Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas
Abstract
1. Introduction
2. A Parallel World of Multicellular Complexity: The Evolution of Social Amoebas
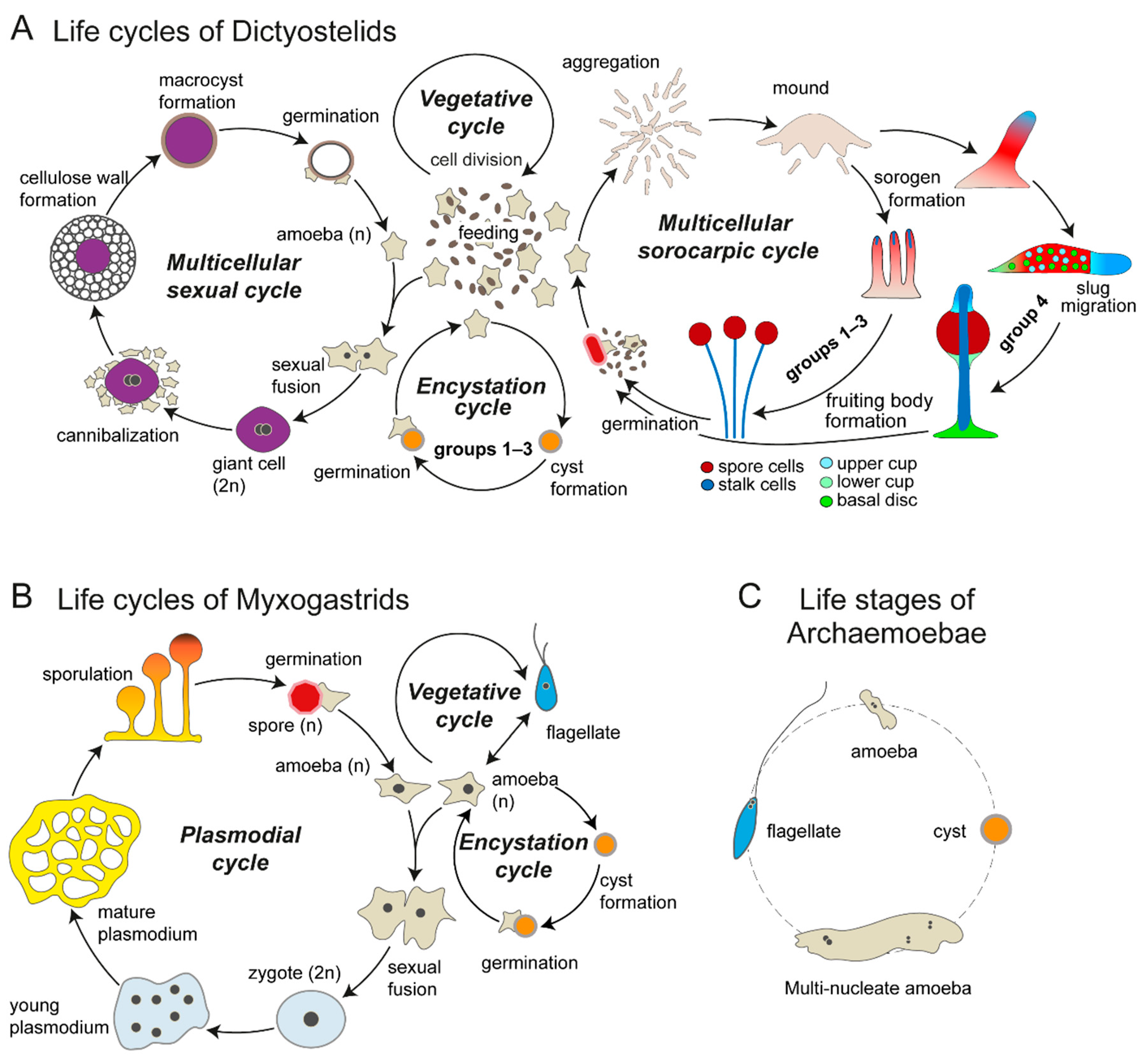
2.1. Social, Sexual, and Solitary Life Cycles of the Social Amoebas
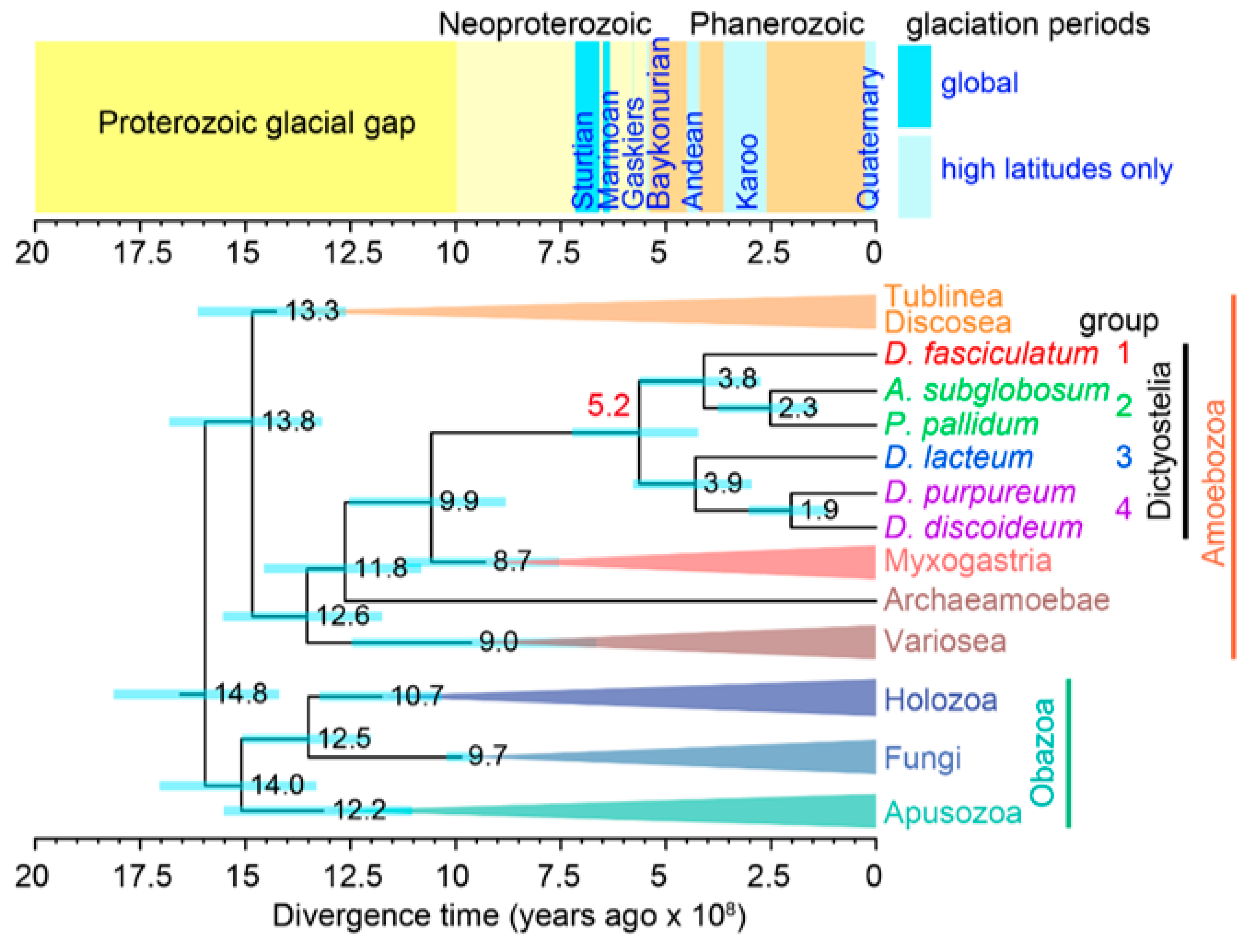
2.2. “Social amoebas” in Amoebozoa—The Phylogenetic Context
2.3. Ecological Advantages of Spores over Cysts: Increased Dispersal or Frost Resistance?
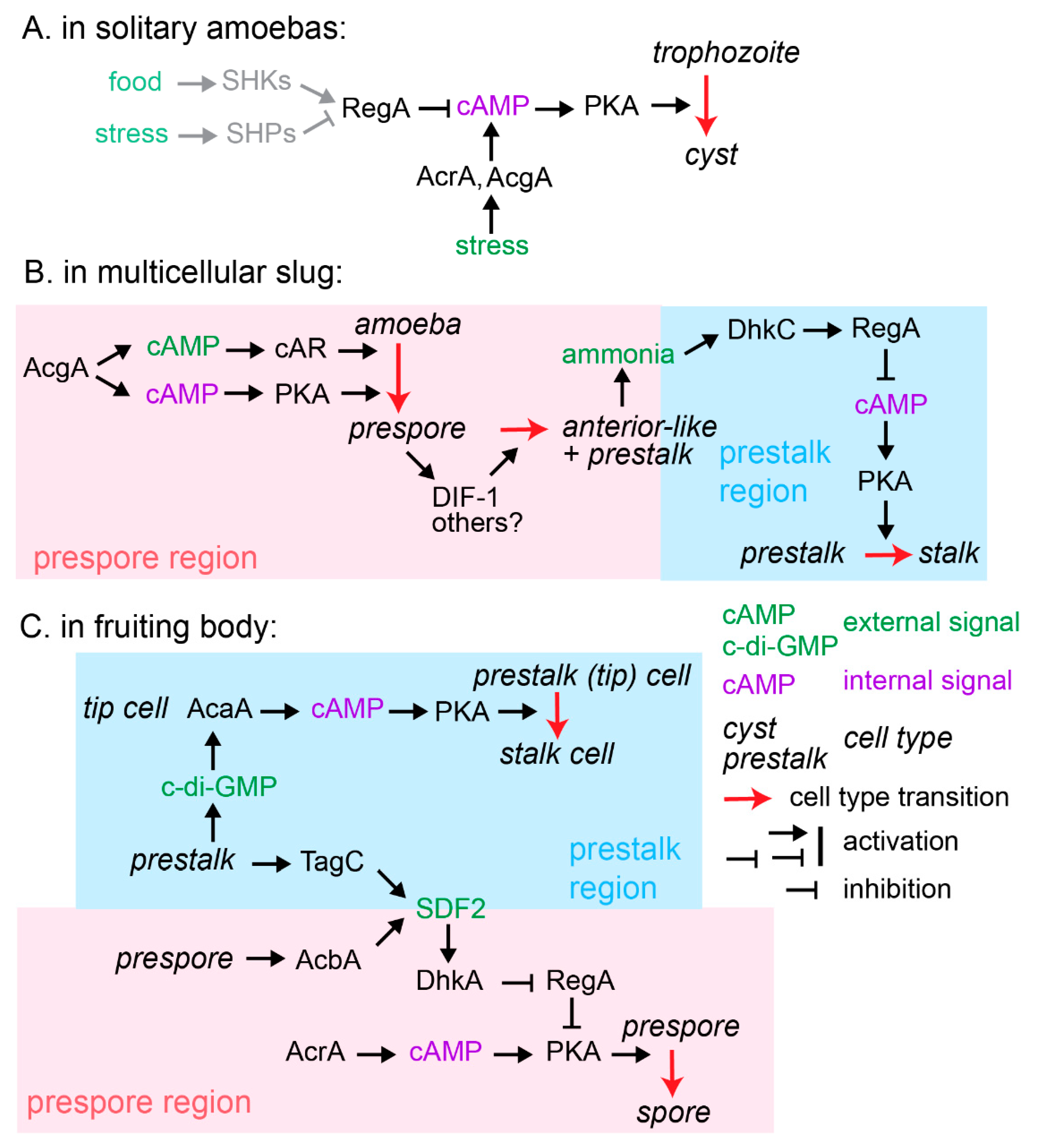
2.4. Evolution of Spore and Stalk Differentiation Pathways from an Ancestral Encystation Pathway
2.5. Major Developmental Innovations and Cell Type Evolution in Group 4 Dictyostelids
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- El Albani, A.; Bengtson, S.; Canfield, D.E.; Bekker, A.; Macchiarelli, R.; Mazurier, A.; Hammarlund, E.U.; Boulvais, P.; Dupuy, J.J.; Fontaine, C.; et al. Large colonial organisms with coordinated growth in oxygenated environments 2.1 Gyr ago. Nature 2010, 466, 100–104. [Google Scholar] [CrossRef] [PubMed]
- El Albani, A.; Mangano, M.G.; Buatois, L.A.; Bengtson, S.; Riboulleau, A.; Bekker, A.; Konhauser, K.; Lyons, T.; Rollion-Bard, C.; Bankole, O.; et al. Organism motility in an oxygenated shallow-marine environment 2.1 billion years ago. Proc. Natl. Acad. Sci. USA 2019, 116, 3431–3436. [Google Scholar] [CrossRef]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Sogaard-Andersen, L.; Van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef]
- Grosberg, R.K.; Strathmann, R.R. The evolution of multicellularity: A minor major transition? Annu. Rev. Ecol. Evol. Syst. 2007, 38, 621–654. [Google Scholar] [CrossRef]
- Knoll, A.H. The Multiple Origins of Complex Multicellularity. Annu. Rev. Earth Planet. Sci. 2011, 39, 217–239. [Google Scholar] [CrossRef]
- Sebe-Pedros, A.; Degnan, B.M.; Ruiz-Trillo, I. The origin of Metazoa: A unicellular perspective. Nat. Rev. Genet. 2017, 18, 498–512. [Google Scholar] [CrossRef]
- Frols, S. Archaeal biofilms: Widespread and complex. Biochem. Soc. Trans. 2013, 41, 393–398. [Google Scholar] [CrossRef]
- Ratcliff, W.C.; Denison, R.F.; Borrello, M.; Travisano, M. Experimental evolution of multicellularity. Proc. Natl. Acad. Sci. USA 2012, 109, 1595–1600. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, W.C.; Herron, M.D.; Howell, K.; Pentz, J.T.; Rosenzweig, F.; Travisano, M. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas Reinhardtii. Nat. Commun. 2013, 4, 2742. [Google Scholar] [CrossRef]
- Herron, M.D.; Borin, J.M.; Boswell, J.C.; Walker, J.; Chen, I.K.; Knox, C.A.; Boyd, M.; Rosenzweig, F.; Ratcliff, W.C. De novo origins of multicellularity in response to predation. Sci. Rep. 2019, 9, 2328. [Google Scholar] [CrossRef]
- Umen, J.G. Green algae and the origins of multicellularity in the plant kingdom. Cold Spring Harb. Perspect. Biol. 2014, 6, a016170. [Google Scholar] [CrossRef]
- O’Malley, M.A.; Wideman, J.G.; Ruiz-Trillo, I. Losing Complexity: The Role of Simplification in Macroevolution. Trends Ecol. Evol. 2016, 31, 608–621. [Google Scholar] [CrossRef]
