DBtRend: A Web-Server of tRNA Expression Profiles from Small RNA Sequencing Data in Humans
Abstract
1. Introduction
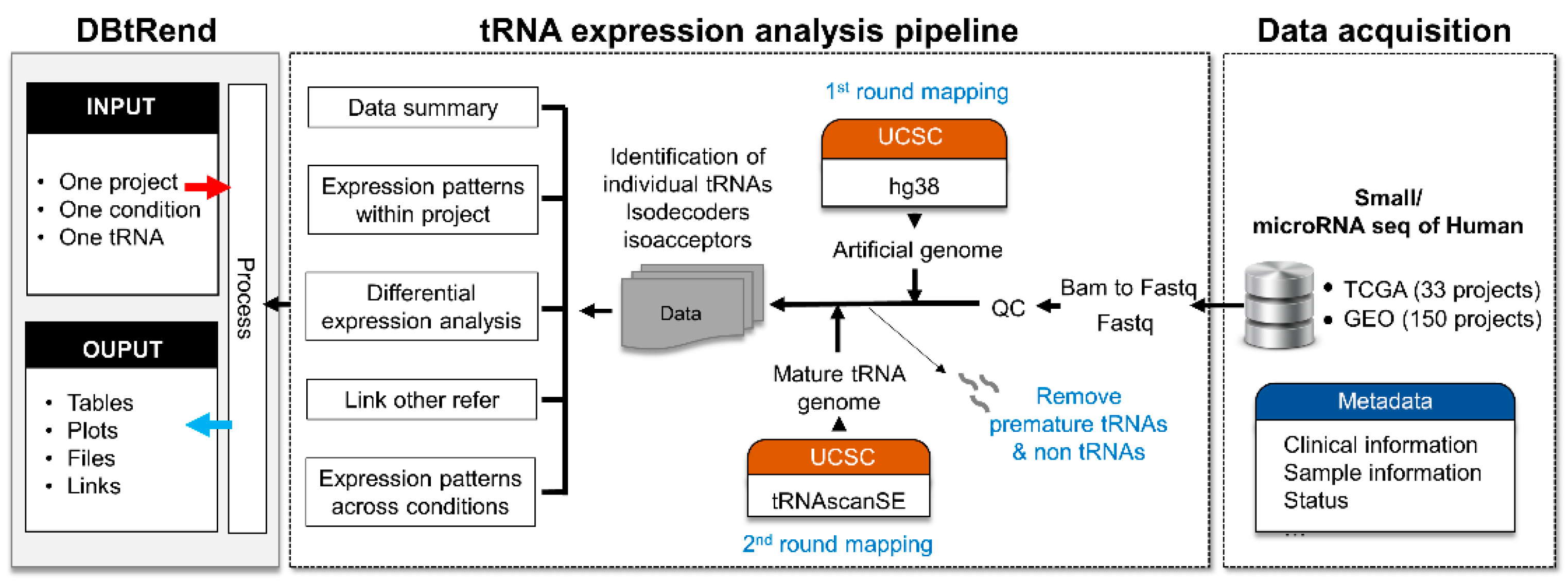
2. Materials and Methods
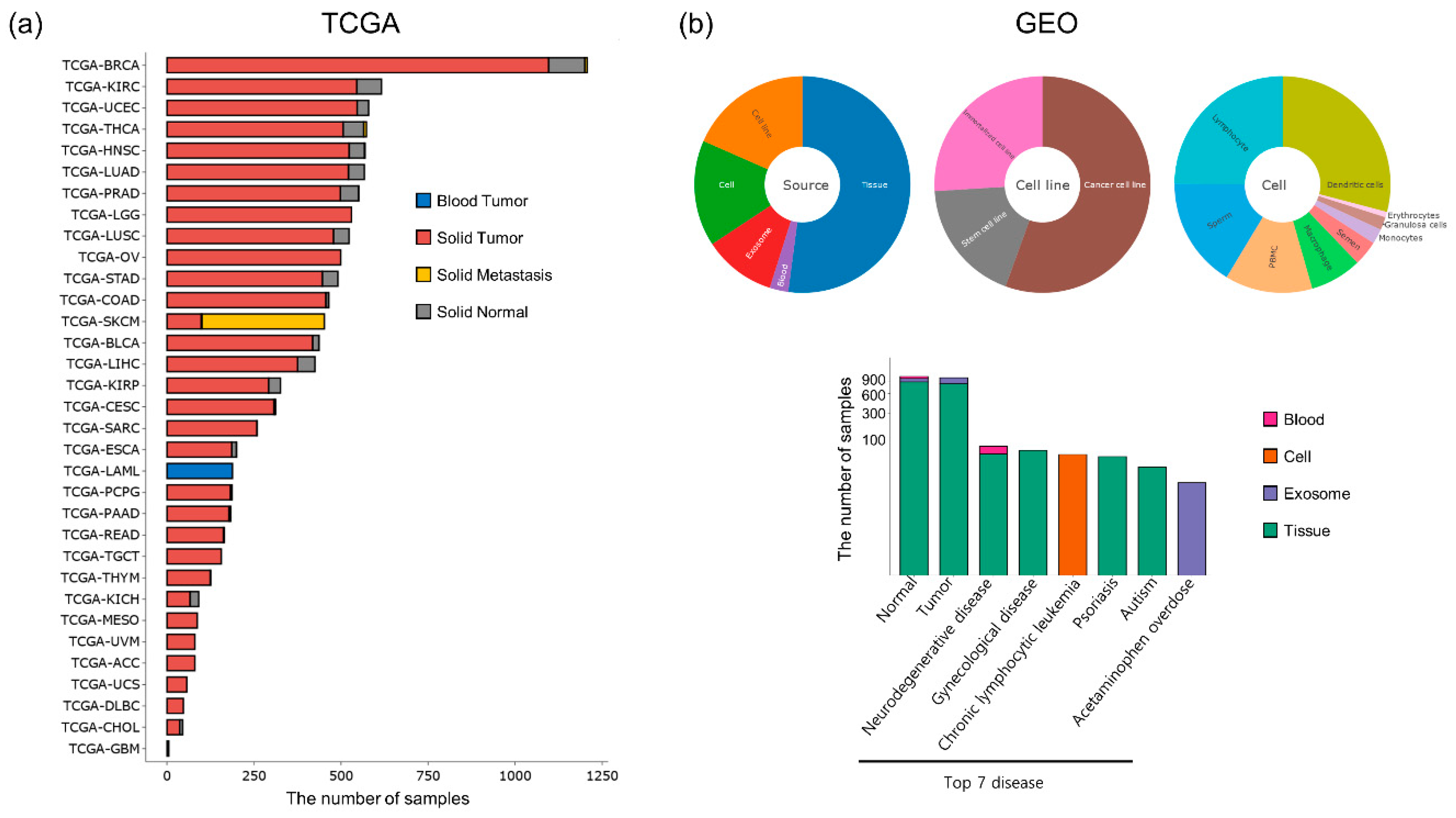
2.1. Data Collection
2.2. tRNA Identification
2.3. Differential Expression Analysis
3. Results
3.1. Data Statistics in DBtRend
3.2. Database Infrastructure
3.2.1. ‘Home’ Page
3.2.2. Query on the ‘Explore Projects’ Page
3.2.3. Query on the ‘Explore DEtRNAs’ Page
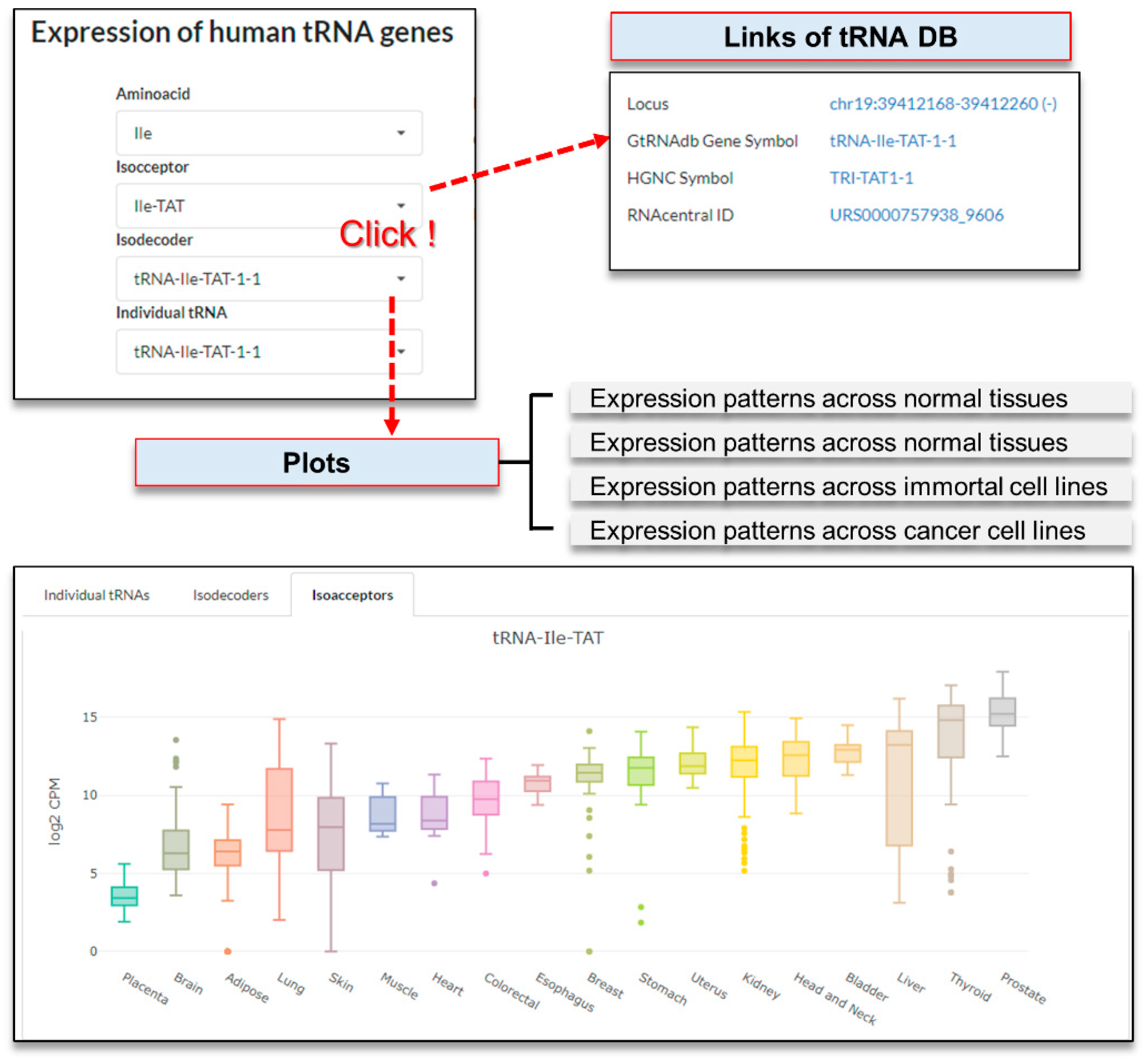
3.2.4. Query on the ‘Explore tRNAs’ Page
3.2.5. An Example Use: Alzheimer’s Disease
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sagi, D.; Rak, R.; Gingold, H.; Adir, I.; Maayan, G.; Dahan, O.; Broday, L.; Pilpel, Y.; Rechavi, O. Tissue- and Time-Specific Expression of Otherwise Identical tRNA Genes. PLoS Genet. 2016, 12, e1006264. [Google Scholar] [CrossRef] [PubMed]
- Frenkel-Morgenstern, M.; Danon, T.; Christian, T.; Igarashi, T.; Cohen, L.; Hou, Y.M.; Jensen, L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012, 8, 572. [Google Scholar] [CrossRef] [PubMed]
- Van Bortle, K.; Nichols, M.H.; Ramos, E.; Corces, V.G. Integrated tRNA, transcript, and protein profiles in response to steroid hormone signaling. RNA 2015, 21, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Krokowski, D.; Han, J.; Saikia, M.; Majumder, M.; Yuan, C.L.; Guan, B.J.; Bevilacqua, E.; Bussolati, O.; Broer, S.; Arvan, P.; et