Integrated Full-Length Transcriptome and RNA-Seq to Identify Immune System Genes from the Skin of Sperm Whale (Physeter macrocephalus)
Abstract
:1. Introduction
2. Materials and Methods
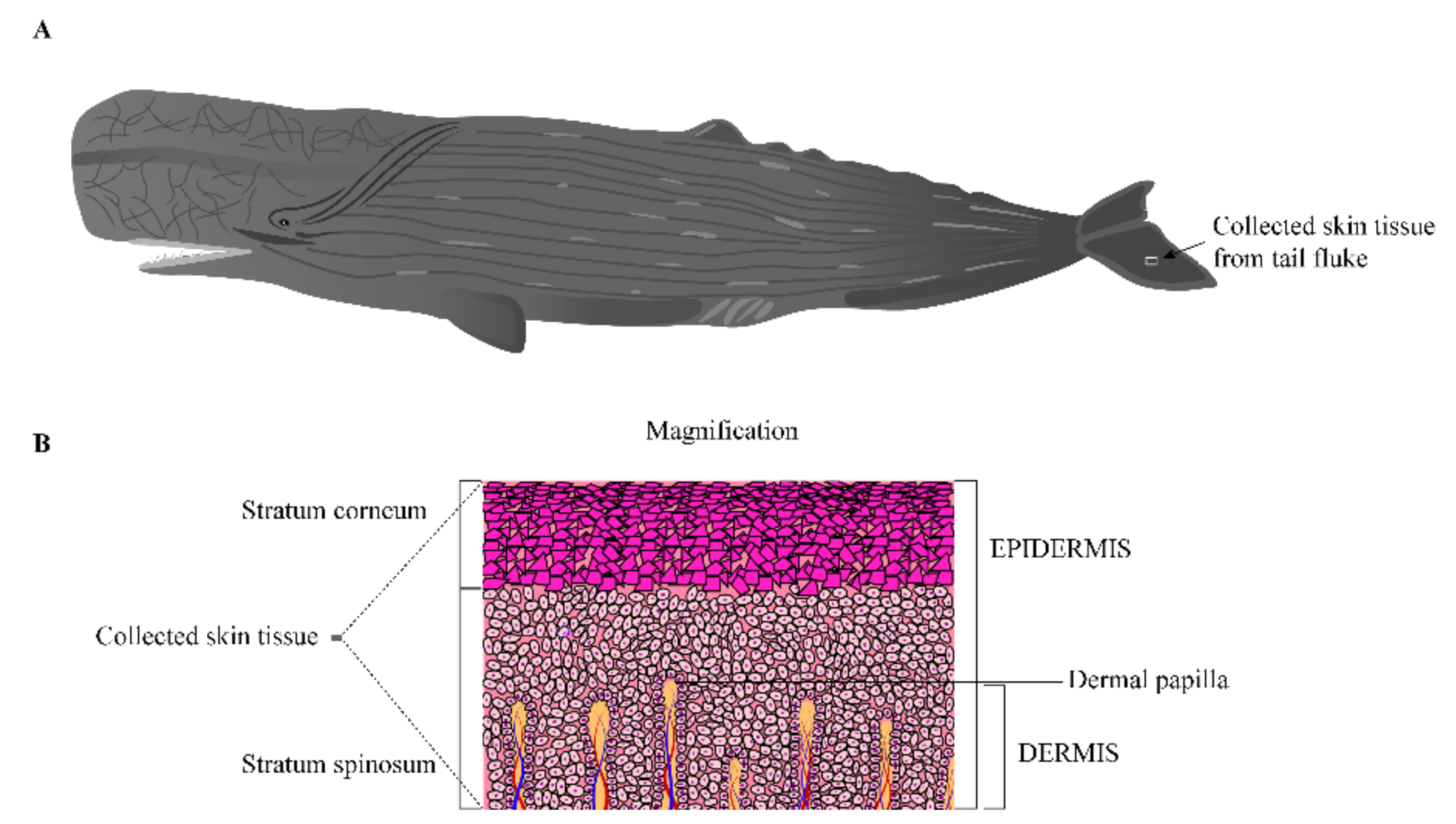
2.1. Animal and Tissue Collection
2.2. RNA Extraction and Quantification
2.3. PacBio Iso-Seq Library Preparation and Sequencing
2.4. RNA-seq Library Preparation and Sequencing
2.5. Bioinformatics Analysis of PacBio Data and RNA-seq Data
2.6. Quantification and Annotation of Gene Expression Levels
2.7. Sequence Analysis
3. Results
3.1. Full-Length Transcripts from the Skin of Sperm Whale
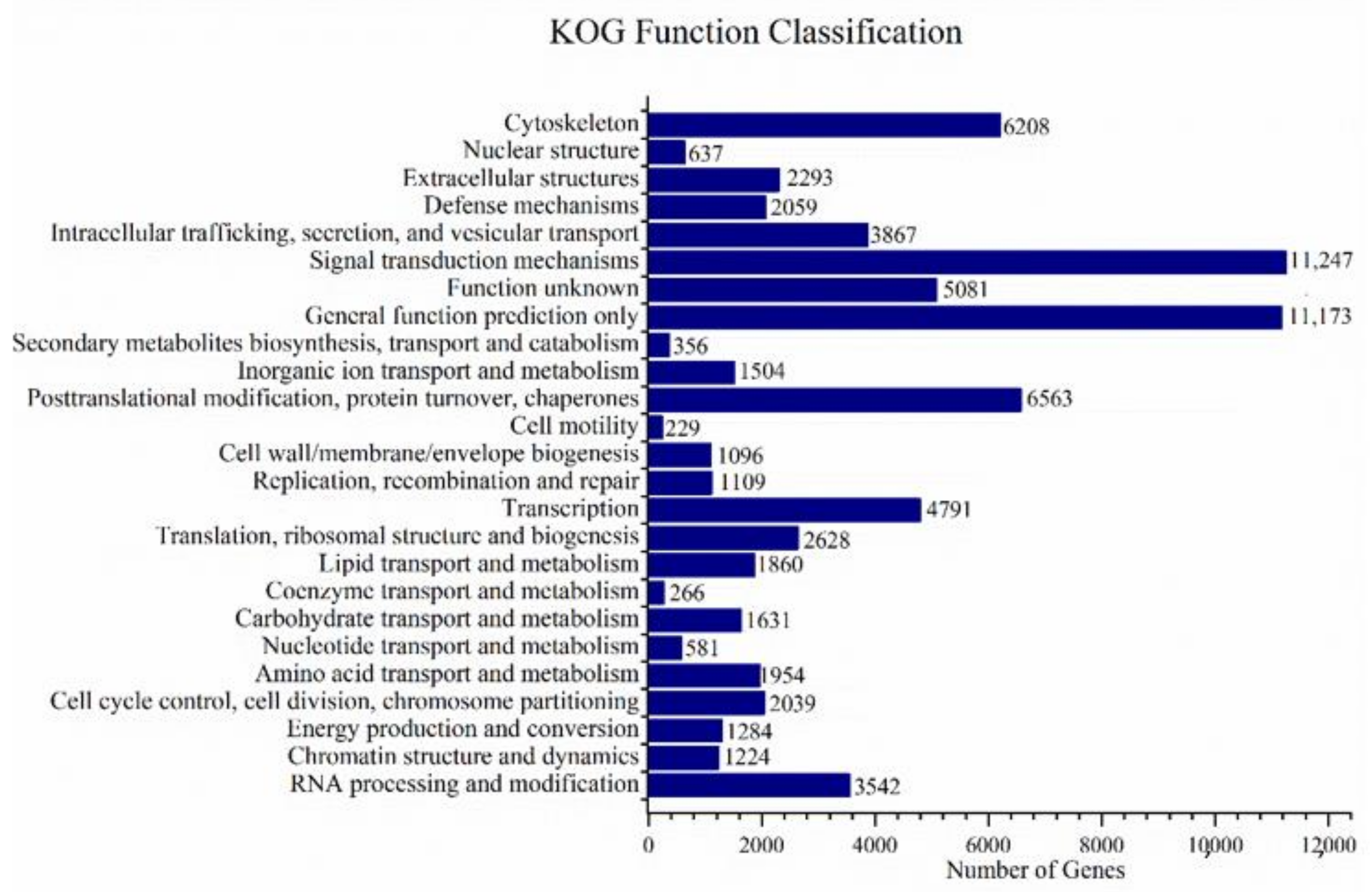
3.2. Annotations and Analysis of Full-Length Transcriptome
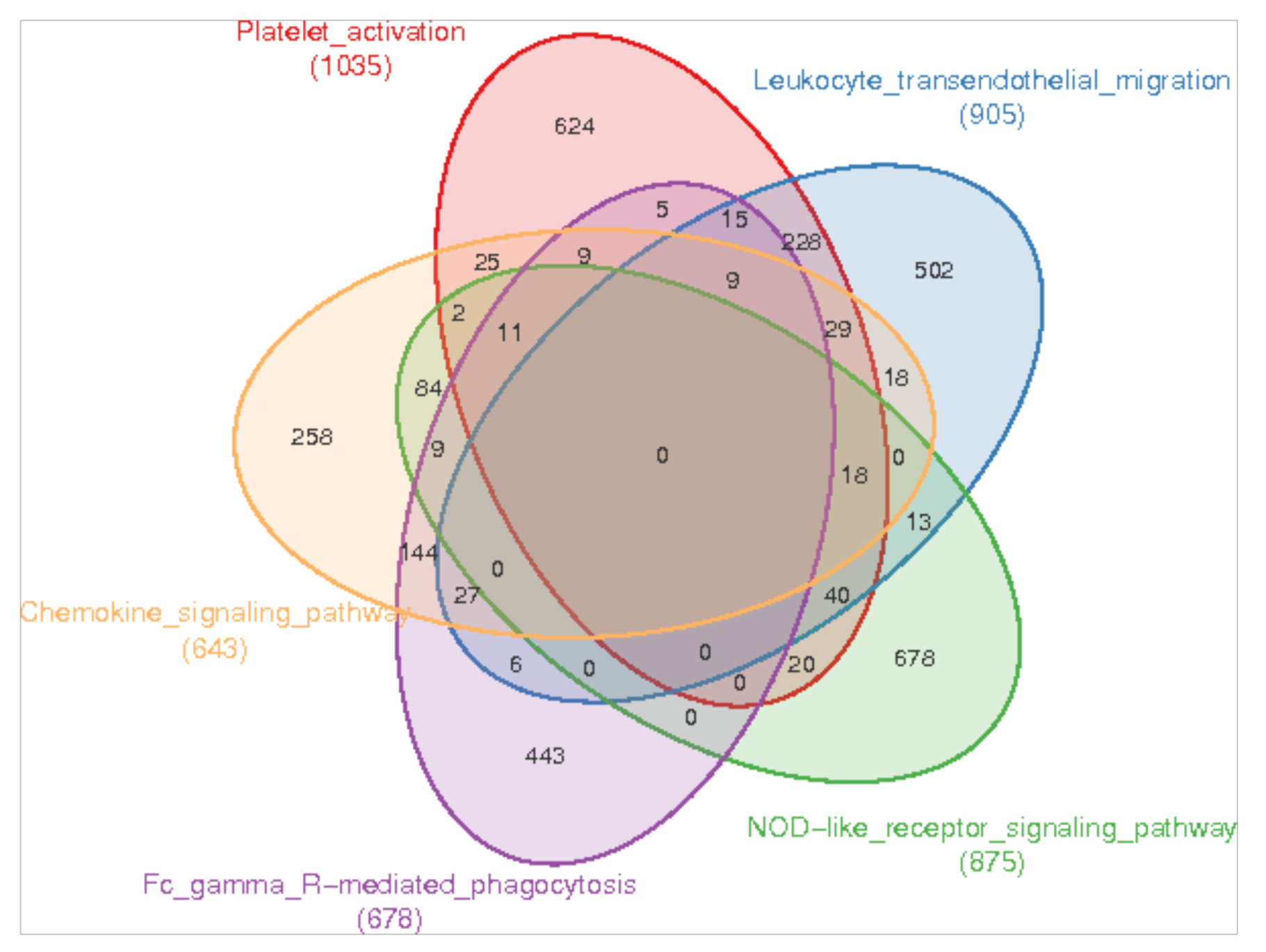
3.3. Enrichment of Immune-Related Pathways in the Skin of Sperm Whale
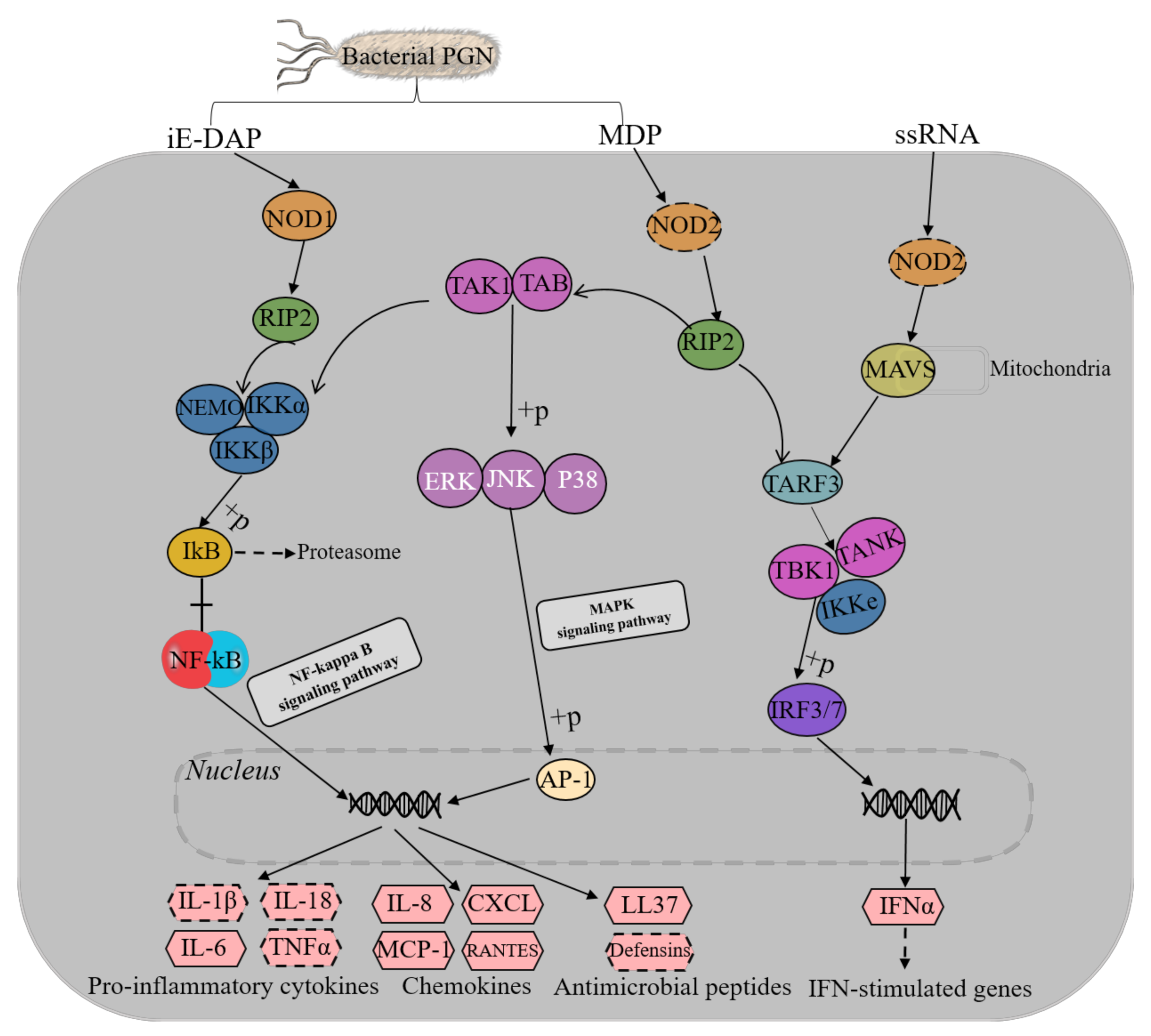
3.4. NOD-Like Receptor Signaling Pathway
3.5. Structural and Evolutional Analysis of NOD1 Gene in Sperm Whale
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

| Species Name | Protein ID | Length (Amino Acids) |
|---|---|---|
| Tursiops truncatus | XP_019781619.1 | 953 |
| Lagnorhynchus obliquidens | XP_026958076.1 | 953 |
| Globicephla melas | XP_030713315.1 | 953 |
| Orcinus orca | XP_033284214.1 | 953 |
| Phocoena sinus | XP_032499084.1 | 972 |
| Neophocaena asiaeorientalis | XP_024610766.1 | 963 |
| Monodon monoceros | XP_029089062.1 | 953 |
| Delphinapterus leucas | XP_022439858.1 | 953 |
| Lipotes vexillifer | XP_007466161.1 | 953 |
| Physeter macrocephalus | Isoform_6383 | 958 |
| Odocoileus virginianus texanus | XP_020748129.1 | 954 |
| Balaenoptera acutorostrata | XP_028020573.1 | 818 |
| Balaenoptera musculus | XP_036718877.1 | 953 |
| Ovis aries | XP_004007979.2 | 954 |
| Bubalus bubalis | XP_006055719.1 | 954 |
| Bos taurus | XP_005205565.2 | 954 |
| Camelus bactrianus | XP_010969658.1 | 953 |
| Sus scrofa | NP_001107749.1 | 953 |
| Anolis carolinensis | XP_003222248.2 | 950 |
| Equus caballus | XP_023495227.1 | 897 |
| Tupaia chinensis | ELW67927.1 | 1109 |
| Homo sapiens | AAD28350.1 | 953 |
| Oryctolagus cuniculus | XP_002713781.1 | 953 |
| Mus musculus | AAN52479.1 | 953 |
| Ornithorhynchus anatinus | XP_001512159.2 | 950 |
| Gallus gallus | NP_001305367.1 | 951 |
| Danio rerio | XP_002665106.3 | 940 |
| Xenopus tropicalis | XP_031759856.1 | 945 |
| Canis lupus familiaris | XP_013974568.1 | 964 |
| Vulpes vulpes | XP_025874889.1 | 953 |
| Proteins/Enzymes Name | Genes ID in This Transcriptome | *** FPKM (Min–Max) | Sequence Alignment Identity Percentage (%) | ||
|---|---|---|---|---|---|
| Physeter macrocephalus | Lipotes vexillifer | Tursiops truncatus | |||
| Nucleotide-binding oligomerization domain-containing protein 1 (NOD1) | Isoform_6383 Isoform_44218 Isoform_24661 Isoform_40056 | 0.41–7.56 | 100.00 (XP_023984584.1) 100.00 (XP_023984584.1) 100.00 (XP_023984584.1) 92.05 (XP_023984584.1) | 96.41 (XP_007466161.1) 98.94 (XP_007466161.1) 98.52 (XP_007466161.1) 90.73 (XP_007466161.1) | 96.73 (XP_019781619.1) 97.88 (XP_033719017.1) 97.78 (XP_019781638.1) 90.07 (XP_033719017.1) |
| Interleukin 6(IL-6) | Isoform_60901 Isoform_23835 | 1.32–1.59 | 100.00 (XP_007105488.3) 100.00 (XP_007105488.3) | NM NM | 100.00 (XP_019778462.1) 100.00 (XP_019778462.1) |
| Interleukin 8 (IL-8) | Isoform_34284 | 2.71 | 100.00 (XP_007126317.1) | 98.02 (XP_007467625.1) | 96.04 (NP_001267571.1) |