- Lozada-Chávez, I.; Stadler, P.F.; Prohaska, S.J. Genome-wide features of introns are evolutionary decoupled among themselves and from genome size throughout Eukarya. bioRxiv 2018, 283549. [Google Scholar] [CrossRef]
- Kirk, D.L. A twelve-step program for evolving multicellularity and a division of labor. Bioessays 2005, 27, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Hanschen, E.R.; Davison, D.R.; Grochau-Wright, Z.I.; Michod, R.E. Evolution of individuality: A case study in the volvocine green algae. Philos. Theory Pract. Biol. 2017, 9, 3. [Google Scholar] [CrossRef][Green Version]
- Bell, G.; Mooers, A. Size and complexity among multicellular organisms. Biol. J. Linn. Soc. 1997, 60, 345–363. [Google Scholar] [CrossRef]
- Niklas, K.J.; Cobb, E.D.; Dunker, A.K. The number of cell types, information content, and the evolution of complex multicellularity. Acta Soc. Bot. Pol. 2014, 83, 337–347. [Google Scholar] [CrossRef]
- Valentine, J.W.; Collins, A.G.; Meyer, C.P. Morphological complexity increase in metazoans. Paleobiology 1994, 20, 131–142. [Google Scholar] [CrossRef]
- Clarrke, E. Multiple realizability of biological individuals. J. Philos. 2013, 8, 413–435. [Google Scholar] [CrossRef]
- Fisher, R.M.; Cornwallis, C.K.; West, S.A. Group formation, relatedness, and the evolution of multicellularity. Curr. Biol. CB 2013, 23, 1120–1125. [Google Scholar] [CrossRef]
- Brown, M.W.; Silberman, J.D. The Non-dictyostelid Sorocarpic Amoebae. In Dictyostelids; Romeralo, M., Baldauf, S.L., Escalante, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Du, Q.; Kawabe, Y.; Schilde, C.; Chen, Z.H.; Schaap, P. The Evolution of Aggregative Multicellularity and Cell-Cell Communication in the Dictyostelia. J. Mol. Biol. 2015, 427, 3722–3733. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, B.E.; De Vos, J.M.; Antonelli, A.; Bagheri, H.C. Evolution of multicellularity coincided with increased diversification of cyanobacteria and the Great Oxidation Event. Proc. Natl. Acad. Sci. USA 2013, 110, 1791–1796. [Google Scholar] [CrossRef] [PubMed]
- Knoll, A.H.; Hewitt, D. Phylogenetic, Functional, and Geological Perspectives on Complex Multicellularity. In The Major Transitions in Evolution Revisited; Calcott, B., Sterelny, K., Eds.; The MIT Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Hoffman, P.F.; Kaufman, A.J.; Halverson, G.P.; Schrag, D.P. A neoproterozoic snowball earth. Science 1998, 281, 1342–1346. [Google Scholar] [CrossRef]
- Lawal, H.M.; Schilde, C.; Kin, K.; Brown, M.W.; James, J.; Prescott, A.R.; Schaap, P. Cold climate adaptation is a plausible cause for evolution of multicellular sporulation in Dictyostelia. Sci. Rep. 2020, 10, 8797. [Google Scholar] [CrossRef]
- Stanley, S.M. An ecological theory for the sudden origin of multicellular life in the late precambrian. Proc. Natl. Acad. Sci. USA 1973, 70, 1486–1489. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, K.V.; Konstantinova, A.V.; Nikitin, M.A.; Troshin, P.V.; Rusin, L.Y.; Lyubetsky, V.A.; Panchin, Y.V.; Mylnikov, A.P.; Moroz, L.L.; Kumar, S.; et al. The origin of Metazoa: A transition from temporal to spatial cell differentiation. Bioessays 2009, 31, 758–768. [Google Scholar] [CrossRef]
