Maternal Vitamin and Mineral Supplementation and Rate of Maternal Weight Gain Affects Placental Expression of Energy Metabolism and Transport-Related Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals, Experimental Design, and Tissue Collection
2.2. Total RNA Isolation, Library Preparation, Sequencing, and Data Analysis
2.3. Differential Expression and Functional Over-Representation Analyses
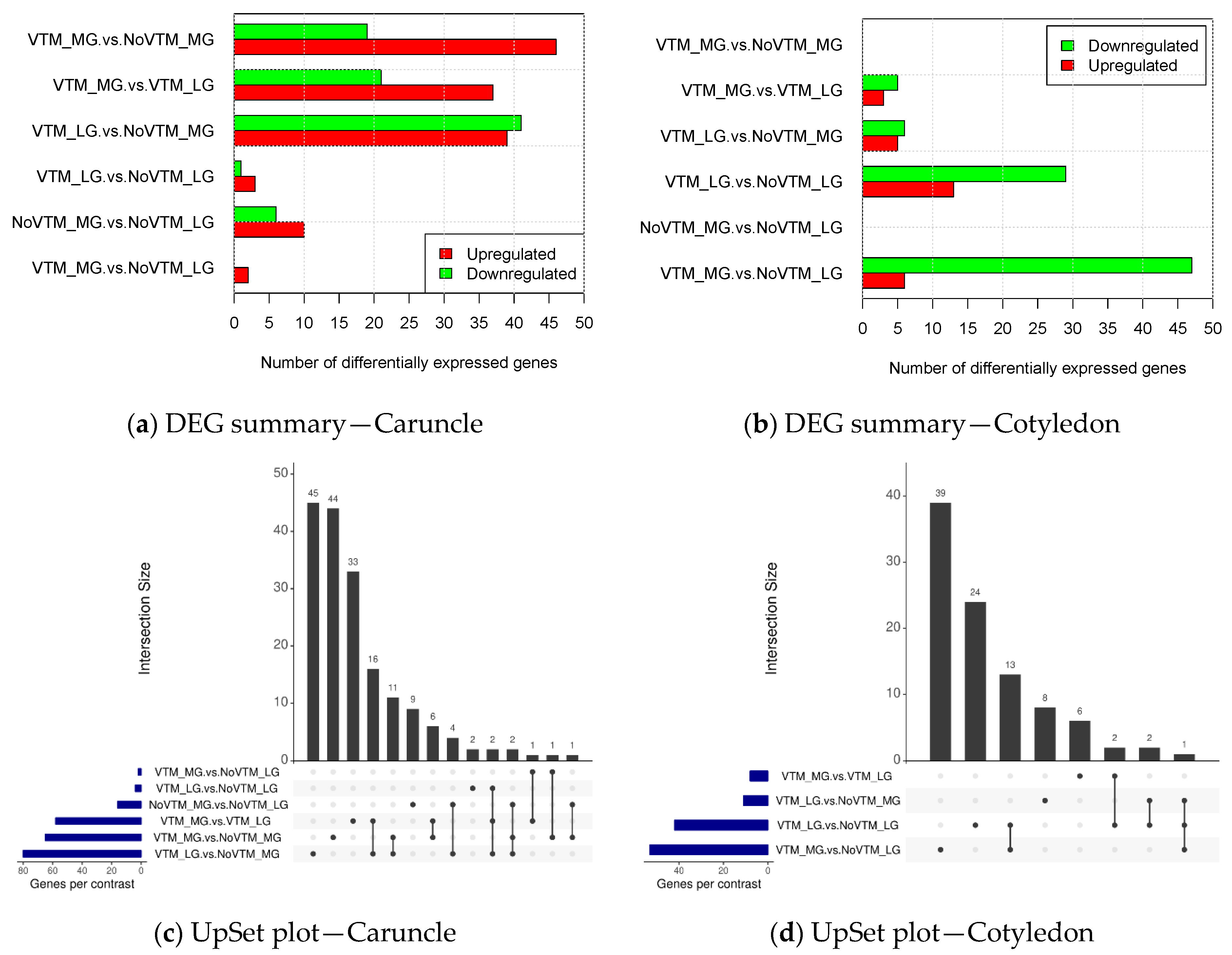
3. Results
3.1. Differentially Expressed Genes
3.2. Functional Over-Representation Analysis
4. Discussion
4.1. Pathways Underlying Caruncular Differential Gene Expression
4.2. Pathways Underlying Cotyledonary Differential Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- King, J.C. Physiology of pregnancy and nutrient metabolism. Am. J. Clin. Nutrit. 2000, 71, 1218S–1225S. [Google Scholar] [CrossRef] [PubMed]
- Khayat, S.; Fanaei, H.; Ghanbarzehi, A. Minerals in pregnancy and lactation: A review article. J. Clin. Diagn. Res. 2017, 11, QE01–QE05. [Google Scholar] [CrossRef]
- Caton, J.S.; Crouse, M.S.; Reynolds, L.P.; Neville, T.L.; Dahlen, C.R.; Ward, A.K.; Swanson, K.C. Maternal nutrition and programming of offspring energy requirements. Transl. Anim. Sci. 2019, 3, 976–990. [Google Scholar] [CrossRef]
- Reynolds, L.P.; Ward, A.K.; Caton, J.S. Epigenetics and developmental programming in ruminants: Long-term impacts on growth and development. In Biology of Domestic Animals; A science publisher’s book; Scanes, C.G., Hill, R.A., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 85–120. ISBN 9781315152080. [Google Scholar]
- Thayer, Z.M.; Rutherford, J.; Kuzawa, C.W. The maternal nutritional buffering model: An evolutionary framework for pregnancy nutritional intervention. Evol. Med. Public Health 2020, 2020, 14–27. [Google Scholar] [CrossRef]
- Sandovici, I.; Hoelle, K.; Angiolini, E.; Constância, M. Placental adaptations to the maternal-fetal environment: Implications for fetal growth and developmental programming. Reprod. Biomed. Online 2012, 25, 68–89. [Google Scholar] [CrossRef] [PubMed]