| C–X–C motif chemokine 2 (CXCL2) C–X–C motif chemokine 3 (CXCL3) | Isoform_67128 Isoform_70740 Isoform_34284 Isoform_69403 Isoform_45444 | 0.79–7.18 2.29-3.46 2.29-3.46 | 98.53 (XP_023971193.1) 98.53 (XP_023971193.1) 98.44 (XP_023971204.1) 98.41 (XP_007126322.2) 98.44 (XP_007126322.2) | 96.88 (NP_007463281.1) 96.88 (NP_007463281.1) 96.88 (NP_007463281.1) NM NM | 94.12 (NP_004331677.2) 94.12 (NP_004331677.2) 93.75 (NP_004331677.2) NM NM |
| C–C motif chemokine 2 (MCP-1) | Isoform_85283 Isoform_14905 Isoform_68475 | 0.08–3.03 | 100.00 (XP_007467625.1) 91.11 (XP_023983766.1) 98.44 (XP_023983766.1) | 90.36 (XP_007458436.1) 82.22 (XP_007458436.1) 89.80 (XP_007458436.1) | NM NM NM |
| Cathelicidin antimicrobial peptide (LL37) | Isoform_27979 Isoform_90183 Isoform_92246 | 0.11–15.39 | 100.00 (XP_007101317.2) 100.00 (XP_007101317.2) 100.00 (XP_007101317.2) | 90.91 (XP_007446211.1) 89.94 (XP_007446211.1) 77.99 (XP_007446211.1) | 93.94 (XP_033720747.1) 88.89 (XP_033720747.1) 90.35 (XP_033720747.1) |
| transcription factor AP-1 (AP-1) Interferon α (IFNα) | Isoform_38241 Isoform_63047 Isoform_56853 Isoform_75661 | 0.25–264.37 1.04 | 100.00 (XP_023975690.1) 100.00 (XP_023975690.1) 80.97 (XP_023975690.1) 100.00 (XP_023982688.1) | 100.00 (XP_007462636.1) 100.00 (XP_007462636.1) NM 58.90 (XP_007455007.1) | 100.00 (XP_019797942.2) 100.00 (XP_019797942.2) NM 97.22 (AFH89765.1) |
| Receptor-interacting serine/threonine–protein kinase 2 (RIP2) | Isoform_20477 Isoform_25948 Isoform_60840 Isoform_78887 | 0.71–1.52 | 100.00 (XP_023982836.1) 100.00 (XP_023982836.1) 100.00 (XP_023982837.1) 100.00 (XP_023982837.1) | 98.33 (XP_007446375.1) 98.33 (XP_007446375.1) 97.07 (XP_007446375.1) 98.39 (XP_007446375.1) | 97.95 (XP_033699058.1) 97.95 (XP_033699058.1) 96.31 (XP_033699060.1) 98.39 (XP_033699059.1) |
| transcription factor p65(RELA) | Isoform_11257 Isoform_15246 Isoform_14067 Isoform_35442 | 0.08–10.86 | 100.00 (XP_023989099.1) 87.06 (XP_023989099.1) 99.64 (XP_007128247.1) 100.00 (XP_007128247.1) | 96.01 (XP_007462051.1) 87.06 (XP_007462053.1) 96.01 (XP_007462052.1) 94.46 (XP_007462051.1) | NM NM NM NM |
| Nuclear factor NF-kappa-B p105 subunit (NFKB1) | Isoform_2285 Isoform_78940 | 1.34–3.73 | 100.00 (XP_023982362.1) 100.00 (XP_023982361.1) | 95.21 (XP_007463876.1) 97.52 (XP_007463874.1) | 91.78 (XP_019782050.1) 96.88 (XP_019782049.1) |
| Inhibitor of nuclear factor kappa-B kinase subunit β (Ikkβ) | Isoform_16029 Isoform_3742 | 0.64–2.16 | 99.44 (XP_028341955.1) 99.73 (XP_028341955.1) | 97.32 (XP_007455533.1) 98.02 (XP_007455533.1) | 96.78 (XP_033704813.1) 98.02 (XP_033704810.1) |
| Inhibitor of nuclear factor kappa-B kinase subunit α (Ikkα) | Isoform_69638 Isoform_28559 | 0.25–3.37 | 100.00 (XP_028337608.1) 100.00 (XP_028337608.1) | 100.00 (XP_007117682.1) 97.84 (XP_007470397.1) | 100.00 (XP_033697982.1) 99.27 (XP_033697983.1) |
| NF-kappa-B inhibitor α (NFKBIA) | Isoform_44406 Isoform_62842 Isoform_57121 Isoform_5319 | 0.03–66.12 | 100.00 (XP_007119015.1) 100.00 (XP_007119015.1) 100.00 (XP_007119015.1) 89.70 (XP_007119015.1) | 95.54 (XP_007464888.1) 93.87 (XP_007464888.1) 95.37 (XP_007464888.1) NM | NM NM NM NM |
| NF-kappa-B inhibitor β(NFKBIB) | Isoform_64970 Isoform_49218 Isoform_61453 Isoform_30066 | 0.36–16.21 | 100.00 (XP_007114641.1) 99.46 (XP_007114641.2) 100.00 (XP_007114641.2) 100.00 (XP_007114641.2) | 98.58 (XP_007464742.1) 97.27 (XP_007464742.1) 99.08 (XP_007464742.1) 98.71 (XP_007464742.1) | 98.58 (XP_019796701.1) 97.28 (XP_019796701.1) 98.18 (XP_019796701.1) 98.39 (XP_019796701.1) |
| Mitogen-activated protein kinase kinase kinase 7 (TAK1) | Isoform_11038 Isoform_3956 Isoform_6441 | 0.39–2.85 | 100.00 (XP_007126290.1) 100.00 (XP_007126290.1) 100.00 (XP_007126290.1) | NM NM NM | NM NM NM |
| TAK1-binding protein 3(TAB3) | Isoform_24688 | 1.38 | 99.81 (XP_023972426.1) | 98.85 (XP_007456911.1) | NM |
