Ocular Phenotype Associated with DYRK1A Variants
Abstract
1. Introduction
2. Materials and Methods
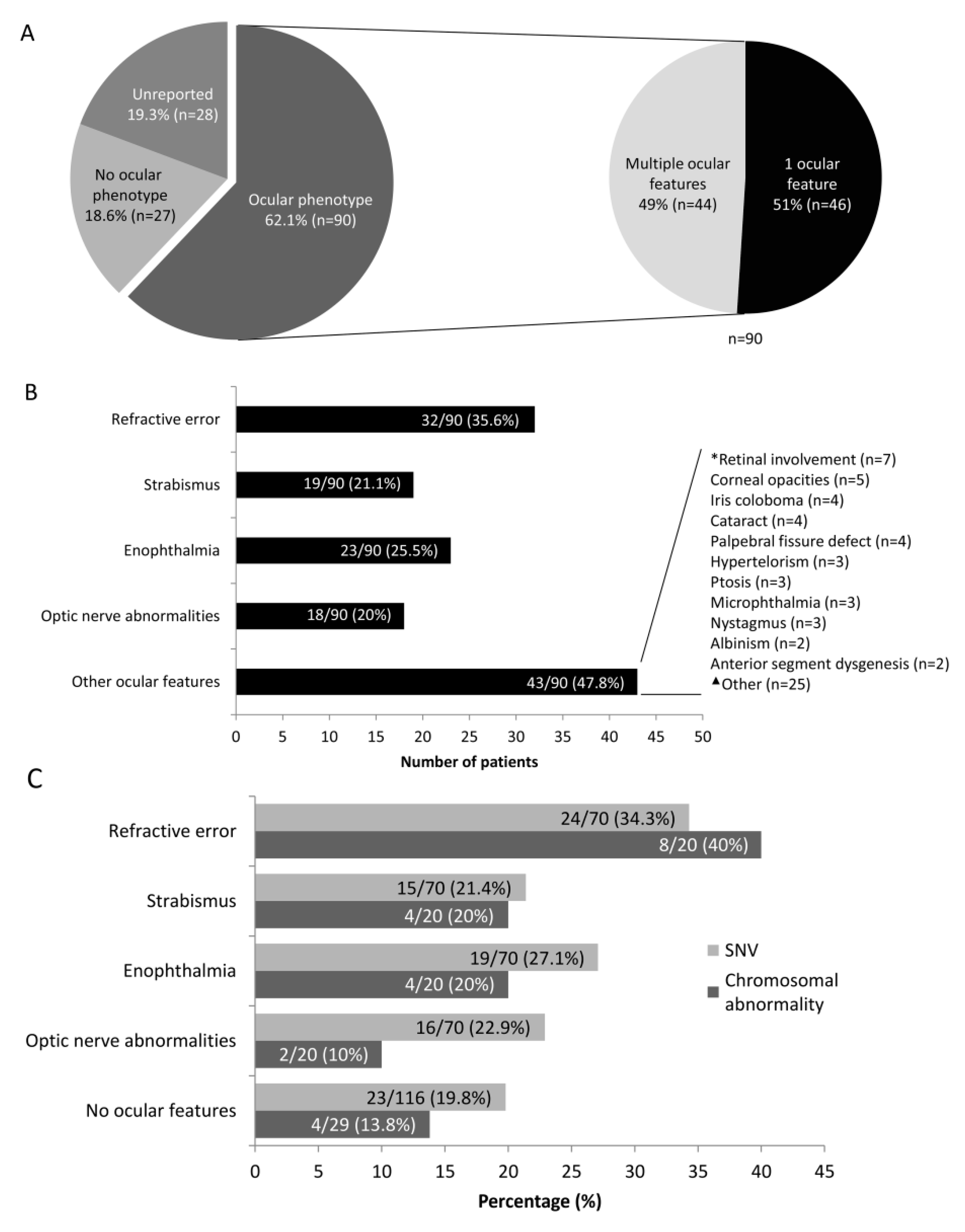
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martí, E.; Altafaj, X.; Dierssen, M.; De La Luna, S.; Fotaki, V.; Alvarez, M.; Pérez-Riba, M.; Ferrer, I.; Estivill, X. Dyrk1A expression pattern supports specific roles of this kinase in the adult central nervous system. Brain Res. 2003, 964, 250–263. [Google Scholar] [CrossRef]
- Arque, G.; Casanovas, A.; Dierssen, M. Dyrk1A is dynamically expressed on subsets of motor neurons and in the neuromuscular junction: Possible role in down syndrome. PLoS ONE 2013, 8, e54285. [Google Scholar] [CrossRef]
- Kentrup, H.; Becker, W.; Heukelbach, J.; Wilmes, A.; Schürmann, A.; Huppertz, C.; Kainulainen, H.; Joost, H.G. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J. Biol. Chem. 1996, 271, 3488–3495. [Google Scholar] [CrossRef] [PubMed]
- Himpel, S.; Tegge, W.; Frank, R.; Leder, S.; Joost, H.G.; Becker, W. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J. Biol. Chem. 2000, 275, 2431–2438. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Song, W.J.; Chung, K.C. Function and regulation of Dyrk1A: Towards understanding Down syndrome. Cell. Mol. Life Sci. 2009, 66, 3235–3240. [Google Scholar] [CrossRef]
- Gwack, Y.; Sharma, S.; Nardone, J.; Tanasa, B.; Iuga, A.; Srikanth, S.; Okamura, H.; Bolton, D.; Feske, S.; Hogan, P.G.; et al. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature 2006, 441, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Parrish, J.Z.; Kim, M.D.; Jan, L.Y.; Yuh, N.J. Genome-wide analyses identify transcription factors required for proper morphogenesis of Drosophila sensory neuron dendrites. Genes Dev. 2006, 20, 820–835. [Google Scholar] [CrossRef] [PubMed]
- Kinstrie, R.; Lochhead, P.A.; Sibbet, G.; Morrice, N.; Cleghon, V. dDYRK2 and Minibrain interact with the chromatin remodelling factors SNR1 and TRX. Biochem. J. 2006, 398, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Arron, J.R.; Winslow, M.M.; Polleri, A.; Chang, C.-P.; Wu, H.; Gao, X.; Neilson, J.R.; Chen, L.; Heit, J.J.; Kim, S.K.; et al. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature 2006, 441, 595–600. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.J.; Ahn, Y.S.; Chung, K.C. Protein kinase Dyrk1 activates cAMP response element-binding protein during neuronal differentiation in hippocampal progenitor cells. J. Biol. Chem. 2001, 276, 39819–39824. [Google Scholar] [CrossRef]
- Nguyen, T.; Di Giovanni, S. NFAT signaling in neural development and axon growth. Int. J. Dev. Neurosci. Int. J. Dev. Neurosci. 2008, 26, 141–145. [Google Scholar] [CrossRef]