- Vonnahme, K.A.; Lemley, C.O.; Caton, J.S.; Meyer, A.M. Impacts of maternal nutrition on vascularity of nutrient transferring tissues during gestation and lactation. Nutrients 2015, 7, 3497–3523. [Google Scholar] [CrossRef] [PubMed]
- Lemley, C.O.; Hart, C.G.; Lemire, R.L.; Heath King, E.; Hopper, R.M.; Park, S.B.; Rude, B.J.; Burnett, D.D. Maternal nutrient restriction alters uterine artery hemodynamics and placentome vascular density in Bos indicus and Bos taurus. J. Anim. Sci. 2018, 96, 4823–4834. [Google Scholar] [CrossRef]
- Grazul-Bilska, A.T.; Gaton, J.S.; Arndt, W.; Burchill, K.; Thorson, C.; Borowczyk, E.; Bilski, J.J.; Redmer, D.A.; Reynolds, L.P.; Vonnahme, K.A. Cellular proliferation and vascularization in ovine fetal ovaries: Effects of undernutrition and selenium in maternal diet. Reproduction 2009, 137, 699–707. [Google Scholar] [CrossRef]
- Grazul-Bilska, A.T.; Borowicz, P.P.; Johnson, M.L.; Minten, M.A.; Bilski, J.J.; Wroblewski, R.; Redmer, D.A.; Reynolds, L.P. Placental development during early pregnancy in sheep: Vascular growth and expression of angiogenic factors in maternal placenta. Reproduction 2010, 140, 165–174. [Google Scholar] [CrossRef]
- Crouse, M.S.; Caton, J.S.; Cushman, R.A.; McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Ward, A.K. Moderate nutrient restriction of beef heifers alters expression of genes associated with tissue metabolism, accretion, and function in fetal liver, muscle, and cerebrum by day 50 of gestation. Transl. Anim. Sci. 2019, 3, 855–866. [Google Scholar] [CrossRef]
- Long, N.M.; Vonnahme, K.A.; Hess, B.W.; Nathanielsz, P.W.; Ford, S.P. Effects of early gestational undernutrition on fetal growth, organ development, and placentomal composition in the bovine. J. Anim. Sci. 2009, 87, 1950–1959. [Google Scholar] [CrossRef] [PubMed]
- Paradis, F.; Wood, K.M.; Swanson, K.C.; Miller, S.P.; McBride, B.W.; Fitzsimmons, C. Maternal nutrient restriction in mid-to-late gestation influences fetal mRNA expression in muscle tissues in beef cattle. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Suttle, N. Mineral Nutrition of Livestock, 4th ed.; Suttle, N., Ed.; CABI: Wallingford, UK, 2010; ISBN 9781845934729. [Google Scholar]