| Mitogen-activated protein kinase 3(ERK) | Isoform_43270 Isoform_52694 Isoform_78038 | 2.06–23.34 | NM NM NM | 98.87 (XP_007452842.1) 100.00 (XP_007452842.1) 100.00 (XP_007452842.1) | NM NM NM |
| Mitogen-activated protein kinase 9 (MAPK9) (JNK) | Isoform_51119 Isoform_55328 | 0.43–1.36 | 100.00 (XP_023972057.1) 99.57 (XP_023972058.1) | 99.59 (XP_007465571.1) 99.13 (XP_007465571.1) | NM NM |
| Mitogen-activated protein kinase 8 (MAPK8) (JNK) | Isoform_76485 | 0.05 | NM | 98.82 (XP_007446292.1) | NM |
| P38 MAP kinase 11 (P38 β) P38 MAP kinase 12 (P38 γ) P38 MAP kinase 13 (P38 δ) | Isoform_54378 Isoform_28186 Isoform_36890 Isoform_46157 Isoform_9786 Isoform_5589 Isoform_11611 Isoform_12832 Isoform_13409 | 1.77 0.36–3.83 0.06–20.27 | 75.34 (XP_023983249.1) 100.00 (XP_028346617.1) 59.13 (XP_028346617.1) 96.18 (XP_028346617.1) 100.00 (XP_023977640.1) 100.00 (XP_023977640.1) 95.60 (XP_023977640.1) 100.00 (XP_023977640.1) 100.00 (XP_023977640.1) | 73.97 (XP_007454143.1) 97.90 (XP_007453950.1) 56.15 (XP_007467328.1) 94.24 (XP_007453950.1) 96.06 (XP_007467328.1) 96.17 (XP_007467328.1) 93.41 (XP_007467328.1) 96.06 (XP_007467328.1) 94.17 (XP_007467328.1) | NM 98.51 (XP_033721676.1) 56.92 (XP_019805222.1) 94.96 (XP_033721674.1) 95.07 (XP_004313866.1) 95.08 (XP_004313866.1) 92.31 (XP_004313866.1) 95.07 (XP_004313866.1) 95.15 (XP_019805222.1) |
| Mitochondrial antiviral-signaling protein (MAVS) | Isoform_11457 Isoform_8429 | 0.19–1.83 | NM NM | 94.08 (XP_007454896.1) 94.08 (XP_007454896.1) | 92.91 (XP_033695234.1) 92.01 (XP_033695233.1) |
| TAK1-binding protein 3 (TAB3) | Isoform_24688 | 0.99 | 99.81 (XP_023972426.1) | 98.85 (XP_007456911.1) | NM |
| Inhibitor of nuclear factor kappa-B kinase subunit epsilon (IKKe) | Isoform_49506 Isoform_42897 Isoform_44117 Isoform_39392 Isoform_73428 | 0.51–1.29 | NM NM NM NM 100.00 (XP_028342232.1) | 78.84 (XP_007470765.1) 95.02 (XP_007470765.1) 93.63 (XP_007470765.1) 94.96 (XP_007470765.1) 94.32 (XP_007470765.1) | NM NM NM NM NM |
| TRAF family member-associated NF-kappa-B activator (TANK) | Isoform_44877 Isoform_30925 Isoform_67916 Isoform_35515 | 0.03–1.95 | NM 98.86 (XP_007107800.2) 100.00 (XP_028334198.1) 100.00 (XP_028334197.1) | 96.77 (XP_019778469.1) 96.80 (XP_019778470.1) 96.77 (XP_007457066.1) 95.12 (XP_007457065.1) | NM 94.65 (XP_019778476.1) 97.42 (XP_019778476.1) 95.89 (XP_019778469.1) |
| Interferon regulatory factor 3 (IRF3) | Isoform_25349 Isoform_45963 Isoform_65550 Isoform_73641 Isoform_51357 | 0.6–3.42 | 93.54 (XP_007112499.1) 100.00 (XP_007112499.1) 100.00 (XP_007112499.1) 95.52 (XP_007112501.1) 100.00 (XP_007112498.1) | 87.76 (XP_007463434.1) 94.78 (XP_007463434.1) 95.08 (XP_007463433.1) 96.22 (XP_007463435.1) 95.93 (XP_007463434.1) | 87.41 (XP_033701337.1) NM95.04 (XP_033701337.1) 84.82 (XP_033701337.1) 95.69 (XP_033701337.1) |
| Interferon regulatory factor 7 (IRF7) Inhibitor of nuclear factor kappa-B kinase subunit γ (NEMO) C–C motif chemokine 5 (CCL5/RANTES) TNF receptor-associated factor 3 (TRAF3) | Isoform_16065 Isoform_57550 Isoform_32564 Isoform_44904 Isoform_30772 Isoform_32236 Isoform_33819 Isoform_86381 Isoform_14574 | 0.4–1.62 0.13–2.14 0.85 0.1 | 100.00 (XP_028341713.1) 87.47 (XP_028341711.1) 87.47 (XP_028341711.1) 100.00 (XP_028341713.1) NM NM NM 100.00 (XP_028355421.1) NM | 95.04 (XP_007465980.1) 84.88 (XP_007465980.1) 93.44 (XP_007465980.1) 86.14 (XP_007465980.1) 92.79 (XP_007457510.1) 93.45 (XP_007457510.1) 93.45 (XP_007457510.1) 96.83 (XP_007447107.1) 93.04 (XP_007455372.1) | 96.17 (XP_033717457.1) NM95.29 (XP_033717454.1) 95.32 (XP_033717457.1) 92.48 (XP_033705598.1) 93.45 (XP_033705598.1) 93.45 (XP_033705598.1) 95.24 (XP_004322904.1) 94.39 (XP_019776547.2) |