- Ahn, K.-J.; Jeong, H.K.; Choi, H.-S.; Ryoo, S.-R.; Kim, Y.J.; Goo, J.-S.; Choi, S.-Y.; Han, J.-S.; Ha, I.; Song, W.-J. DYRK1A BAC transgenic mice show altered synaptic plasticity with learning and memory defects. Neurobiol. Dis. 2006, 22, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Altafaj, X.; Dierssen, M.; Baamonde, C.; Martí, E.; Visa, J.; Guimerà, J.; Oset, M.; González, J.R.; Flórez, J.; Fillat, C.; et al. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down’s syndrome. Hum. Mol. Genet. 2001, 10, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Stevens, M.E.; Sudanagunta, S.P.; Bronson, R.T.; Makhinson, M.; Watabe, A.M.; O’Dell, T.J.; Fung, J.; Weier, H.U.; Cheng, J.F.; et al. Functional screening of 2 Mb of human chromosome 21q22.2 in transgenic mice implicates minibrain in learning defects associated with Down syndrome. Nat. Genet. 1997, 16, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Burger, P.C.; Vogel, F.S. The development of the pathologic changes of Alzheimer’s disease and senile dementia in patients with Down’s syndrome. Am. J. Pathol. 1973, 73, 457–476. [Google Scholar] [PubMed]
- Wisniewski, K.E.; Wisniewski, H.M.; Wen, G.Y. Occurrence of neuropathological changes and dementia of Alzheimer’s disease in Down’s syndrome. Ann. Neurol. 1985, 17, 278–282. [Google Scholar] [CrossRef]
- Olson, M.I.; Shaw, C.M. Presenile dementia and Alzheimer’s disease in mongolism. Brain 1969, 92, 147–156. [Google Scholar] [CrossRef]
- Chettouh, Z.; Croquette, M.F.; Delobel, B.; Gilgenkrants, S.; Leonard, C.; Maunoury, C.; Prieur, M.; Rethoré, M.O.; Sinet, P.M.; Chery, M. Molecular mapping of 21 features associated with partial monosomy 21: Involvement of the APP-SOD1 region. Am. J. Hum. Genet. 1995, 57, 62–71. [Google Scholar]
- Matsumoto, N.; Ohashi, H.; Tsukahara, M.; Kim, K.C.; Soeda, E.; Niikawa, N. Possible narrowed assignment of the loci of monosomy 21-associated microcephaly and intrauterine growth retardation to a 1.2-Mb segment at 21q22.2. Am. J. Hum. Genet. 1997, 60, 997–999. [Google Scholar]
- Bartsch, O.; Hinkel, G.K.; Petersen, M.B.; König, U.; Bugge, M.; Mikkelsen, M.; Avramopoulos, D.; Morris, M.; Antonarakis, S.E. A large family with subtelomeric translocation t(18;21)(q23;q22.1) and molecular breakpoint in the Down syndrome critical region. Hum. Genet. 1997, 100, 669–675. [Google Scholar] [CrossRef]
- Fujita, H.; Torii, C.; Kosaki, R.; Yamaguchi, S.; Kudoh, J.; Hayashi, K.; Takahashi, T.; Kosaki, K. Microdeletion of the Down syndrome critical region at 21q22. Am. J. Med. Genet. Part A 2010, 152A, 950–953. [Google Scholar] [CrossRef]