al. A self-defeating anabolic program leads to beta-cell apoptosis in endoplasmic reticulum stress-induced diabetes via regulation of amino acid flux. J. Biol. Chem. 2013, 288, 17202–17213. [Google Scholar] [CrossRef]
- Landwehrmeyer, G.B.; McNeil, S.M.; Dure, L.S.T.; Ge, P.; Aizawa, H.; Huang, Q.; Ambrose, C.M.; Duyao, M.P.; Bird, E.D.; Bonilla, E.; et al. Huntington’s disease gene: Regional and cellular expression in brain of normal and affected individuals. Ann. Neurol. 1995, 37, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated Expression of Specific tRNAs Drives Gene Expression and Cancer Progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef]
- Guo, Y.; Bosompem, A.; Mohan, S.; Erdogan, B.; Ye, F.; Vickers, K.C.; Sheng, Q.; Zhao, S.; Li, C.I.; Su, P.F.; et al. Transfer RNA detection by small RNA deep sequencing and disease association with myelodysplastic syndromes. BMC Genomics 2015, 16, 727. [Google Scholar] [CrossRef]
- Pundhir, S.; Gorodkin, J. Differential and coherent processing patterns from small RNAs. Sci. Rep. 2015, 5, 12062. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torres, A.G.; Pineyro, D.; Rodriguez-Escriba, M.; Camacho, N.; Reina, O.; Saint-Leger, A.; Filonava, L.; Batlle, E.; Ribas de Pouplana, L. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015, 43, 5145–5157. [Google Scholar] [CrossRef]