References
- Uhen, M.D. The Origin (s) of Whales. Annu. Rev. Earth Planet. Sci. 2010, 38, 189–219. [Google Scholar] [CrossRef]
- Whitehead, H. Sperm Whales: Social Evolution in the Ocean; The University of Chicago Press: Chicago, IL, USA, 2003. [Google Scholar]
- Jaquet, N.; Whitehead, H. Scale-dependent correlation of sperm whale distribution with environmental features and productivity in the South Pacific. Mar. Ecol. Prog. Ser. 1996, 135, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Thewissen, J.G.M.; Cooper, L.N.; George, J.C.; Bajpai, S. From Land to Water: The Origin of Whales, Dolphins, and Porpoises. Evol. Educ. Outreach 2009, 2, 272–288. [Google Scholar] [CrossRef] [Green Version]
- Schreer, J.F.; Kovacs, K.M. Allometry of diving capacity in air-breathing vertebrates. Can. J. Zool. 1997, 75, 339–358. [Google Scholar] [CrossRef]
- Steeman, M.E.; Hebsgaard, M.B.; Fordyce, R.E.; Ho, S.Y.W.; Rabosky, D.L.; Nielsen, R.; Rahbek, C.; Glenner, H.; Sørensen, M.V.; Willerslev, E. Radiation of Extant Cetaceans Driven by Restructuring of the Oceans. Syst. Biol. 2009, 58, 573–585. [Google Scholar] [CrossRef] [Green Version]
- Warren, W.C.; Kuderna, L.; Alexander, A.; Catchen, J.; Pérez-Silva, J.G.; López-Otín, C.; Quesada, V.; Minx, P.; Tomlinson, C.; Montague, M.J.; et al. The Novel Evolution of the Sperm Whale Genome. Genome Biol. Evol. 2017, 9, 3260–3264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, B.; Baird, R.; Barlow, J.; Dawson, S.; Ford, J.; Mead, J. Physeter Macrocephalus (Amended version of 2008 Assessment). IUCN Red List Threat. Species 2019, 2307–8235. [Google Scholar] [CrossRef]
- Ellis, A.E. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Long, Y.; Li, Q.; Zhou, B.; Song, G.; Li, T.; Bin, C.Z. De Novo Assembly of Mud Loach (Misgurnus anguillicaudatus) skin transcriptome to identify putative genes involved in immunity and epidermal mucus secretion. PLoS ONE 2013, 8, e56998. [Google Scholar] [CrossRef] [Green Version]
- Reidenberg, J.S. Anatomical adaptations of aquatic mammals. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2007, 290, 507–513. [Google Scholar] [CrossRef]
- Davies, K.T.J.; Cotton, J.A.; Kirwan, J.D.; Teeling, E.C.; Rossiter, S.J. Parallel signatures of sequence evolution among hearing genes in echolocating mammals: An emerging model of genetic convergence. Heredity 2012, 108, 480–489. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, X.; Hu, B.; Zheng, J.; Xiao, W.; Hao, Y.; Liu, W.; Wang, D. Physicochemical Evolution and Molecular Adaptation of the Cetacean Osmoregulation-related Gene UT-A2 and Implications for Functional Studies. Sci. Rep. 2015, 5, 8795. [Google Scholar] [CrossRef] [Green Version]
- De Guise, S.; Ross, P.S.; Osterhaus, A.D.; Martineau, D.; Beland, P.; Fournier, M. Immune functions in beluga whales (Delphinapterus leucas): Evaluation of natural killer cell activity. Vet. Immunol. Immunopathol. 1997, 58, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Beineke, A.; Siebert, U.; Wohlsein, P.; Baumgärtner, W. Immunology of whales and dolphins. Vet. Immunol. Immunopathol. 2010, 133, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Zabka, T.S.; Romano, T.A. Distribution of MHC II (+) cells in skin of the Atlantic bottlenose dolphin (Tursiops truncatus): An initial investigation of dolphin dendritic cells. Anat. Rec. 2003, 273A, 636–647. [Google Scholar] [CrossRef] [PubMed]