- Oegema, R.; De Klein, A.; Verkerk, A.J.; Schot, R.; Dumee, B.; Douben, H.; Eussen, B.; Dubbel, L.; Poddighe, P.J.; Van Der Laar, I.; et al. Distinctive phenotypic abnormalities associated with submicroscopic 21q22 deletion including DYRK1A. Mol. Syndromol. 2010, 1, 113–120. [Google Scholar] [CrossRef] [PubMed]
- van Bon, B.W.M.; Hoischen, A.; Hehir-Kwa, J.; de Brouwer, A.P.M.; Ruivenkamp, C.; Gijsbers, A.C.J.; Marcelis, C.L.; de Leeuw, N.; Veltman, J.A.; Brunner, H.G.; et al. Intragenic deletion in DYRK1A leads to mental retardation and primary microcephaly. Clin. Genet. 2011, 79, 296–299. [Google Scholar] [CrossRef] [PubMed]
- Møller, R.S.; Kübart, S.; Hoeltzenbein, M.; Heye, B.; Vogel, I.; Hansen, C.P.; Menzel, C.; Ullmann, R.; Tommerup, N.; Ropers, H.-H.; et al. Truncation of the Down syndrome candidate gene DYRK1A in two unrelated patients with microcephaly. Am. J. Hum. Genet. 2008, 82, 1165–1170. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Shimojima, K.; Nishizawa, T.; Matsuo, M.; Ito, M.; Imai, K. Clinical manifestations of the deletion of Down syndrome critical region including DYRK1A and KCNJ6. Am. J. Med. Genet. Part A 2011, 155, 113–119. [Google Scholar] [CrossRef]
- Valetto, A.; Orsini, A.; Bertini, V.; Toschi, B.; Bonuccelli, A.; Simi, F.; Sammartino, I.; Taddeucci, G.; Simi, P.; Saggese, G. Molecular cytogenetic characterization of an interstitial deletion of chromosome 21 (21q22.13q22.3) in a patient with dysmorphic features, intellectual disability and severe generalized epilepsy. Eur. J. Med. Genet. 2012, 55, 362–366. [Google Scholar] [CrossRef]
- Ji, J.; Lee, H.; Argiropoulos, B.; Dorrani, N.; Mann, J.; Martinez-Agosto, J.A.; Gomez-Ospina, N.; Gallant, N.; Bernstein, J.A.; Hudgins, L.; et al. DYRK1A haploinsufficiency causes a new recognizable syndrome with microcephaly, intellectual disability, speech impairment, and distinct facies. Eur. J. Hum. Genet. 2015, 23, 1473–1481. [Google Scholar] [CrossRef]
- Bronicki, L.M.; Redin, C.; Drunat, S.; Piton, A.; Lyons, M.; Passemard, S.; Baumann, C.; Faivre, L.; Thevenon, J.; Rivière, J.-B.; et al. Ten new cases further delineate the syndromic intellectual disability phenotype caused by mutations in DYRK1A. Eur. J. Hum. Genet. 2015, 23, 1482–1487. [Google Scholar] [CrossRef]
- Redin, C.; Gérard, B.; Lauer, J.; Herenger, Y.; Muller, J.; Quartier, A.; Masurel-Paulet, A.; Willems, M.; Lesca, G.; El-Chehadeh, S.; et al. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J. Med. Genet. 2014, 51, 724–736. [Google Scholar] [CrossRef]