- Christian, P.; Stewart, C.P. Maternal micronutrient deficiency, fetal development, and the risk of chronic disease. J. Nutr. 2010, 140, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Gernand, A.D.; Schulze, K.J.; Stewart, C.P.; West, K.P.; Christian, P. Micronutrient deficiencies in pregnancy worldwide: Health effects and prevention HHS Public Access. Nat. Rev. Endocrinol. 2016, 12, 274–289. [Google Scholar] [CrossRef] [PubMed]
- Keshri, A.; Bashir, Z.; Kumari, V.; Prasad, K.; Joysowal, M.; Singh, M.; Singh, D.; Tarun, A.; Shukla, S. Role of micronutrients during peri-parturient period of dairy animals–A review. Biol. Rhythm Res. 2019. [Google Scholar] [CrossRef]
- Lekatz, L.A.; Caton, J.S.; Taylor, J.B.; Reynolds, L.P.; Redmer, D.A.; Vonnahme, K.A. Maternal selenium supplementation and timing of nutrient restriction in pregnant sheep: Effects on maternal endocrine status and placental characteristics. J. Anim. Sci. 2010, 88, 955–971. [Google Scholar] [CrossRef] [PubMed]
- Orr, J.P.; Blakley, B.R. Investigation of the selenium status of aborted calves with cardiac failure and myocardial necrosis. J. Vet. Diagn. Investig. 1997, 9, 172–179. [Google Scholar] [CrossRef]
- Jacometo, C.B.; Osorio, J.S.; Socha, M.; Corrêa, M.N.; Piccioli-Cappelli, F.; Trevisi, E.; Loor, J.J. Maternal consumption of organic trace minerals alters calf systemic and neutrophil mRNA and microRNA indicators of inflammation and oxidative stress. J. Dairy Sci. 2015, 98, 7717–7729. [Google Scholar] [CrossRef]
- Davy, J.S.; Forero, L.C.; Shapero, M.W.K.; Rao, D.R.; Becchetti, T.A.; Koopman Rivers, C.; Stackhouse, J.W.; Deatley, K.L.; McNabb, B.R. Mineral status of California beef cattle. Transl. Anim. Sci. 2019, 3, 1–8. [Google Scholar] [CrossRef]
- Diniz, W.J.S.; Banerjee, P.; Regitano, L.C.A. Cross talk between mineral metabolism and meat quality: A systems biology overview. Physiol. Genom. 2019, 51, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Lalman, D.; Richards, C. Nutrient Requirements of Beef Cattle. Available online: http://pods.dasnr.okstate.edu/docushare/dsweb/Get/Document-1921/E-974web.pdf (accessed on 4 November 2020).