- Torres, A.G.; Reina, O.; Stephan-Otto Attolini, C.; Ribas de Pouplana, L. Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 8451–8456. [Google Scholar] [CrossRef]
- Hoffmann, A.; Fallmann, J.; Vilardo, E.; Morl, M.; Stadler, P.F.; Amman, F. Accurate mapping of tRNA reads. Bioinformatics 2018, 34, 1116–1124. [Google Scholar] [CrossRef]
- Hernandez-Alias, X.; Benisty, H.; Schaefer, M.H.; Serrano, L. Translational efficiency across healthy and tumor tissues is proliferation-related. Mol. Syst. Biol. 2020, 16, e9275. [Google Scholar] [CrossRef]
- Zhang, Z.; Ye, Y.; Gong, J.; Ruan, H.; Liu, C.J.; Xiang, Y.; Cai, C.; Guo, A.Y.; Ling, J.; Diao, L.; et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Commun. Biol. 2018, 1, 234. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wu, L.; Wang, A.; Tang, W.; Zhao, Y.; Zhao, H.; Teschendorff, A.E. dbDEMC 2.0: Updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2017, 45, D812–D818. [Google Scholar] [CrossRef] [PubMed]
- Papatheodorou, I.; Fonseca, N.A.; Keays, M.; Tang, Y.A.; Barrera, E.; Bazant, W.; Burke, M.; Fullgrabe, A.; Fuentes, A.M.; George, N.; et al. Expression Atlas: Gene and protein expression across multiple studies and organisms. Nucleic Acids Res. 2018, 46, D246–D251. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Sun, X.; Zhou, L.; Amanullah, M.; Pan, X.; Liu, Y.; Liang, M.; Liu, P.; Lu, Y. OncotRF: An online resource for exploration of tRNA-derived fragments in human cancers. RNA Biol. 2020, 