- Lunardi, D.; Abelli, L.; Panti, C.; Marsili, L.; Fossi, M.C.; Mancia, A. Transcriptomic analysis of bottlenose dolphin (Tursiops truncatus) skin biopsies to assess the effects of emerging contaminants. Mar. Environ. Res. 2016, 114, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.; Jia, K.; Xia, J.; Yang, L.; Chen, J.; Wu, Y.; Yi, M. De novo Assembly of the Indo-Pacific Humpback Dolphin Leucocyte Transcriptome to Identify Putative Genes Involved in the Aquatic Adaptation and Immune Response. PLoS ONE 2013, 8, e72417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishengoma, E.; Agaba, M. Evolution of toll-like receptors in the context of terrestrial ungulates and cetaceans diversification. BMC Evol. Biol. 2017, 17, 54. [Google Scholar] [CrossRef] [Green Version]
- Tian, R.; Seim, I.; Zhang, Z.; Yang, Y.; Ren, W.; Xu, S.; Yang, G. Distinct evolution of toll-like receptor signaling pathway genes in cetaceans. Genes Genom. 2019, 41, 1417–1430. [Google Scholar] [CrossRef]
- Delbridge, L.M.; O’Riordan, M.X.D. Innate recognition of intracellular bacteria. Curr. Opin. Immunol. 2006, 19, 10–17. [Google Scholar] [CrossRef]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [Green Version]
- Bi, D.; Gao, Y.; Chu, Q.; Cui, J.; Xu, T. NOD1 is the innate immune receptor for iE-DAP and can activate NF-κB pathway in teleost fish. Dev. Comp. Immunol. 2017, 76, 238–246. [Google Scholar] [CrossRef]
- Shaw, M.H.; Reimer, T.; Kim, Y.-G.; Nuñez, G. NOD-like receptors (NLRs): Bona fide intracellular microbial sensors. Curr. Opin. Immunol. 2008, 20, 377–382. [Google Scholar] [CrossRef] [Green Version]
- Jacobs, S.R.; Blossom, D. NLRs, inflammasomes, and viral infection. J. Leukoc. Biol. 2012, 92, 469–477. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, M.; Yang, K.; Hashimoto, M.; Park, J.-H.; Kim, Y.-G.; Fujimoto, Y.; Nuñez, G.; Fukase, K.; Inohara, N. Differential Release and Distribution of Nod1 and Nod2 Immunostimulatory Molecules among Bacterial Species and Environments. J. Biol. Chem. 2006, 281, 29054–29063. [Google Scholar] [CrossRef] [Green Version]
- Kanneganti, T.D.; Lamkanfi, M.; Núñez, G. Intracellular NOD-like Receptors in Host Defense and Disease. Immunity 2007, 27, 549–559. [Google Scholar] [CrossRef]
- Zhan, F.; Yu, X.; Zhang, X.; Chen, L.; Sun, X.; Yu, R.-Q.; Wu, Y. Tissue distribution of organic contaminants in stranded pregnant sperm whale (Physeter microcephalus) from the Huizhou coast of the South China Sea. Mar. Pollut. Bull. 2019, 144, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Tang, Q.; Wu, M.; Mou, D.; Liu, H.; Wang, S.; Zhang, C.; Ding, L.; Luo, J. Comparative transcriptomics provides novel insights into the mechanisms of selenium tolerance in the hyperaccumulator plant Cardamine hupingshanensis. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, F.; Gish, W.; Miller, W.; Myers, W. Basic local alignment search tool. J. Mol. Biol. 1990, 3, 403–410. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Quevillon, E.; Silventoinen, V.; Pillai, S.; Harte, N.; Mulder, N.; Apweiler, R.; Lopez, R. InterProScan: Protein domains identifier. Nucleic Acids Res. 2005, 33, W116–W120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Ciclitira, P.; Messing, J. PacBio sequencing of gene families—A case study with wheat gluten genes. Gene 2014, 533, 541–546. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Ponting, C.P.; Schultz, J.; Milpetz, F.; Bork, P. SMART: Identification and annotation of domains from signalling and extracellular protein sequences. Nucleic Acids Res. 1999, 27, 229–232. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Inohara, N.; Koseki, T.; Del, P.L.; Hu, Y.; Yee, C.; Chen, S.; Carrio, R.; Merino, J.; Liu, D.; Ni, J.; et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 1999, 274, 14560–14567. [Google Scholar] [CrossRef] [Green Version]