- Nedelcu, A.M.; Michod, R.E. The evolutionary origin of an altruistic gene. Mol. Biol. Evol. 2006, 23, 1460–1464. [Google Scholar] [CrossRef]
- Schaap, P.; Winckler, T.; Nelson, M.; Alvarez-Curto, E.; Elgie, B.; Hagiwara, H.; Cavender, J.; Milano-Curto, A.; Rozen, D.E.; Dingermann, T.; et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science 2006, 314, 661–663. [Google Scholar] [CrossRef]
- Singh, R.; Schilde, C.; Schaap, P. A core phylogeny of Dictyostelia inferred from genomes representative of the eight major and minor taxonomic divisions of the group. BMC Evol. Biol. 2016, 16, 251. [Google Scholar] [CrossRef]
- Schilde, C.; Lawal, H.M.; Kin, K.; Shibano-Hayakawa, I.; Inouye, K.; Schaap, P. A well supported multi gene phylogeny of 52 dictyostelia. Mol. Phylogenet. Evol. 2019, 134, 66–73. [Google Scholar] [CrossRef]
- Sheikh, S.; Gloeckner, G.; Kuwayama, H.; Schaap, P.; Urushihara, H.; Baldauf, S.L. Root of Dictyostelia based on 213 universal proteins. Mol. Phylogenet. Evol. 2015, 92, 53–62. [Google Scholar] [CrossRef]
- Sheikh, S.; Thulin, M.; Cavender, J.C.; Escalante, R.; Kawakami, S.I.; Lado, C.; Landolt, J.C.; Nanjundiah, V.; Queller, D.C.; Strassmann, J.E.; et al. A New Classification of the Dictyostelids. Protist 2018, 169, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Olive, L.S. The Mycetozoa: A revised classification. Bot. Rev. 1970, 36, 59–89. [Google Scholar] [CrossRef]
- Simpson, A.G.B.; Bernard, C.; Fenchel, T.; Patterson, D.J. The Organisation of Mastigamoeba schizophrenia n. sp.: More Evidence of Ultrastructural Idiosyncrasy and Simplicity in Pelobiont Protists. Eur. J. Protistol. 1997, 33, 87–98. [Google Scholar] [CrossRef]
- Bloomfield, G. Sex in Dictyostelia. In Dictyostelids; Romeralo, M., Baldauf, S., Escalante, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 129–148. [Google Scholar]
- Bloomfield, G.; Skelton, J.; Ivens, A.; Tanaka, Y.; Kay, R.R. Sex determination in the social amoeba Dictyostelium discoideum. Science 2010, 330, 1533–1536. [Google Scholar] [CrossRef] [PubMed]
- Schaap, P.; Schilde, C. Encystation: The most prevalent and underinvestigated differentiation pathway of eukaryotes. Microbiology 2018, 164, 727–739. [Google Scholar] [CrossRef]
- Kang, S.; Tice, A.K.; Spiegel, F.W.; Silberman, J.D.; Panek, T.; Cepicka, I.; Kostka, M.; Kosakyan, A.; Alcantara, D.M.C.; Roger, A.J.; et al. Between a Pod and a Hard Test: The Deep Evolution of Amoebae. Mol. Biol. Evol. 2017, 34, 2258–2270. [Google Scholar] [CrossRef]
- Schilde, C.; Schaap, P. The Amoebozoa. Methods Mol. Biol. 2013, 983, 1–15. [Google Scholar] [CrossRef]
- Burki, F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 2014, 6, a016147. [Google Scholar] [CrossRef]
- Stephenson, S.L.; Shchnittler, M. Myxomycetes. In Handbook of the Protists; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1405–1431. [Google Scholar]
- Bailey, J. Plasmodium development in the myxomycete Physarum polycephalum: Genetic control and cellular events. Microbiology 1995, 141 Pt 10, 2355–2365. [Google Scholar] [CrossRef]