- van Bon, B.W.M.; Coe, B.P.; Bernier, R.; Green, C.; Gerdts, J.; Witherspoon, K.; Kleefstra, T.; Willemsen, M.H.; Kumar, R.; Bosco, P.; et al. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol. Psychiatry 2016, 21, 126–132. [Google Scholar] [CrossRef]
- Hartman, P.; Beckman, K.; Silverstein, K.; Yohe, S.; Schomaker, M.; Henzler, C.; Onsongo, G.; Lam, H.C.; Munro, S.; Daniel, J.; et al. Next generation sequencing for clinical diagnostics: Five year experience of an academic laboratory. Mol. Genet. Metab. Rep. 2019, 19, 100464. [Google Scholar] [CrossRef] [PubMed]
- Luco, S.M.; Pohl, D.; Sell, E.; Wagner, J.D.; Dyment, D.A.; Daoud, H. Case report of novel DYRK1A mutations in 2 individuals with syndromic intellectual disability and a review of the literature. BMC Med Genet. 2016. [Google Scholar] [CrossRef]
- Tejedor, F.; Zhu, X.R.; Kaltenbach, E.; Ackermann, A.; Baumann, A.; Canal, I.; Heisenberg, M.; Fischbach, K.F.; Pongs, O. minibrain: A new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 1995, 14, 287–301. [Google Scholar] [CrossRef]
- Laguna, A.; Aranda, S.; Barallobre, M.J.; Barhoum, R.; Fernández, E.; Fotaki, V.; Delabar, J.M.; de la Luna, S.; de la Villa, P.; Arbonés, M.L. The protein kinase DYRK1A regulates caspase-9-mediated apoptosis during retina development. Dev. Cell 2008, 15, 841–853. [Google Scholar] [CrossRef]
- Laguna, A.; Barallobre, M.-J.; Marchena, M.-Á.; Mateus, C.; Ramírez, E.; Martínez-Cue, C.; Delabar, J.M.; Castelo-Branco, M.; de la Villa, P.; Arbonés, M.L. Triplication of DYRK1A causes retinal structural and functional alterations in Down syndrome. Hum. Mol. Genet. 2013, 22, 2775–2784. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.; Alabek, M.L.; Eldib, A.; Madan-Khetarpal, S.; Sebastian, J.; Bhatia, A.; Liasis, A.; Nischal, K.K. Ocular findings of albinism in DYRK1A-related intellectual disability syndrome. Ophthalmic Genet. 2020, 41, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Scott, R.H.; Thomas, E.; Jones, L.; Murugaesu, N.; Pretty, F.B.; Halai, D.; Baple, E.; Craig, C.; Hamblin, A.; et al. The 100 000 Genomes Project: Bringing whole genome sequencing to the NHS. BMJ 2018, 361, k1687. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Lindstrand, A.; Malmgren, H.; Sahlén, S.; Schoumans, J.; Nordgren, A.; Ergander, U.; Holm, E.; Anderlid, B.M.; Blennow, E. Detailed molecular and clinical characterization of three patients with 21q deletions. Clin. Genet. 2010, 77, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Iossifov, I.; Ronemus, M.; Levy, D.; Wang, Z.; Hakker, I.; Rosenbaum, J.; Yamrom, B.; Lee, Y.-H.; Narzisi, G.; Leotta, A.; et al. De novo gene disruptions in children on the autistic spectrum. Neuron 2012, 74, 285–299. [Google Scholar] [CrossRef] [PubMed]
