RANK-C Expression Sensitizes ER-Negative, EGFR-Positive Breast Cancer Cells to EGFR-Tyrosine Kinase Inhibitors (TKIs)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Plasmids and Transfection
2.2. Immunoblotting
2.3. Antibodies and Reagents
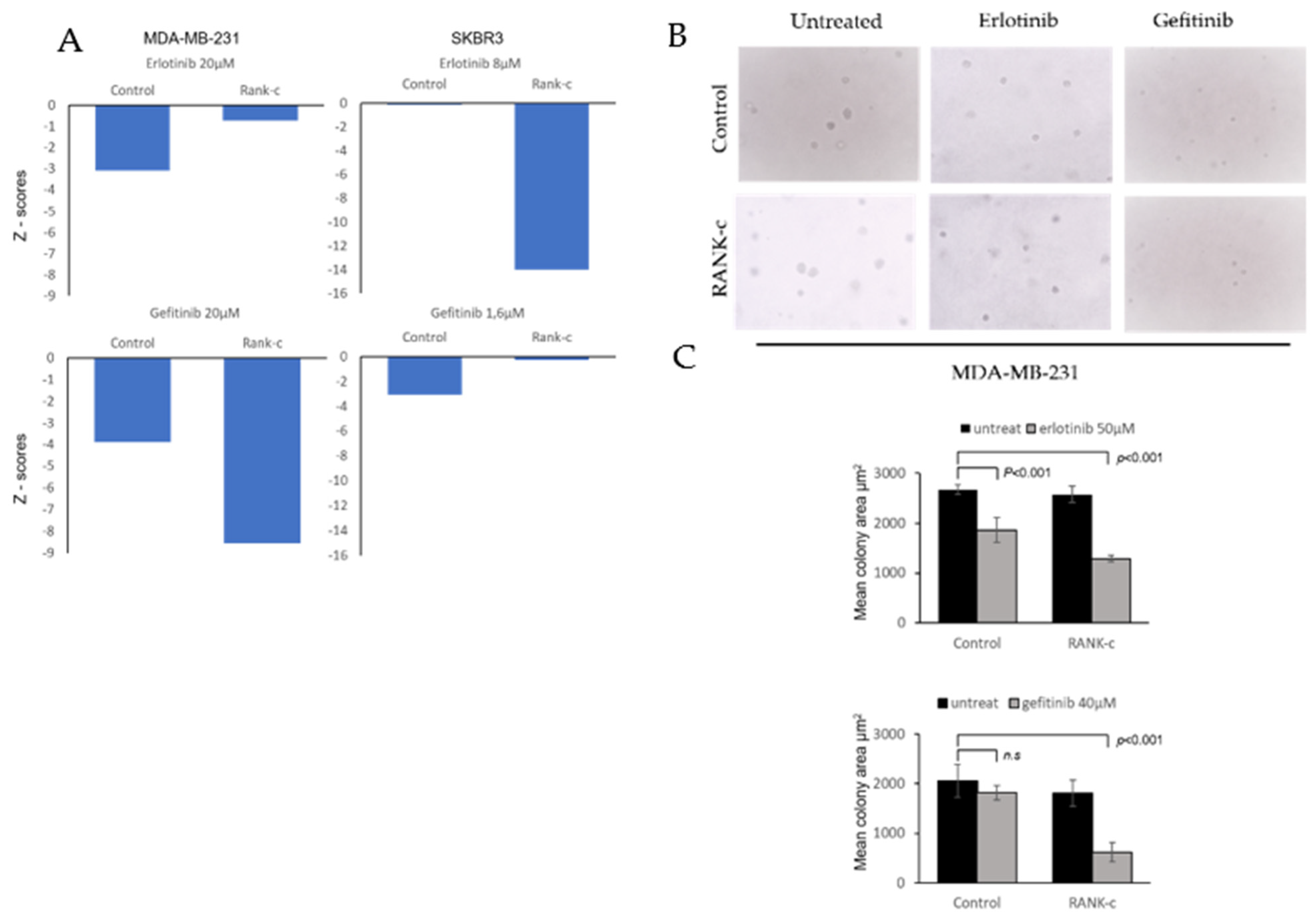
2.4. Colony Formation Assay
2.5. Proliferation Assay and Cell Counting
2.6. Statistical Analysis
3. Results
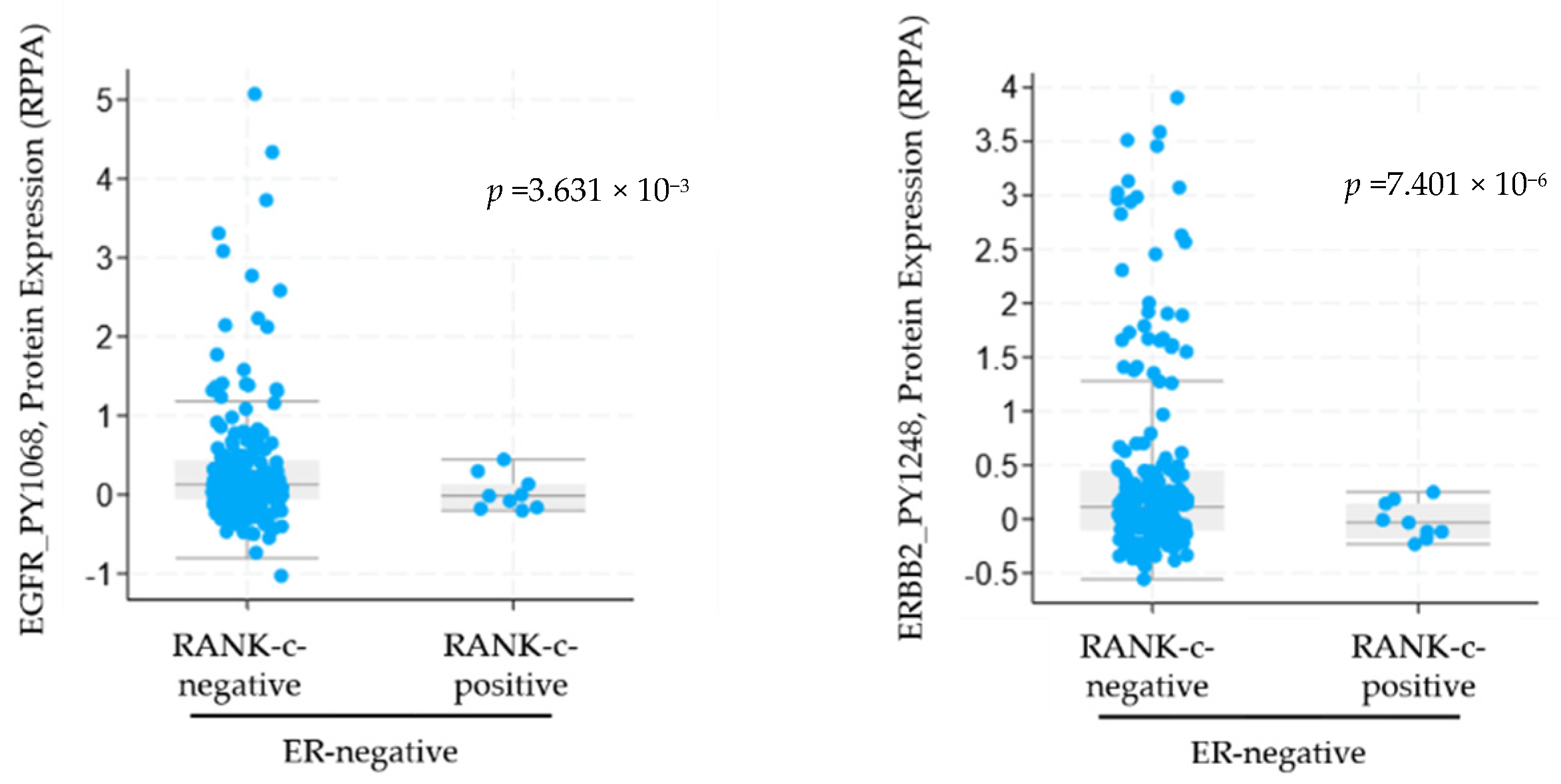
3.1. RANK-C Expression in ER-Negative Breast Cancer Samples Affects ERBB Phosphorylation Status
3.2. EGFR Tyrosine Kinase Inhibitors Affects Heterogeneously ER-Negative, RANK-C Expressing Breast Cancer Cell Lines
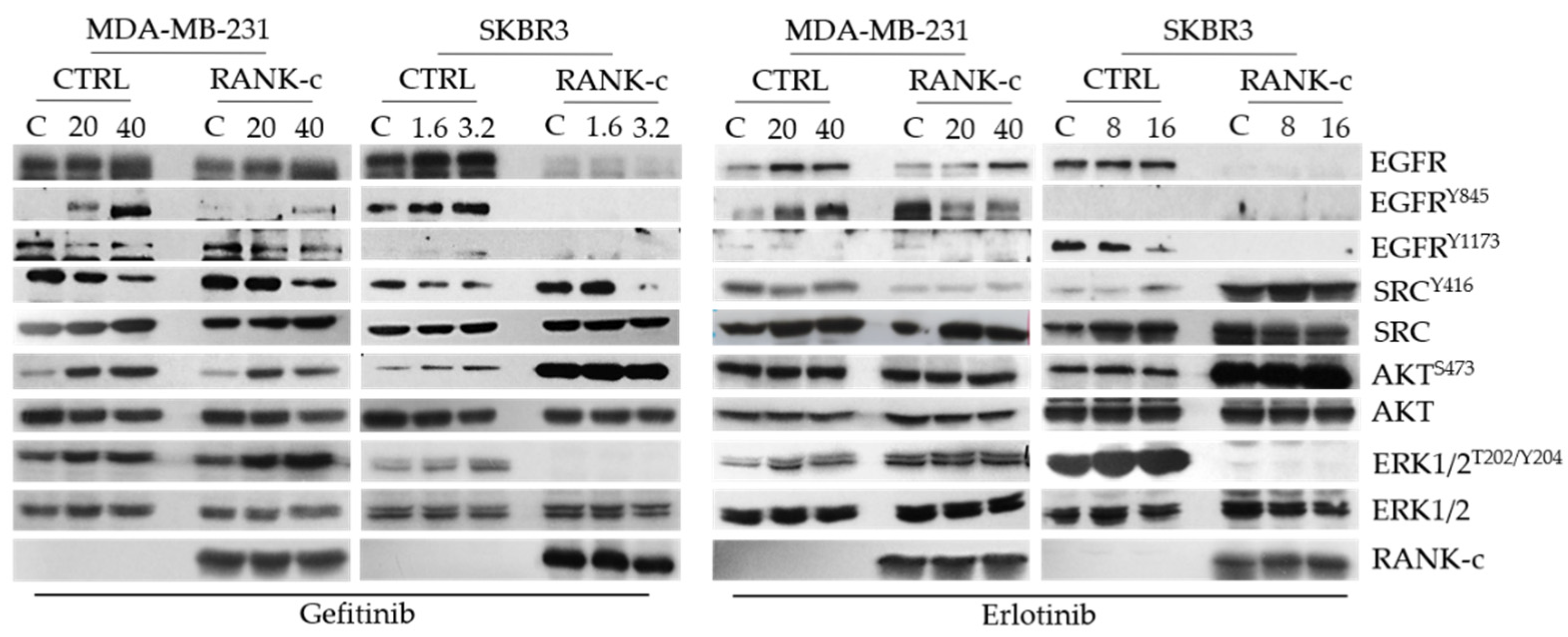
3.3. TKIs Affects EGFR Downstream Signaling in RANK-C Expressing Breast Cancer Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Marchio, C.; Geyer, F.C.; Reis-Filho, J.S. Pathology and Molecular Pathology of Breast Cancer. In Pathology and Epidemiology of Cancer; Loda, M., Mucci, L., Mittelstadt, M., Van Hemelrijck, M., Cotter, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 173–231. [Google Scholar] [CrossRef]
- Lopez-Garcia, M.A.; Geyer, F.C.; Lacroix-Triki, M.; Marchio, C.; Reis-Filho, J.S. Breast cancer precursors revisited: Molecular features and progression pathways. Histopathology 2010, 57, 171–192. [Google Scholar] [CrossRef]
- Marchio, C.; Reis-Filho, J.S. Molecular diagnosis in breast cancer. Diagn. Mol. Pathol. 2008, 14, 202–213. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Nakai, K.; Hung, M.C.; Yamaguchi, H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am. J. Cancer Res. 2016, 6, 1609–1623. [Google Scholar] [PubMed]