- Hehmeyer, J. Two potential evolutionary origins of the fruiting bodies of the dictyostelid slime molds. Biol. Rev. 2019. [Google Scholar] [CrossRef]
- Bloomfield, G. The evolution of ogres: Cannibalistic growth in giant phagotrophs. bioRxiv 2018, 262378. [Google Scholar] [CrossRef]
- Walker, G.; Zadrobílková, E.; Čepička, I. Archamoebae. In Handbook of the Protists; Archibald, J.M., Simpson, A.G.B., Slamovits, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar]
- Chavez, L.A.; Balamuth, W.; Gong, T. A light and electron microscopical study of a new, polymorphic free-living amoeba, Phreatamoeba balamuthi n. g., n. sp. J. Protozool. 1986, 33, 397–404. [Google Scholar] [CrossRef]
- Hofstatter, P.G.; Brown, M.W.; Lahr, D.J.G. Comparative Genomics Supports Sex and Meiosis in Diverse Amoebozoa. Genome Biol. Evol. 2018, 10, 3118–3128. [Google Scholar] [CrossRef] [PubMed]
- Tekle, Y.I.; Wood, F.C.; Katz, L.A.; Ceron-Romero, M.A.; Gorfu, L.A. Amoebozoans Are Secretly but Ancestrally Sexual: Evidence for Sex Genes and Potential Novel Crossover Pathways in Diverse Groups of Amoebae. Genome Biol. Evol. 2017, 9, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Heidel, A.; Lawal, H.; Felder, M.; Schilde, C.; Helps, N.; Tunggal, B.; Rivero, F.; John, U.; Schleicher, M.; Eichinger, L.; et al. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 2011, 21, 1882–1891. [Google Scholar] [CrossRef] [PubMed]
- Fiz-Palacios, O.; Romeralo, M.; Ahmadzadeh, A.; Weststrand, S.; Ahlberg, P.E.; Baldauf, S. Did terrestrial diversification of amoebas (amoebozoa) occur in synchrony with land plants? PLoS ONE 2013, 8, e74374. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, S.C.; Eme, L.; Brown, M.W.; Roger, A.J. Timing the Origins of Multicellular Eukaryotes Through Phylogenomics and Relaxed Molecular Clock Analyses. In Evolutionary Transitions to Multicellular Life; Ruiz-Trillo, I., Nedelcu, A.M., Eds.; Springer: Dordrecht, The Netherlands, 2015. [Google Scholar]
- Bonner, J.T. Evolutionary strategies and developmental constraints in the cellular slime molds. Am. Nat. 1982, 119, 530–552. [Google Scholar] [CrossRef]
- Smith, J.; Queller, D.C.; Strassmann, J.E. Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evol. Biol. 2014, 14, 105. [Google Scholar] [CrossRef]
- Pu, J.P.; Bowring, S.A.; Ramezani, J.; Myrow, P.; Raub, T.D.; Landing, E.; Mills, A.; Hodgin, E.; Macdonald, F.A. Dodging snowballs: Geochronology of the Gaskiers glaciation and the first appearance of the Ediacaran biota. Geology 2016, 44, 955–958. [Google Scholar] [CrossRef]
- Rooney, A.D.; Strauss, J.V.; Brandon, A.D.; Macdonald, F.A. A Cryogenian chronology: Two long-lasting synchronous Neoproterozoic glaciations. Geology 2015, 43, 459–462. [Google Scholar] [CrossRef]
- Kay, R.R.; Thompson, C.R. Cross-induction of cell types in Dictyostelium: Evidence that DIF-1 is made by prespore cells. Development 2001, 128, 4959–4966. [Google Scholar]
- Kawabe, Y.; Schilde, C.; Du, Q.; Schaap, P. A conserved signalling pathway for amoebozoan encystation that was co-opted for multicellular development. Sci. Rep. 2015, 5, 9644. [Google Scholar] [CrossRef]
