω = 8.049 | |||||||||
| M7 | 10 | −9124.83 | p= | 0.673 | q= | 1.922 | Not Allowed | |||
| TPS-e | M3 | 39 | −6467.88 | p: | 0.300 | 0.539 | 0.160 | M0 vs. M3 | 0.000 | [] |
| ω: | 0.077 | 0.351 | 0.785 | |||||||
| M0 | 35 | −6537.92 | ω0: | 0.310 | Not Allowed | |||||
| M2a | 38 | −6492.46 | p: | 0.739 | 0.167 | 0.095 | M1a vs. M2a | 1.000 | [] | |
| ω: | 0.231 | 1.000 | 1.000 | |||||||
| M1a | 36 | −6492.46 | p: | 0.739 | 0.261 | Not Allowed | ||||
| ω: | 0.231 | 1.000 | ||||||||
| M8 | 38 | −6468.70 | p0 = 0.966 | p = 1.035 | q = 2.155 | M7 vs. M8 | 0.858 | 45 R 0.514,234 V 0.633 | ||
| p1 = 0.034 | ω = 1.000 | |||||||||
| M7 | 36 | −6468.86 | p= | 0.962 | q= | 1.829 | Not Allowed | |||
| TPS-g | M3 | 11 | −5784.14 | p: | 0.284 | 0.560 | 0.156 | M0 vs. M3 | 0.000 | [] |
| ω: | 0.046 | 0.296 | 24.257 | |||||||
| M0 | 7 | −5866.96 | ω0: | 0.202 | Not Allowed | |||||
| M2a | 10 | −5795.20 | p: | 0.652 | 0.232 | 0.117 | M1a vs. M2a | 1.000 | [] | |
| ω: | 0.134 | 1.000 | 1.000 | |||||||
| M1a | 8 | −5795.20 | p: | 0.652 | 0.348 | Not Allowed | ||||
| ω: | 0.134 | 1.000 | ||||||||
| M8 | 10 | −5784.63 | p0 = 0.869 | p = 0.935 | q = 2.849 | M7 vs. M8 | 0.008 | 15 K 0.532,141 C 0.547,177 N 0.551,294 R 0.510,299 W 0.517,363 R 0.524,423 D 0.501 | ||
| p1 = 0.131 | ω = 31.804 | |||||||||
| M7 | 8 | −5789.50 | p= | 0.716 | q= | 1.590 | Not Allowed | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Vining, K.J.; Qi, X.; Yu, X.; Zheng, Y.; Liu, Z.; Fang, H.; Li, L.; Bai, Y.; Liang, C.; et al. Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase. Genes 2021, 12, 518. https://doi.org/10.3390/genes12040518
Chen Z, Vining KJ, Qi X, Yu X, Zheng Y, Liu Z, Fang H, Li L, Bai Y, Liang C, et al. Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase. Genes. 2021; 12(4):518. https://doi.org/10.3390/genes12040518
Chicago/Turabian StyleChen, Zequn, Kelly J. Vining, Xiwu Qi, Xu Yu, Ying Zheng, Zhiqi Liu, Hailing Fang, Li Li, Yang Bai, Chengyuan Liang, and et al. 2021. "Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase" Genes 12, no. 4: 518. https://doi.org/10.3390/genes12040518
APA StyleChen, Z., Vining, K. J., Qi, X., Yu, X., Zheng, Y., Liu, Z., Fang, H., Li, L., Bai, Y., Liang, C., Li, W., & Lange, B. M. (2021). Genome-Wide Analysis of Terpene Synthase Gene Family in Mentha longifolia and Catalytic Activity Analysis of a Single Terpene Synthase. Genes, 12(4), 518. https://doi.org/10.3390/genes12040518