17, 1081–1091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ruan, H.; Liu, C.J.; Ye, Y.; Gong, J.; Diao, L.; Guo, A.Y.; Han, L. tRic: A user-friendly data portal to explore the expression landscape of tRNAs in human cancers. RNA Biol. 2020, 17, 1674–1679. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Davis, S.; Stephens, R.; Meltzer, P.S.; Chen, Y. GEOmetadb: Powerful alternative search engine for the Gene Expression Omnibus. Bioinformatics 2008, 24, 2798–2800. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol. 2019, 1962, 1–14. [Google Scholar] [CrossRef]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-O.; Lee, M.; Chung, Y.-J. DBtRend: A Web-Server of tRNA Expression Profiles from Small RNA Sequencing Data in Humans. Genes 2021, 12, 1576. https://doi.org/10.3390/genes12101576
Lee J-O, Lee M, Chung Y-J. DBtRend: A Web-Server of tRNA Expression Profiles from Small RNA Sequencing Data in Humans. Genes. 2021; 12(10):1576. https://doi.org/10.3390/genes12101576
Chicago/Turabian StyleLee, Jin-Ok, Minho Lee, and Yeun-Jun Chung. 2021. "DBtRend: A Web-Server of tRNA Expression Profiles from Small RNA Sequencing Data in Humans" Genes 12, no. 10: 1576. https://doi.org/10.3390/genes12101576
APA StyleLee, J.-O., Lee, M., & Chung, Y.-J. (2021). DBtRend: A Web-Server of tRNA Expression Profiles from Small RNA Sequencing Data in Humans. Genes, 12(10), 1576. https://doi.org/10.3390/genes12101576