- Kawabe, Y.; Morio, T.; James, J.L.; Prescott, A.R.; Tanaka, Y.; Schaap, P. Activated cAMP receptors switch encystation into sporulation. Proc. Natl. Acad. Sci. USA 2009, 106, 7089–7094. [Google Scholar] [CrossRef]
- Kesbeke, F.; Van Haastert, P.J.M. Reduced cAMP secretion in Dictyostelium discoideum mutant HB3. Dev. Biol. 1988, 130, 464–470. [Google Scholar] [CrossRef]
- Yamada, Y.; Schaap, P. Cyclic AMP induction of Dictyostelium prespore gene expression requires autophagy. Dev. Biol. 2019, 452, 114–126. [Google Scholar] [CrossRef]
- Schaap, P. Quantitative analysis of the spatial distribution of ultrastructural differentiation markers during development of Dictyostelium discoideum. Roux’s Arch. Dev. Biol. 1983, 192, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Loomis, W.F. Cell signaling during development of Dictyostelium. Dev. Biol. 2014, 391, 1–16. [Google Scholar] [CrossRef]
- Kessin, R.H. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity; Cambridge University Press: Cambridge, UK, 2001. [Google Scholar] [CrossRef]
- Alvarez-Curto, E.; Rozen, D.E.; Ritchie, A.V.; Fouquet, C.; Baldauf, S.L.; Schaap, P. Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc. Natl. Acad. Sci. USA 2005, 102, 6385–6390. [Google Scholar] [CrossRef]
- Ritchie, A.V.; Van Es, S.; Fouquet, C.; Schaap, P. From drought sensing to developmental control: Evolution of cyclic AMP signaling in social amoebas. Mol. Biol. Evol. 2008, 25, 2109–2118. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Schilde, C.; Birgersson, E.; Chen, Z.H.; McElroy, S.; Schaap, P. The cyclic AMP phosphodiesterase RegA critically regulates encystation in social and pathogenic amoebas. Cell. Signal. 2014, 26, 453–459. [Google Scholar] [CrossRef]
- Hillmann, F.; Forbes, G.; Novohradska, S.; Ferling, I.; Riege, K.; Groth, M.; Westermann, M.; Marz, M.; Spaller, T.; Winckler, T.; et al. Multiple Roots of Fruiting Body Formation in Amoebozoa. Genome Biol. Evol. 2018, 10, 591–606. [Google Scholar] [CrossRef]
- Schaap, P.; Barrantes, I.; Minx, P.; Sasaki, N.; Anderson, R.W.; Benard, M.; Biggar, K.K.; Buchler, N.E.; Bundschuh, R.; Chen, X.; et al. The Physarum polycephalum Genome Reveals Extensive Use of Prokaryotic Two-Component and Metazoan-Type Tyrosine Kinase Signaling. Genome Biol. Evol. 2015, 8, 109–125. [Google Scholar] [CrossRef]
- Fritz-Laylin, L.K.; Prochnik, S.E.; Ginger, M.L.; Dacks, J.B.; Carpenter, M.L.; Field, M.C.; Kuo, A.; Paredez, A.; Chapman, J.; Pham, J.; et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 2010, 140, 631–642. [Google Scholar] [CrossRef]
- Schaap, P.; Van Driel, R. Induction of post-aggregative differentiation in Dictyostelium discoideum by cAMP. Evidence of involvement of the cell surface cAMP receptor. Exp. Cell Res. 1985, 159, 388–398. [Google Scholar] [CrossRef]