- Burns, B.M.; Fordyce, G.; Holroyd, R.G. A review of factors that impact on the capacity of beef cattle females to conceive, maintain a pregnancy and wean a calf—Implications for reproductive efficiency in northern Australia. Anim. Reprod. Sci. 2010, 122, 1–22. [Google Scholar] [CrossRef]
- Menezes, A.C.B.; McCarthy, K.L.; Kassetas, C.J.; Baumgaertner, F.; Kirsch, J.D.; Dorsam, S.; Neville, T.L.; Ward, A.K.; Borowicz, P.P.; Reynolds, L.P.; et al. Vitamin and mineral supplementation and rate of gain during the first trimester of gestation affect concentrations of amino acids in maternal serum and allantoic fluid of beef heifers. J. Anim. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Galyean, M.L.; Beauchemin, K.A.; Caton, J.; Cole, N.A.; Eisemann, J.H.; Engle, T.; Erickson, G.E.; Krehbiel, C.R.; Lemenager, R.P.; Tedeschi, L.O. National Academies of Sciences and Engineering and Medicine. In Nutrient Requirements of Beef Cattle, 8th ed.; National Academies Press: Washington, DC, USA, 2016; ISBN 978-0-309-31702-3. [Google Scholar]
- McCarthy, K.L.; Nestande, J.; Kassetas, C.J.; Menezes, A.C.B.; Baumgaertner, F.; Kirsch, J.D.; Dorsam, S.T.; Neville, T.L.; Ward, A.K.; Borowicz, P.P.; et al. Effects of Feeding a Vitamin and Mineral Supplement and/or an Energy Supplement to Beef Heifers During the First 84 Days of Pregnancy on Heifer Performance, Concentrations of Progesterone, and Corpus Luteum Size and Fetal Body Measurements. 2020. Available online: https://www.ag.ndsu.edu/publications/livestock/2020-north-dakota-beef-and-sheep-report#section-10 (accessed on 2 November 2020).
- McLean, K.J.; Dahlen, C.R.; Borowicz, P.P.; Reynolds, L.P.; Crosswhite, M.R.; Neville, B.W.; Walden, S.D.; Caton, J.S. Technical note: A new surgical technique for ovariohysterectomy during early pregnancy in beef heifers. J. Anim. Sci. 2016, 94, 5089–5096. [Google Scholar] [CrossRef] [PubMed]
- Grazul-Bilska, A.T.; Johnson, M.L.; Borowicz, P.P.; Minten, M.; Bilski, J.J.; Wroblewski, R.; Velimirovich, M.; Coupe, L.R.; Redmer, D.A.; Reynolds, L.P. Placental development during early pregnancy in sheep: Cell proliferation, global methylation, and angiogenesis in the fetal placenta. Reproduction 2011, 141, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FASTQC. A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 October 2020).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Rosen, B.D.; Bickhart, D.M.; Schnabel, R.D.; Koren, S.; Elsik, C.G.; Tseng, E.; Rowan, T.N.; Low, W.Y.; Zimin, A.; Couldrey, C.; et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience 2020, 9, 1–9. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Di Pietro, A.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Leek, J.T.; Storey, J.D. Capturing heterogeneity in gene expression studies by Surrogate Variable Analysis. PLoS Genet. 2007, 3, e161. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, K.M.; Barker, D.J.P. Fetal nutrition and adult disease. Am. J. Clin. Nutr. 2000, 71, 1344–1352. [Google Scholar] [CrossRef] [PubMed]
- Caton, J.S.; Crouse, M.S.; McLean, K.J.; Dahlen, C.R.; Ward, A.K.; Cushman, R.A.; Grazul-Bilska, A.T.; Neville, B.W.; Borowicz, P.P.; Reynolds, L.P. Maternal periconceptual nutrition, early pregnancy, and developmental outcomes in beef cattle. J. Anim. Sci. 2020, 98. [Google Scholar] [CrossRef]
- Sibley, C.P. Understanding placental nutrient transfer—Why bother? New biomarkers of fetal growth. J. Physiol. 2009, 587, 3431–3440. [Google Scholar] [CrossRef]
- Shrestha, N.; Holland, O.J.; Kent, N.L.; Perkins, A.V.; McAinch, A.J.; Cuffe, J.S.M.; Hryciw, D.H. Maternal high linoleic acid alters placental fatty acid composition. Nutrients 2020, 12, 2183. [Google Scholar] [CrossRef]