- O’Roak, B.J.; Vives, L.; Fu, W.; Egertson, J.D.; Stanaway, I.B.; Phelps, I.G.; Carvill, G.; Kumar, A.; Lee, C.; Ankenman, K.; et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 2012, 338, 1619–1622. [Google Scholar] [CrossRef]
- Iglesias, A.; Anyane-Yeboa, K.; Wynn, J.; Wilson, A.; Truitt Cho, M.; Guzman, E.; Sisson, R.; Egan, C.; Chung, W.K. The usefulness of whole-exome sequencing in routine clinical practice. Genet. Med. 2014, 16, 922–931. [Google Scholar] [CrossRef]
- Okamoto, N.; Miya, F.; Tsunoda, T.; Kato, M.; Saitoh, S.; Yamasaki, M.; Shimizu, A.; Torii, C.; Kanemura, Y.; Kosaki, K. Targeted next-generation sequencing in the diagnosis of neurodevelopmental disorders. Clin. Genet. 2015, 88, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Farwell, K.D.; Shahmirzadi, L.; El-Khechen, D.; Powis, Z.; Chao, E.C.; Tippin Davis, B.; Baxter, R.M.; Zeng, W.; Mroske, C.; Parra, M.C.; et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model-based analysis: Results from 500 unselected families with undiagnosed genetic conditions. Genet. Med. 2015, 17, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Ruaud, L.; Mignot, C.; Guët, A.; Ohl, C.; Nava, C.; Héron, D.; Keren, B.; Depienne, C.; Benoit, V.; Maystadt, I.; et al. DYRK1A mutations in two unrelated patients. Eur. J. Med. Genet. 2015, 58, 168–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kong, W.; Gao, Y.; Liu, X.; Gao, K.; Xie, H.; Wu, Y.; Zhang, Y.; Wang, J.; Gao, F.; et al. Gene mutation analysis in 253 Chinese children with unexplained epilepsy and intellectual/developmental disabilities. PLoS ONE 2015, 10, e0141782. [Google Scholar] [CrossRef]
- Helbig, K.L.; Farwell Hagman, K.D.; Shinde, D.N.; Mroske, C.; Powis, Z.; Li, S.; Tang, S.; Helbig, I. Diagnostic exome sequencing provides a molecular diagnosis for a significant proportion of patients with epilepsy. Genet. Med. 2016, 18, 898–905. [Google Scholar] [CrossRef]
- Rump, P.; Jazayeri, O.; van Dijk-Bos, K.K.; Johansson, L.F.; van Essen, A.J.; Verheij, J.B.G.M.; Veenstra-Knol, H.E.; Redeker, E.J.W.; Mannens, M.M.A.M.; Swertz, M.A.; et al. Whole-exome sequencing is a powerful approach for establishing the etiological diagnosis in patients with intellectual disability and microcephaly. BMC Med. Genom. 2016, 9, 7. [Google Scholar] [CrossRef]