- Merino Bonilla, J.A.; Torres Tabanera, M.; Ros Mendoza, L.H. Breast cancer in the 21st century: From early detection to new therapies. Radiologia 2017, 59, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; Siewertsz van Reesema, L.L.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers 2020, 12, 2392. [Google Scholar] [CrossRef] [PubMed]
- Papanastasiou, A.D.; Sirinian, C.; Kalofonos, H.P. Identification of novel human receptor activator of nuclear factor-kB isoforms generated through alternative splicing: Implications in breast cancer cell survival and migration. Breast Cancer Res. 2012, 14, R112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirinian, C.; Papanastasiou, A.D.; Schizas, M.; Spella, M.; Stathopoulos, G.T.; Repanti, M.; Zarkadis, I.K.; King, T.A.; Kalofonos, H.P. RANK-c attenuates aggressive properties of ER-negative breast cancer by inhibiting NF-κB activation and EGFR signaling. Oncogene 2018, 37, 5101–5114. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ades, F.; Zardavas, D.; Bozovic-Spasojevic, I.; Pugliano, L.; Fumagalli, D.; De Azambuja, E.; Viale, G.; Sotiriou, C.; Piccart, M. Luminal B Breast Cancer: Molecular Characterization, Clinical Management, and Future Perspectives. J. Clin. Oncol. 2014, 32, 2794–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Guerrab, A.; Bamdad, M.; Kwiatkowski, F.; Bignon, Y.J.; Penault-Llorca, F.; Aubel, C. Anti-EGFR monoclonal antibodies and EGFR tyrosine kinase inhibitors as combination therapy for triple-negative breast cancer. Oncotarget 2016, 7, 73618–73637. [Google Scholar] [CrossRef] [Green Version]