- Cerri, R.; Thompson, I.; Kim, I.; Ealy, A.; Hansen, P.; Staples, C.; Li, J.; Santos, J.; Thatcher, W. Effects of lactation and pregnancy on gene expression of endometrium of Holstein cows at day 17 of the estrous cycle or pregnancy. J. Dairy Sci. 2012, 95, 5657–5675. [Google Scholar] [CrossRef]
- Madison, B.B. Srebp2: A master regulator of sterol and fatty acid synthesis1. J. Lipid Res. 2016, 57, 333–335. [Google Scholar] [CrossRef]
- Chatuphonprasert, W.; Jarukamjorn, K.; Ellinger, I. Physiology and pathophysiology of steroid biosynthesis, transport and metabolism in the human placenta. Front. Pharmacol. 2018, 9, 1027. [Google Scholar] [CrossRef]
- Schuler, G.; Greven, H.; Kowalewski, M.P.; Döring, B.; Özalp, C.R.; Hoffmann, B. Placental steroids in cattle: Hormones, placental growth factors or by-products of trophoblast giant cell differentiation? Exp. Clin. Endocrinol. Diabetes 2008, 116, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Baczyk, D.; Kingdom, J.C.P.; Uhlén, P. Calcium signaling in placenta. Cell Calcium 2011, 49, 350–356. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Strauss, J.F.; Hansel, W.; Shore, L.S.; Izhar, M. Control of bovine placental progesterone synthesis: Roles of cholesterol availability and calcium-activated systems. J. Steroid Biochem. 1988, 29, 21–25. [Google Scholar] [CrossRef]
- Sellak, H.; Choi, C.S.; Dey, N.B.; Lincoln, T.M. Transcriptional and post-transcriptional regulation of cGMP-dependent protein kinase (PKG-I): Pathophysiological significance. Cardiovasc. Res. 2013, 97, 200–207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dordea, A.C.; Sweeney, M.; Taggart, J.; Lartey, J.; Wessel, H.; Robson, S.C.; Taggart, M.J. Differential vasodilation of human placental and myometrial arteries related to myofilament Ca2+-desensitization and the expression of Hsp20 but not MYPT1. Mol. Hum. Reprod. 2013, 19, 727–736. [Google Scholar] [CrossRef]
- Shin, J.S.; Choi, M.Y.; Longtine, M.S.; Nelson, D.M. Vitamin D effects on pregnancy and the placenta. Placenta 2010, 31, 1027–1034. [Google Scholar] [CrossRef] [PubMed]
- Zingg, J.-M.; Meydani, M.; Azzi, A. α-Tocopheryl phosphate-An activated form of vitamin E important for angiogenesis and vasculogenesis? BioFactors 2012, 38, 24–33. [Google Scholar] [CrossRef]
- Narlis, M.; Grote, D.; Gaitan, Y.; Boualia, S.K.; Bouchard, M. Pax2 and Pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J. Am. Soc. Nephrol. 2007, 18, 1121–1129. [Google Scholar] [CrossRef]
- Ferretti, E.; Arturi, F.; Mattei, T.; Scipioni, A.; Tell, G.; Tosi, E.; Presta, I.; Morisi, R.; Lacroix, L.; Gulino, A.; et al. Expression, regulation, and function of PAX8 in the human placenta and placental cancer cell lines. Endocrinology 2005, 146, 4009–4015. [Google Scholar] [CrossRef][Green Version]
- Mao, J.; Zhang, X.; Sieli, P.T.; Falduto, M.T.; Torres, K.E.; Rosenfeld, C.S. Contrasting effects of different maternal diets on sexually dimorphic gene expression in the murine placenta. Proc. Natl. Acad. Sci. USA 2010, 107, 5557–5562. [Google Scholar] [CrossRef]
- Reynolds, L.P.; Biondini, M.E.; Borowicz, P.P.; Vonnahme, K.A.; Caton, J.S.; Grazul-Bilska, A.T.; Redmer, D.A. Functional significance of developmental changes in placental microvascular architecture: The sheep as a model. Endothel. J. Endothel. Cell Res. 2005, 12, 11–19. [Google Scholar] [CrossRef]