- Qiao, F.; Shao, B.; Wang, C.; Wang, Y.; Zhou, R.; Liu, G.; Meng, L.; Hu, P.; Xu, Z. A de novo mutation in DYRK1A causes syndromic intellectual disability: A Chinese case report. Front. Genet. 2019, 10, 1194. [Google Scholar] [CrossRef]
- Courcet, J.-B.; Faivre, L.; Malzac, P.; Masurel-Paulet, A.; Lopez, E.; Callier, P.; Lambert, L.; Lemesle, M.; Thevenon, J.; Gigot, N.; et al. The DYRK1A gene is a cause of syndromic intellectual disability with severe microcephaly and epilepsy. J. Med. Genet. 2012, 49, 731–736. [Google Scholar] [CrossRef]
- Evers, J.M.G.; Laskowski, R.A.; Bertolli, M.; Clayton-Smith, J.; Deshpande, C.; Eason, J.; Elmslie, F.; Flinter, F.; Gardiner, C.; Hurst, J.A.; et al. Structural analysis of pathogenic mutations in the DYRK1A gene in patients with developmental disorders. Hum. Mol. Genet. 2017, 26, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, M.; Estivill, X.; de la Luna, S. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 2003, 116, 3099–3107. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.J.; Anderson, M.G. Genetic modifiers as relevant biological variables of eye disorders. Hum. Mol. Genet. 2017, 26, R58–R67. [Google Scholar] [CrossRef] [PubMed]
- Božanić, D.; Tafra, R.; Saraga-Babić, M. Role of apoptosis and mitosis during human eye development. Eur. J. Cell Biol. 2003, 82, 421–429. [Google Scholar] [CrossRef]
- Cellerino, A.; Bähr, M.; Isenmann, S. Apoptosis in the developing visual system. Cell Tissue Res. 2000, 301, 53–69. [Google Scholar] [CrossRef]
- Bähr, M. Live or let die—Retinal ganglion cell death and survival during development and in the lesioned adult CNS. Trends Neurosci. 2000, 23, 483–490. [Google Scholar] [CrossRef]
- Thomas, C.N.; Berry, M.; Logan, A.; Blanch, R.J.; Ahmed, Z. Caspases in retinal ganglion cell death and axon regeneration. Cell Death Discov. 2017, 3, 1–13. [Google Scholar] [CrossRef]
- Da Cunha, R.P.; Moreira, J.B.D.C. Ocular findings in Down’s syndrome. Am. J. Ophthalmol. 1996, 122, 236–244. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Fang, P.-C. Sensory evoked potentials in infants with Down syndrome. Acta Paediatr. 2005, 94, 1615–1618. [Google Scholar] [CrossRef]
- Warburg, M. Visual impairment in adult people with moderate, severe, and profound intellectual disability. Acta Ophthalmol. Scand. 2001, 79, 450–454. [Google Scholar] [CrossRef]
- Krinsky-McHale, S.J.; Silverman, W.; Gordon, J.; Devenny, D.A.; Oley, N.; Abramov, I. Vision deficits in adults with Down syndrome. J. Appl. Res. Intellect. Disabil. 2014, 27, 247–263. [Google Scholar] [CrossRef]
- Fletcher, M.C.; Thompson, M.M. Eye abnormalities in the mentally defective. Am. J. Ment. Defic. 1961, 66, 242–244. [Google Scholar]