- Ogura, Y.; Inohara, N.; Benito, A.; Chen, F.F.; Yamaoka, S.; Núñez, G. Nod2, a Nod1/Apaf-1 Family Member That Is Restricted to Monocytes and Activates NF-κB. J. Biol. Chem. 2001, 276, 4812–4818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guang, Y.; Ran, T.; ShiXia, X.U.; WenHua, R. Molecular adaptation mechanism of secondary aquatic life in cetaceans. Sci. China Ser. C 2019, 49, 380–391. [Google Scholar]
- Matejuk, A. Skin Immunity. Arch. Immunol. Ther. Exp. 2018, 66, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, G.K.; Grayson, S.; Brown, B.E.; Elias, P.M. Lipokeratinocytes of the epidermis of a cetacean (Phocena phocena). Cell Tissue Res. 1986, 246, 227. [Google Scholar] [CrossRef] [Green Version]
- Montie, E.W.; Fair, P.A.; Bossart, G.D.; Mitchum, G.B.; Houde, M.; Muir, D.C.G.; Letcher, R.J.; McFee, W.E.; Starczak, V.R.; Stegeman, J.J.; et al. Cytochrome P4501A1 expression, polychlorinated biphenyls and hydroxylated metabolites, and adipocyte size of bottlenose dolphins from the Southeast United States. Aquat. Toxicol. 2008, 86, 397–412. [Google Scholar] [CrossRef]
- Seegers, U.; Meyer, W. A preliminary approach to epidermal antimicrobial defense in the Delphinidae. Mar. Biol. 2004, 144, 841–844. [Google Scholar] [CrossRef]
- Theissinger, K.; Falckenhayn, C.; Blande, D.; Toljamo, A.; Gutekunst, J.; Makkonen, J.; Jussila, J.; Lyko, F.; Schrimpf, A.; Schulz, R.; et al. De Novo assembly and annotation of the freshwater crayfish Astacus astacus transcriptome. Mar. Genom. 2016, 28, 7–10. [Google Scholar] [CrossRef]
- Gomes, F.; Watanabe, L.; Vianez, J.; Nunes, M.; Cardoso, J.; Lima, C.; Schneider, H.; Sampaio, I. Comparative analysis of the transcriptome of the Amazonian fish species Colossoma macropomum (tambaqui) and hybrid tambacu by next generation sequencing. PLoS ONE 2019, 14, e0212755. [Google Scholar] [CrossRef]
- Scannapieco, A.C.; Conte, C.A.; Rivarola, M.; Wulff, J.P.; Muntaabski, I.; Ribone, A.; Milla, F.; Cladera, J.L.; Lanzavecchia, S.B. Transcriptome analysis of Anastrepha fraterculus sp. 1 males, females, and embryos: Insights into development, courtship, and reproduction. BMC Genet. 2020, 21, 136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, B.; Nie, X.; Liu, Q.; Xie, F.; Shang, D. Transcriptome Analysis and Identification of Genes Related to Immune Function in Skin of the Chinese Brown Frog. Zool. Sci. 2009, 26, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Sueiro, M.C.; Bagnato, E.; Palacios, M.G. Parasite infection and immune and health-state in wild fish exposed to marine pollution. Mar. Pollut. Bull. 2017, 119, 320–324. [Google Scholar] [CrossRef]
- Zhang, W.; Lv, Z.; Li, C.; Sun, Y.; Jiang, H.; Zhao, M.; Zhao, X.; Shao, Y.; Chang, Y. Transcriptome profiling reveals key roles of phagosome and NOD-like receptor pathway in spotting diseased Strongylocentrotus intermedius. Fish Shellfish Immunol. 2019, 84, 521–531. [Google Scholar] [CrossRef]
- Sahoo, B.R. Structure of fish Toll-like receptors (TLR) and NOD-like receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602–1617. [Google Scholar] [CrossRef]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- Botos, I.; Segal, D.M.; Davies, D.R. The Structural Biology of Toll-like Receptors. Structure 2011, 19, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Marc, G.; Veronica, S.; Peter, D.E. Human and chicken TLR pathways: Manual curation and computer-based orthology analysis. Mamm. Genome 2011, 22, 130–138. [Google Scholar] [CrossRef] [Green Version]