- Van Eetvelde, M.; Kamal, M.M.; Hostens, M.; Vandaele, L.; Fiems, L.O.; Opsomer, G. Evidence for placental compensation in cattle. Animal 2016, 10, 1342–1350. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Anderle, P.; Hostettler, L.; Baumann, M.U.; Surbek, D.V.; Ontsouka, E.C.; Albrecht, C. Identification of placental nutrient transporters associated with intrauterine growth restriction and pre-eclampsia. BMC Genom. 2018, 19, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Gaccioli, F.; Lager, S.; Powell, T.L.; Jansson, T. Placental transport in response to altered maternal nutrition. J. Dev. Orig. Health Dis. 2013, 4, 101–115. [Google Scholar] [CrossRef]
- Dunlap, K.A.; Brown, J.D.; Keith, A.B.; Satterfield, M.C. Factors controlling nutrient availability to the developing fetus in ruminants. J. Anim. Sci. Biotechnol. 2015, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Palacios, M.; Ruiz-Alcaraz, A.J.; Sanchez-Campillo, M.; Larqué, E. Role of Insulin in placental transport of nutrients in gestational diabetes mellitus. Ann. Nutr. Metab. 2017, 70, 16–25. [Google Scholar] [CrossRef]
- Hoffman, N.J.; Penque, B.A.; Habegger, K.M.; Sealls, W.; Tackett, L.; Elmendorf, J.S. Chromium Enhances Insulin Responsiveness via AMPK. J. Nutr. Biochem. 2015, 25, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Batistel, F.; Alharthi, A.S.; Wang, L.; Parys, C.; Pan, Y.X.; Cardoso, F.C.; Loor, J.J. Placentome nutrient transporters and mammalian target of rapamycin signaling proteins are altered by the methionine supply during late gestation in dairy cows and are associated with newborn birth weight. J. Nutr. 2017, 147, 1640–1647. [Google Scholar] [CrossRef]
- Yang, T.; Espenshade, P.J.; Wright, M.E.; Yabe, D.; Gong, Y.; Aebersold, R.; Goldstein, J.L.; Brown, M.S. Crucial step in cholesterol homeostasis: Sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 2002, 110, 489–500. [Google Scholar] [CrossRef]
- Matsuda, S.; Kobayashi, M.; Kitagishi, Y. Expression and Function of PPARs in Placenta. PPAR Res. 2013, 2013, 1–7. [Google Scholar] [CrossRef]
- Lewis, R.M.; Wadsack, C.; Desoye, G. Placental fatty acid transfer. Curr. Opin. Clin. Nutr. Metab. Care 2018, 21, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Weedon-Fekjaer, M.S.; Dalen, K.T.; Solaas, K.; Staff, A.C.; Duttaroy, A.K.; Nebb, H.I. Activation of LXR increases acyl-CoA synthetase activity through direct regulation of ACSL3 in human placental trophoblast cells. J. Lipid Res. 2010, 51, 1886–1896. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.; Ohkuni, A.; Kitamura, T.; Abe, K.; Naganuma, T.; Ohno, Y.; Zoeller, R.A.; Kihara, A. The Sjögren-Larsson syndrome gene encodes a hexadecenal dehydrogenase of the sphingosine 1-phosphate degradation pathway. Mol. Cell 2012, 46, 461–471. [Google Scholar] [CrossRef]
- Bildirici, I.; Schaiff, W.T.; Chen, B.; Morizane, M.; Oh, S.Y.; O’Brien, M.; Sonnenberg-Hirche, C.; Chu, T.; Barak, Y.; Nelson, D.M.; et al. PLIN2 is essential for trophoblastic lipid droplet accumulation and cell survival during hypoxia. Endocrinology 2018, 159, 3937–3949. [Google Scholar] [CrossRef]
- Ohi, K.; Ursini, G.; Li, M.; Shin, J.H.; Ye, T.; Chen, Q.; Tao, R.; Kleinman, J.E.; Hyde, T.M.; Hashimoto, R.; et al. DEGS2 polymorphism associated with cognition in schizophrenia is associated with gene expression in brain. Transl. Psychiatry 2015, 5, 550. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, R.N.; Johnson, C.O.; Hesselson, S.; Sotoodhenia, N.; McKnight, B.; Sitlani, C.M.; Rea, T.D.; King, I.B.; Kwok, P.Y.; Mak, A.; et al. Common variation in fatty acid metabolic genes and risk of incident sudden cardiac arrest. Hear. Rhythm 2014, 11, 471–477. [Google Scholar] [CrossRef]
- Putzer, H.; Laalami, S. Regulation of the Expression of Aminoacyl-tRNA Synthetases and Translation Factors. Available online: https://www.ncbi.nlm.nih.gov/books/NBK6026/ (accessed on 2 November 2020).