- Martelotto, L.G.; Ng, C.K.; Piscuoglio, S.; Weigelt, B.; Reis-Filho, J.S. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014, 16, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freelander, A.; Brown, L.J.; Parker, A.; Segara, D.; Portman, N.; Lau, B.; Lim, E. Molecular Biomarkers for Contemporary Therapies in Hormone Receptor-Positive Breast Cancer. Genes 2021, 12, 285. [Google Scholar] [CrossRef]
- McLaughlin, R.P.; He, J.; van der Noord, V.E.; Redel, J.; Foekens, J.A.; Martens, J.W.M.; Smid, M.; Zhang, Y.; van de Water, B. A kinase inhibitor screen identifies a dual cdc7/CDK9 inhibitor to sensitise triple-negative breast cancer to EGFR-targeted therapy. Breast Cancer Res. 2019, 21, 77. [Google Scholar] [CrossRef]
- Diluvio, G.; Del Gaudio, F.; Giuli, M.V.; Franciosa, G.; Giuliani, E.; Palermo, R.; Besharat, Z.M.; Pignataro, M.G.; Vacca, A.; d’Amati, G.; et al. NOTCH3 inactivation increases triple negative breast cancer sensitivity to gefitinib by promoting EGFR tyrosine dephosphorylation and its intracellular arrest. Oncogenesis 2018, 7, 42. [Google Scholar] [CrossRef]
- House, C.D.; Grajales, V.; Ozaki, M.; Jordan, E.; Wubneh, H.; Kimble, D.C.; James, J.M.; Kim, M.K.; Annunziata, C.M. IΚΚε cooperates with either MEK or non-canonical NF-kB driving growth of triple-negative breast cancer cells in different contexts. BMC Cancer 2018, 18, 595. [Google Scholar] [CrossRef] [Green Version]
- Abo-Zeid, M.A.M.; Abo-Elfadl, M.T.; Gamal-Eldeen, A.M. Evaluation of lapatinib cytotoxicity and genotoxicity on MDA-MB-231 breast cancer cell line. Environ. Toxicol. Pharmacol. 2019, 71, 103207. [Google Scholar] [CrossRef]