- Shen, T.; Xu, S.; Wang, X.; Yu, W.; Zhou, K.; Yang, G. Adaptive evolution and functional constraint at TLR4 during the secondary aquatic adaptation and diversification of cetaceans. BMC Evol. Biol. 2012, 12, 39. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Yu, M.; Tong, S.; Jia, K.; Liu, R.; Wang, H.; Li, S.; Ning, Z. Tissue-specific expression of the NOD-like receptor protein 3 in BALB/c mice. J. Vet. Sci. 2014, 15, 173–177. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, K.V.; Zhang, J.; Liu, S.; Kucuktas, H.; Wang, X.; Liu, H.; Sha, Z.; Terhune, J.; Peatman, E.; Liu, Z. Pathogen recognition receptors in channel catfish: I. Identification, phylogeny and expression of NOD-like receptors. Dev. Comp. Immunol. 2012, 37, 77–86. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Kinio, A.; Saleh, M. Function of NOD-like receptors in immunity and disease. Curr. Opin. Investig. Drugs 2010, 11, 1246–1255. [Google Scholar]
- Xiao, G.; Zhuang, W.; Wang, T.; Lian, G.; Luo, L.; Ye, C.; Wang, H.; Xie, L. Transcriptomic analysis identifies Toll-like and Nod-like pathways and necroptosis in pulmonary arterial hypertension. J. Cell. Mol. Med. 2020, 24, 11409–11421. [Google Scholar] [CrossRef] [PubMed]
- Correa, R.G.; Milutinovic, S.; Reed, J.C. Roles of NOD1 (NLRC1) and NOD2 (NLRC2) in innate immunity and inflammatory diseases. Biosci. Rep. 2012, 32, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Sabbah, A.; Chang, T.H.; Harnack, R.; Frohlich, V.; Tominaga, K.; Dube, P.H.; Xiang, Y.; Bose, S. Activation of innate immune antiviral responses by Nod2. Nat. Immunol. 2009, 10, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- Lupfer, C.; Thomas, P.G.; Kanneganti, T.-D. Nucleotide Oligomerization and Binding Domain 2-Dependent Dendritic Cell Activation Is Necessary for Innate Immunity and Optimal CD8+ T Cell Responses to Influenza A Virus Infection. J. Virol. 2014, 88, 8946–8955. [Google Scholar] [CrossRef] [Green Version]
- Nedoszytko, B.; Sokołowska-Wojdyło, M.; Ruckemann-Dziurdzińska, K.; Roszkiewicz, J.; Nowicki, R.J. Chemokines and cytokines network in the pathogenesis of the inflammatory skin diseases: Atopic dermatitis, psoriasis and skin mastocytosis. Postȩpy Dermatol. Alergol. 2014, 2, 84–91. [Google Scholar] [CrossRef]
- McCarthy, J.V.; Ni, J.; Dixit, V.M. RIP2 is a Novel NF-κB-activating and Cell Death-inducing Kinase. J. Biol. Chem. 1998, 273, 16968–16975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kobayashi, K.; Inohara, N.; Hernandez, L.D.; Galán, J.E.; Núñez, G.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 2002, 416, 194–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inohara, N.; Koseki, T.; Lin, J.; Del Peso, L.; Lucas, P.C.; Chen, F.F.; Ogura, Y.; Núñez, G. An Induced Proximity Model for NF-κB Activation in the Nod1/RICK and RIP Signaling Pathways. J. Biol. Chem. 2000, 275, 27823–27831. [Google Scholar] [CrossRef] [Green Version]
- Strober, W.; Murray, P.J.; Kitani, A.; Watanabe, T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat. Rev. Immunol. 2006, 6, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-H.; Kim, Y.-G.; McDonald, C.; Kanneganti, T.-D.; Hasegawa, M.; Body-Malapel, M.; Inohara, N.; Núñez, G. RICK/RIP2 Mediates Innate Immune Responses Induced through Nod1 and Nod2 but Not TLRs. J. Immunol. 2007, 178, 2380–2386. [Google Scholar] [CrossRef]
- Nembrini, C.; Kisielow, J.; Shamshiev, A.T.; Tortola, L.; Coyle, A.J.; Kopf, M.; Marsland, B.J. The Kinase Activity of Rip2 Determines Its Stability and Consequently Nod1- and Nod2-mediated Immune Responses. J. Biol. Chem. 2009, 284, 19183–19188. [Google Scholar] [CrossRef] [Green Version]