- Alvarez-Curto, E.; Saran, S.; Meima, M.; Zobel, J.; Scott, C.; Schaap, P. cAMP production by adenylyl cyclase G induces prespore differentiation in Dictyostelium slugs. Development 2007, 134, 959–966. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hopper, N.A.; Harwood, A.J.; Bouzid, S.; Véron, M.; Williams, J.G. Activation of the prespore and spore cell pathway of Dictyostelium differentiation by cAMP-dependent protein kinase and evidence for its upstream regulation by ammonia. EMBO J. 1993, 12, 2459–2466. [Google Scholar] [CrossRef] [PubMed]
- Schindler, J.; Sussman, M. Ammonia determines the choice of morphogenetic pathways in Dictyostelium discoideum. J. Mol. Biol. 1977, 116, 161–169. [Google Scholar] [CrossRef]
- Singleton, C.K.; Zinda, M.J.; Mykytka, B.; Yang, P. The histidine kinase dhkC regulates the choice between migrating slugs and terminal differentiation in Dictyostelium Discoideum. Dev. Biol. 1998, 203, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Pitt, G.S.; Milona, N.; Borleis, J.; Lin, K.C.; Reed, R.R.; Devreotes, P.N. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell 1992, 69, 305–315. [Google Scholar] [CrossRef]
- Singer, G.; Araki, T.; Weijer, C.J. Oscillatory cAMP cell-cell signalling persists during multicellular Dictyostelium development. Commun. Biol. 2019, 2, 139. [Google Scholar] [CrossRef]
- Verkerke-van Wijk, I.; Fukuzawa, M.; Devreotes, P.N.; Schaap, P. Adenylyl cyclase A expression is tip-specific in Dictyostelium slugs and directs StatA nuclear translocation and CudA gene expression. Dev. Biol. 2001, 234, 151–160. [Google Scholar] [CrossRef][Green Version]
- Chen, Z.H.; Schaap, P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 2012, 488, 680–683. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Singh, R.; Cole, C.; Lawal, H.M.; Schilde, C.; Febrer, M.; Barton, G.J.; Schaap, P. Adenylate cyclase A acting on PKA mediates induction of stalk formation by cyclic diguanylate at the Dictyostelium organizer. Proc. Natl. Acad. Sci. USA 2017, 114, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Anjard, C.; Loomis, W.F. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. USA 2005, 102, 7607–7611. [Google Scholar] [CrossRef]
- Anjard, C.; Zeng, C.J.; Loomis, W.F.; Nellen, W. Signal transduction pathways leading to spore differentiation in dictyostelium discoideum. Dev. Biol. 1998, 193, 146–155. [Google Scholar] [CrossRef]
- Glockner, G.; Lawal, H.M.; Felder, M.; Singh, R.; Singer, G.; Weijer, C.J.; Schaap, P. The multicellularity genes of dictyostelid social amoebas. Nat. Commun. 2016, 7, 12085. [Google Scholar] [CrossRef] [PubMed]
- Morris, H.R.; Taylor, G.W.; Masento, M.S.; Jermyn, K.A.; Kay, R.R. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature 1987, 328, 811–814. [Google Scholar] [CrossRef]
- Saito, T.; Kato, A.; Kay, R.R. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev. Biol. 2008, 317, 444–453. [Google Scholar] [CrossRef]
- Neumann, C.S.; Walsh, C.T.; Kay, R.R. A flavin-dependent halogenase catalyzes the chlorination step in the biosynthesis of Dictyostelium differentiation-inducing factor 1. Proc. Natl. Acad. Sci. USA 2010, 107, 5798–5803. [Google Scholar] [CrossRef]
- Thompson, C.R.; Kay, R.R. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell. 2000, 6, 1509–1514. [Google Scholar] [CrossRef]