- Byron, H.M. Ophthalmic survey of 162 mentally retarded children. N. Y. State J. Med. 1962, 62, 3102–3104. [Google Scholar]
- Akinci, A.; Oner, O.; Bozkurt, O.H.; Guven, A.; Degerliyurt, A.; Munir, K. Refractive errors and strabismus in children with down syndrome: A controlled study. J. Pediatr. Ophthalmol. Strabismus 2009, 46, 83–86. [Google Scholar] [CrossRef]
- Yurdakul, N.S.; Ugurlu, S.; Maden, A. Strabismus in down syndrome. J. Pediatr. Ophthalmol. Strabismus 2006, 43, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K. Visual characteristics of children with Down syndrome. Jpn. J. Ophthalmol. 2017, 61, 271–279. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, H.; Fotouhi, A.; Yekta, A.; Pakzad, R.; Ostadimoghaddam, H.; Khabazkhoob, M. Global and regional estimates of prevalence of refractive errors: Systematic review and meta-analysis. J. Curr. Ophthalmol. 2018, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.C.; van der Lee, R.; Wasserman, W.W.; Study, C.; Friedman, J.M.; Lehman, A. Strabismus in children with intellectual disability: Part of a broader motor control phenotype? Pediatr. Neurol. 2019, 100, 87–91. [Google Scholar] [CrossRef]
- Hashemi, H.; Pakzad, R.; Heydarian, S.; Yekta, A.; Aghamirsalim, M.; Shokrollahzadeh, F.; Khoshhal, F.; Pakbin, M.; Ramin, S.; Khabazkhoob, M. Global and regional prevalence of strabismus: A comprehensive systematic review and meta-analysis. Strabismus 2019, 27, 54–65. [Google Scholar] [CrossRef]
- Li, J.C.; Wong, K.; Park, A.S.; Fricke, T.R.; Jackson, A.J. The challenges of providing eye care for adults with intellectual disabilities. Clin. Exp. Optom. 2015, 98, 420–429. [Google Scholar] [CrossRef]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to Autism Spectrum Disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef] [PubMed]
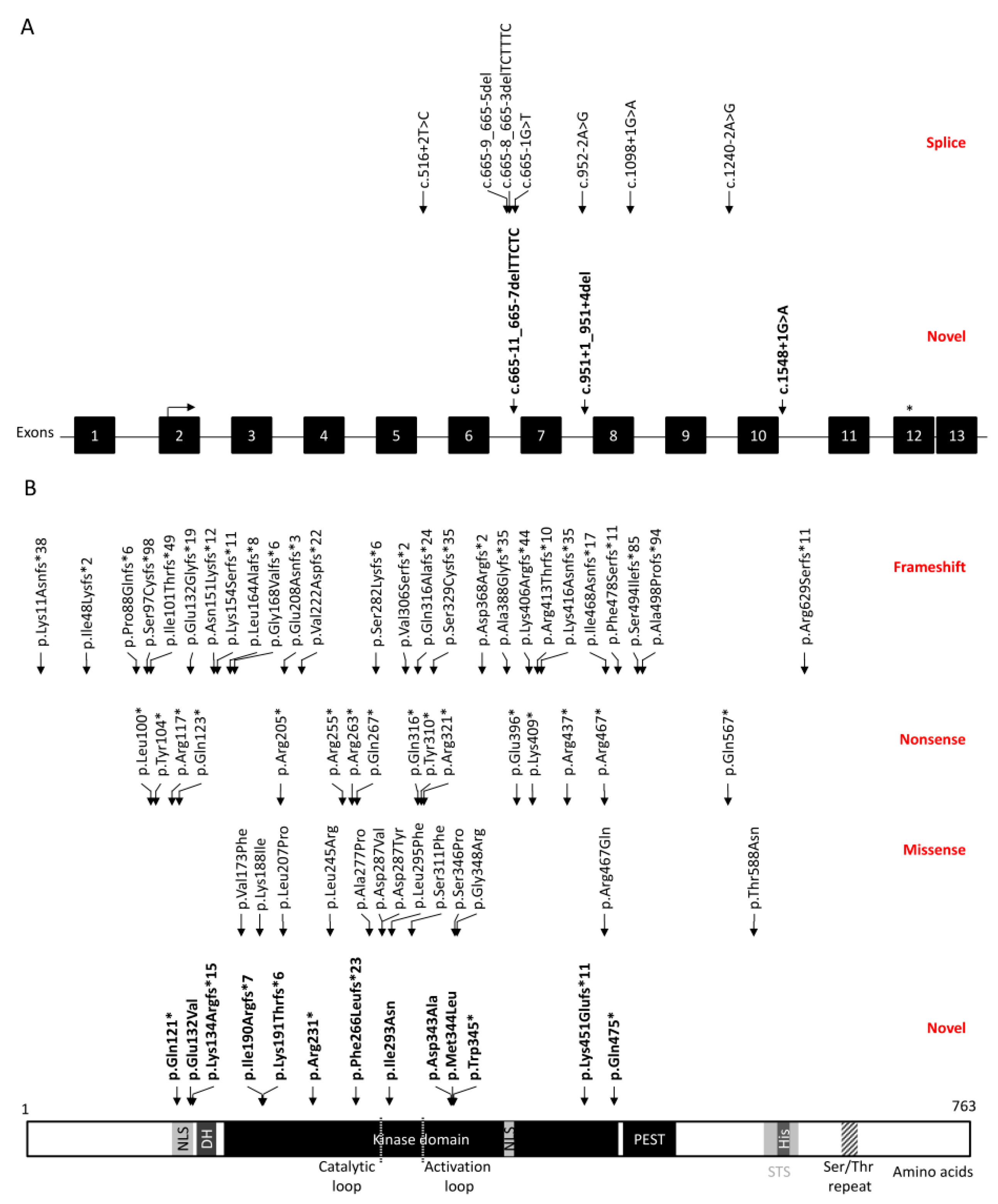
| DYRK1A Families Belonging to the DYRK1A Syndrome International Association (DSIA) | |||||
| Patient ID | DYRK1ADefect | Refractive Error | Strabismus | Optic Nerve Abnormalities | Other Findings |
| 1 | c.569_572delTAAA p.(Ile190Argfs*7) | Amblyopic hyperopia | Congenital nasolacrimal duct obstruction | ||
| 2 | c.572_575del p.(Lys191Thrfs*6) 1 | Exotropia | |||
| 3 | c.613C>T p.(Arg205*) | Hyperopic astigmatism | Superior oblique palsy Intermittent exotropia | ||
| 4 | c.691C>T p.(Arg231*) | Myopia | Iris coloboma (with CHARGE association) | ||
| 5 | c.763C>T p.(Arg255*) | Hypermetropia | Exotropia | Optic nerve hypoplasia | |
| 6 | c.860A>T p.(Asp287Val) | Hyperopia | Optic nerve hypoplasia | Bilateral cataracts | |