- Bellizzi, A.; Greco, M.R.; Rubino, R.; Paradiso, A.; Forciniti, S.; Zeeberg, K.; Cardone, R.A.; Reshkin, S.J. The scaffolding protein NHERF1 sensitizes EGFR-dependent tumor growth, motility and invadopodia function to gefitinib treatment in breast cancer cells. Int. J. Oncol. 2015, 46, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sirinian, C.; Papanastasiou, A.D.; Degn, S.E.; Frantzi, T.; Aronis, C.; Chaniotis, D.; Makatsoris, T.; Koutras, A.; Kalofonos, H.P. RANK-C Expression Sensitizes ER-Negative, EGFR-Positive Breast Cancer Cells to EGFR-Tyrosine Kinase Inhibitors (TKIs). Genes 2021, 12, 1686. https://doi.org/10.3390/genes12111686
Sirinian C, Papanastasiou AD, Degn SE, Frantzi T, Aronis C, Chaniotis D, Makatsoris T, Koutras A, Kalofonos HP. RANK-C Expression Sensitizes ER-Negative, EGFR-Positive Breast Cancer Cells to EGFR-Tyrosine Kinase Inhibitors (TKIs). Genes. 2021; 12(11):1686. https://doi.org/10.3390/genes12111686
Chicago/Turabian StyleSirinian, Chaido, Anastasios D. Papanastasiou, Soren E. Degn, Theodora Frantzi, Christos Aronis, Dimitrios Chaniotis, Thomas Makatsoris, Angelos Koutras, and Haralabos P. Kalofonos. 2021. "RANK-C Expression Sensitizes ER-Negative, EGFR-Positive Breast Cancer Cells to EGFR-Tyrosine Kinase Inhibitors (TKIs)" Genes 12, no. 11: 1686. https://doi.org/10.3390/genes12111686
APA StyleSirinian, C., Papanastasiou, A. D., Degn, S. E., Frantzi, T., Aronis, C., Chaniotis, D., Makatsoris, T., Koutras, A., & Kalofonos, H. P. (2021). RANK-C Expression Sensitizes ER-Negative, EGFR-Positive Breast Cancer Cells to EGFR-Tyrosine Kinase Inhibitors (TKIs). Genes, 12(11), 1686. https://doi.org/10.3390/genes12111686