- Alegado, R.A.; King, N. Bacterial influences on animal origins. Cold Spring Harb. Perspect. Biol. 2014, 6, a016162. [Google Scholar] [CrossRef] [PubMed]
- De Mendoza, A.; Suga, H.; Permanyer, J.; Irimia, M.; Ruiz-Trillo, I. Complex transcriptional regulation and independent evolution of fungal-like traits in a relative of animals. eLife 2015, 4, e08904. [Google Scholar] [CrossRef]
- Romeralo, M.; Skiba, A.; Gonzalez-Voyer, A.; Schilde, C.; Lawal, H.; Kedziora, S.; Cavender, J.C.; Glockner, G.; Urushihara, H.; Schaap, P. Analysis of phenotypic evolution in Dictyostelia highlights developmental plasticity as a likely consequence of colonial multicellularity. Proc. Biol. Sci. 2013, 280, 20130976. [Google Scholar] [CrossRef]
- Schilde, C.; Skiba, A.; Schaap, P. Evolutionary reconstruction of pattern formation in 98 Dictyostelium species reveals that cell-type specialization by lateral inhibition is a derived trait. EvoDevo 2014, 5, 34. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, Y.; Takeuchi, I. Differentiation of various cell types during fruiting body formatio of Dictyostelium discoideum. Develop. Growth Differ. 1981, 23, 533–542. [Google Scholar] [CrossRef]
- Sternfeld, J.; David, C.N. Cell sorting during pattern formation in Dictyostelium. Differentiation 1981, 20, 10–21. [Google Scholar] [CrossRef]
- Sternfeld, J.; David, C.N. Fate and regulation of anterior-like cells in Dictyostelium slugs. Dev. Biol. 1982, 93, 111–118. [Google Scholar] [CrossRef]
- Sternfeld, J. The anterior-like cells in Dictyostelium are required for the elevation of the spores during culmination. Dev. Genes Evol. 1998, 208, 487–494. [Google Scholar] [CrossRef]
- Early, A.E.; Gaskell, M.J.; Traynor, D.; Williams, J.G. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development 1993, 118, 353–362. [Google Scholar] [CrossRef]
- Jermyn, K.A.; Williams, J.G. An analysis of culmination in Dictyostelium using prestalk and stalk-specific cell autonomous markers. Development 1991, 111, 779–787. [Google Scholar]
- Jermyn, K.; Traynor, D.; Williams, J. The initiation of basal disc formation in Dictyostelium discoideum is an early event in culmination. Development 1996, 122, 753–760. [Google Scholar]
- Kin, K.; Forbes, G.; Cassidy, A.; Schaap, P. Cell-type specific RNA-Seq reveals novel roles and regulatory programs for terminally differentiated Dictyostelium cells. BMC Genom. 2018, 19, 764. [Google Scholar] [CrossRef]
- Shimada, N.; Kanno-Tanabe, N.; Minemura, K.; Kawata, T. GBF-dependent family genes morphologically suppress the partially active Dictyostelium STATa strain. Dev. Genes Evol. 2008, 218, 55–68. [Google Scholar] [CrossRef] [PubMed]
- Vicente, J.J.; Galardi-Castilla, M.; Escalante, R.; Sastre, L. Structural and functional studies of a family of Dictyostelium discoideum developmentally regulated, prestalk genes coding for small proteins. BMC Microbiol. 2008, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Kin, K.; Chen, Z.H.; Forbes, G.; Schaap, P. Gene duplication of a cudA-like transcription factor involved in the evolution of a novel cell type in the Dictyostelid social amoebas. Manuscript in preparation.