- Robles, M.; Couturier-Tarrade, A.; Derisoud, E.; Geeverding, A.; Dubois, C.; Dahirel, M.; Aioun, J.; Prezelin, A.; Calvez, J.; Richard, C.; et al. Effects of dietary arginine supplementation in pregnant mares on maternal metabolism, placental structure and function and foal growth. Sci. Rep. 2019, 9, 1–19. [Google Scholar] [CrossRef]
- Bassila, C.; Ghemrawi, R.; Flayac, J.; Froese, D.S.; Baumgartner, M.R.; Guéant, J.L.; Coelho, D. Methionine synthase and methionine synthase reductase interact with MMACHC and with MMADHC. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 103–112. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diniz, W.J.S.; Reynolds, L.P.; Borowicz, P.P.; Ward, A.K.; Sedivec, K.K.; McCarthy, K.L.; Kassetas, C.J.; Baumgaertner, F.; Kirsch, J.D.; Dorsam, S.T.; et al. Maternal Vitamin and Mineral Supplementation and Rate of Maternal Weight Gain Affects Placental Expression of Energy Metabolism and Transport-Related Genes. Genes 2021, 12, 385. https://doi.org/10.3390/genes12030385
Diniz WJS, Reynolds LP, Borowicz PP, Ward AK, Sedivec KK, McCarthy KL, Kassetas CJ, Baumgaertner F, Kirsch JD, Dorsam ST, et al. Maternal Vitamin and Mineral Supplementation and Rate of Maternal Weight Gain Affects Placental Expression of Energy Metabolism and Transport-Related Genes. Genes. 2021; 12(3):385. https://doi.org/10.3390/genes12030385
Chicago/Turabian StyleDiniz, Wellison J. S., Lawrence P. Reynolds, Pawel P. Borowicz, Alison K. Ward, Kevin K. Sedivec, Kacie L. McCarthy, Cierrah J. Kassetas, Friederike Baumgaertner, James D. Kirsch, Sheri T. Dorsam, and et al. 2021. "Maternal Vitamin and Mineral Supplementation and Rate of Maternal Weight Gain Affects Placental Expression of Energy Metabolism and Transport-Related Genes" Genes 12, no. 3: 385. https://doi.org/10.3390/genes12030385
APA StyleDiniz, W. J. S., Reynolds, L. P., Borowicz, P. P., Ward, A. K., Sedivec, K. K., McCarthy, K. L., Kassetas, C. J., Baumgaertner, F., Kirsch, J. D., Dorsam, S. T., Neville, T. L., Forcherio, J. C., Scott, R. R., Caton, J. S., & Dahlen, C. R. (2021). Maternal Vitamin and Mineral Supplementation and Rate of Maternal Weight Gain Affects Placental Expression of Energy Metabolism and Transport-Related Genes. Genes, 12(3), 385. https://doi.org/10.3390/genes12030385