- Windheim, M.; Lang, C.; Peggie, M.; Plater, L.A.; Cohen, P. Molecular mechanisms involved in the regulation of cytokine production by muramyl dipeptide. Biochem. J. 2007, 404, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.; Ghosh, S. NF-κB, An Evolutionarily Conserved Mediator of Immune and Inflammatory Responses. In Mechanisms of Lymphocyte Activation and Immune Regulation X; Gupta, S., Paul, W.E., Steinman, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2005; Volume 560, pp. 41–45. [Google Scholar] [CrossRef]
- Brady, G.; Haas, D.A.; Farrell, P.J.; Pichlmair, A.; Bowie, A.G. Poxvirus Protein MC132 from Molluscum Contagiosum Virus Inhibits NF-κB Activation by Targeting p65 for Degradation. J. Virol. 2015, 89, 8406–8415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glasgow, J.N.; Wood, T.; Perez-Polo, J.R. Identification and characterization of nuclear factor kappaB binding sites in the murine bcl-x promoter. J. Neurochem. 2000, 75, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.B.; Schreck, R.; Baeuerle, P.A. NF-kappa B contacts DNA by a heterodimer of the p50 and p65 subunit. EMBO J. 1991, 10, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Beinke, S. Structure-Function Analysis of NF-κB1 p105. Ph.D. Thesis, University College London, London, UK, 2003. [Google Scholar]
- Perkins, N.D. The Rel/NF-kappa B family: Friend and foe. Trends Biochem. Sci. 2000, 25, 434–440. [Google Scholar] [CrossRef]
- Elewaut, D.; Di Donato, J.A.; Kim, J.M.; Truong, F.; Eckmann, L.; Kagnoff, M.F. NF-κB is a central regulator of the intestinal epithelial cell innate immune response induced by infection with enteroinvasive bacteria. J. Immunol. 1999, 163, 1457–1466. [Google Scholar]
- Schattenberg, J.M.; Czaja, M.J. TNF/TNF Receptors; Springer: Berlin/Heidelberg, Germany, 2009; pp. 161–177. [Google Scholar]
- Inohara, N.; Núñez, G. NODs: Intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003, 3, 371–382. [Google Scholar] [CrossRef]





| Sample | Read of Insert | Total Transcripts | Mean Length (bp) | N50 (bp) |
|---|---|---|---|---|
| Total | 1,056,247 | 96,350 | 1705 | 1996 |
| Sequence Size (bp) | UniGene Number |
|---|---|
| 200–500 | 16,252 |
| 500–1000 | 7251 |
| 1000–2000 | 6444 |
| 2000–3000 | 3192 |
| ≥3000 | 2843 |
| Total | 35,982 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, D.; Li, Y.; Aierken, R.; Kang, Q.; Wang, X.; Zeng, Q.; Fan, Z.; Zhen, Y.; Zhao, L. Integrated Full-Length Transcriptome and RNA-Seq to Identify Immune System Genes from the Skin of Sperm Whale (Physeter macrocephalus). Genes 2021, 12, 233. https://doi.org/10.3390/genes12020233
Wang D, Li Y, Aierken R, Kang Q, Wang X, Zeng Q, Fan Z, Zhen Y, Zhao L. Integrated Full-Length Transcriptome and RNA-Seq to Identify Immune System Genes from the Skin of Sperm Whale (Physeter macrocephalus). Genes. 2021; 12(2):233. https://doi.org/10.3390/genes12020233
Chicago/Turabian StyleWang, Daling, Ying Li, Reyilamu Aierken, Qi Kang, Xianyan Wang, Qianhui Zeng, Zhichang Fan, Yu Zhen, and Liyuan Zhao. 2021. "Integrated Full-Length Transcriptome and RNA-Seq to Identify Immune System Genes from the Skin of Sperm Whale (Physeter macrocephalus)" Genes 12, no. 2: 233. https://doi.org/10.3390/genes12020233
APA StyleWang, D., Li, Y., Aierken, R., Kang, Q., Wang, X., Zeng, Q., Fan, Z., Zhen, Y., & Zhao, L. (2021). Integrated Full-Length Transcriptome and RNA-Seq to Identify Immune System Genes from the Skin of Sperm Whale (Physeter macrocephalus). Genes, 12(2), 233. https://doi.org/10.3390/genes12020233