- Forbes, G.; Chen, Z.H.; Kin, K.; Lawal, H.M.; Schilde, C.; Yamada, Y.; Schaap, P. Phylogeny-wide conservation and change in developmental expression, cell-type specificity and functional domains of the transcriptional regulators of social amoebas. BMC Genom. 2019, 20, 890. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C. Six major steps in animal evolution: Are we derived sponge larvae? Evol. Dev. 2008, 10, 241–257. [Google Scholar] [CrossRef]
- Grochau-Wright, Z.I.; Hanschen, E.R.; Ferris, P.J.; Hamaji, T.; Nozaki, H.; Olson, B.J.S.C.; Michod, R.E. Genetic basis for soma is present in undifferentiated volvocine green algae. J. Evol. Biol. 2017, 30, 1205–1218. [Google Scholar] [CrossRef]
- Konig, S.G.; Nedelcu, A.M. The genetic basis for the evolution of soma: Mechanistic evidence for the co-option of a stress-induced gene into a developmental master regulator. Proc. Biol. Sci. 2020, 287, 20201414. [Google Scholar] [CrossRef]
- Fairclough, S.R.; Chen, Z.; Kramer, E.; Zeng, Q.; Young, S.; Robertson, H.M.; Begovic, E.; Richter, D.J.; Russ, C.; Westbrook, M.J.; et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013, 14, R15. [Google Scholar] [CrossRef] [PubMed]
- King, N.; Westbrook, M.J.; Young, S.L.; Kuo, A.; Abedin, M.; Chapman, J.; Fairclough, S.; Hellsten, U.; Isogai, Y.; Letunic, I.; et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 2008, 451, 783–788. [Google Scholar] [CrossRef]
- Suga, H.; Chen, Z.; De Mendoza, A.; Sebe-Pedros, A.; Brown, M.W.; Kramer, E.; Carr, M.; Kerner, P.; Vervoort, M.; Sanchez-Pons, N.; et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat. Commun. 2013, 4, 2325. [Google Scholar] [CrossRef] [PubMed]
- Ros-Rocher, N.; Perez-Posada, A.; Leger, M.M.; Ruiz-Trillo, I. The origin of animals: An ancestral reconstruction of the unicellular-to-multicellular transition. Open Biol. 2021, 11, 200359. [Google Scholar] [CrossRef]
- Olson, B.J.; Nedelcu, A.M. Co-option during the evolution of multicellular and developmental complexity in the volvocine green algae. Curr. Opin. Genet. Dev. 2016, 39, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Wagner, G.P.; Erkenbrack, E.M.; Love, A.C. Stress-Induced Evolutionary Innovation: A Mechanism for the Origin of Cell Types. Bioessays 2019, 41, e1800188. [Google Scholar] [CrossRef]
- Nedelcu, A.M.; Michod, R.E. Stress Responses Co-Opted for Specialized Cell Types During the Early Evolution of Multicellularity: The Role of Stress in the Evolution of Cell Types Can Be Traced Back to the Early Evolution of Multicellularity. Bioessays News Rev. Mol. Cell. Dev. Biol. 2020, 42, e2000029. [Google Scholar] [CrossRef]
- Schlichting, C.D. Origins of differentiation via phenotypic plasticity. Evol. Dev. 2003, 5, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Arendt, D. The evolution of cell types in animals: Emerging principles from molecular studies. Nat. Rev. Genet. 2008, 9, 868–882. [Google Scholar] [CrossRef]
- Arendt, D.; Musser, J.M.; Baker, C.V.H.; Bergman, A.; Cepko, C.; Erwin, D.H.; Pavlicev, M.; Schlosser, G.; Widder, S.; Laubichler, M.D.; et al. The origin and evolution of cell types. Nat. Rev. Genet. 2016, 17, 744–757. [Google Scholar] [CrossRef]
- Medina, J.M.; Shreenidhi, P.M.; Larsen, T.J.; Queller, D.C.; Strassmann, J.E. Cooperation and conflict in the social amoeba Dictyostelium discoideum. Int. J. Dev. Biol. 2019, 63, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, E.A. Enforcing Cooperation in the Social Amoebae. Curr. Biol. CB 2019, 29, R474–R484. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, O.M.; Foster, K.R.; Mehdiabadi, N.J.; Strassmann, J.E.; Queller, D.C. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc. Natl. Acad. Sci. USA 2007, 104, 8913–8917. [Google Scholar] [CrossRef]
- Benabentos, R.; Hirose, S.; Sucgang, R.; Curk, T.; Katoh, M.; Ostrowski, E.A.; Strassmann, J.E.; Queller, D.C.; Zupan, B.; Shaulsky, G.; et al. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr. Biol. 2009, 19, 567–572. [Google Scholar] [CrossRef]
- Hirose, S.; Benabentos, R.; Ho, H.I.; Kuspa, A.; Shaulsky, G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science 2011, 333, 467–470. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kin, K.; Schaap, P. Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas. Genes 2021, 12, 487. https://doi.org/10.3390/genes12040487
Kin K, Schaap P. Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas. Genes. 2021; 12(4):487. https://doi.org/10.3390/genes12040487
Chicago/Turabian StyleKin, Koryu, and Pauline Schaap. 2021. "Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas" Genes 12, no. 4: 487. https://doi.org/10.3390/genes12040487
APA StyleKin, K., & Schaap, P. (2021). Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas. Genes, 12(4), 487. https://doi.org/10.3390/genes12040487