| 7 | c.1035G>A p.(Trp345*) 1 | Hyperopic astigmatism | |||
| 8 | c.1217_1220del p.(Lys406Argfs*44) | Optic nerve hypoplasia | Schnyder corneal dystrophy (additional mutation to UBIAD1) | ||
| 9 | c.1350dupG p.(Lys451Glufs*11) 1 | Myopic astigmatism | Esotropia | ||
| 10 | c.1400dupG p.(Ile468Asnfs*17) 1 | Refractive amblyopia | Superior oblique palsyIntermittent exotropia | Optic nerve hypoplasia | |
| 11 | del(21)(q22.12q23.3) | Microphthalmia Anterior segment dysgenesis Corneal opacities | |||
| 12 | del(21)(q22.13) | Optic nerve hypoplasia | Fundal pallor | ||
| 13 | del(21)(q22.13) | Myopic astigmatism | Esotropia | Optic nerve hypoplasia | |
| 14 | del(21)(q22.13q22.3) | Microphthalmia Sclerocornea | |||
| DYRK1Apatients from the UK Genomics England 100,000 Genomes Project with ocular features | |||||
| Patient ID | DYRK1ADefect | Refractive Error | Strabismus | Optic Nerve Abnormalities | Other Findings |
| 1 | c.361C>T p.(Gln121*) 1 | Severe visual impairment (HP:0001141) | |||
| 2 | c.613C>T p.(Arg205*) | Abnormality of the eye (HP:0000478) | |||
| 3 | c.763C>T p.(Arg255*) | Abnormality of the eye (HP:0000478) | |||
| 4 | c.878T>A p.(Ile293Asn) 1 | Aplasia/ Hypoplasia of the optic nerve (HP:0008058) | Abnormality of the eye (HP:0000478) | ||
| 5 | c.914_919del p.(Ile305_Asp307delinsAsn) 1 | Optic nerve hypoplasia (HP:0000609) | Abnormality of the eye (HP:0000478) Cataract (HP:0000518) | ||
| 6 | c.691C>T p.(Arg321*) 1 | Anterior segment dysgenesis (HP:0007700) | |||
| 7 | c.1028A>C p.(Asp343Ala) 1 | Hypertelorism (HP:0000316) Nonprogressive visual loss (HP:0200068) | |||
| 8 | c.1030A>T p.(Met344Leu) 1 | Hypertelorism (HP:0000316) Nonprogressive visual loss (HP:0200068) | |||
| 9 | c.1548+1G>A 1 | Optic nerve hypoplasia (HP:0000609) | Downslanted palpebral fissures (HP:0000494) Nystagmus (HP:0000639) Visual impairment (HP:0000505) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Méjécase, C.; Way, C.M.; Owen, N.; Moosajee, M. Ocular Phenotype Associated with DYRK1A Variants. Genes 2021, 12, 234. https://doi.org/10.3390/genes12020234
Méjécase C, Way CM, Owen N, Moosajee M. Ocular Phenotype Associated with DYRK1A Variants. Genes. 2021; 12(2):234. https://doi.org/10.3390/genes12020234
Chicago/Turabian StyleMéjécase, Cécile, Christopher M. Way, Nicholas Owen, and Mariya Moosajee. 2021. "Ocular Phenotype Associated with DYRK1A Variants" Genes 12, no. 2: 234. https://doi.org/10.3390/genes12020234
APA StyleMéjécase, C., Way, C. M., Owen, N., & Moosajee, M. (2021). Ocular Phenotype Associated with DYRK1A Variants. Genes, 12(2), 234. https://doi.org/10.3390/genes12020234







