The Complete Mitochondrial Genome of One Breeding Strain of Asian Swamp Eel (Monopterus albus, Zuiew 1793) Using PacBio and Illumina Sequencing Technologies and Phylogenetic Analysis in Synbranchiformes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling, Sequencing, Genome Assembly, and Annotation
2.2. Phylogenetic Analysis, Topology Testing, and Molecular Dating
3. Results
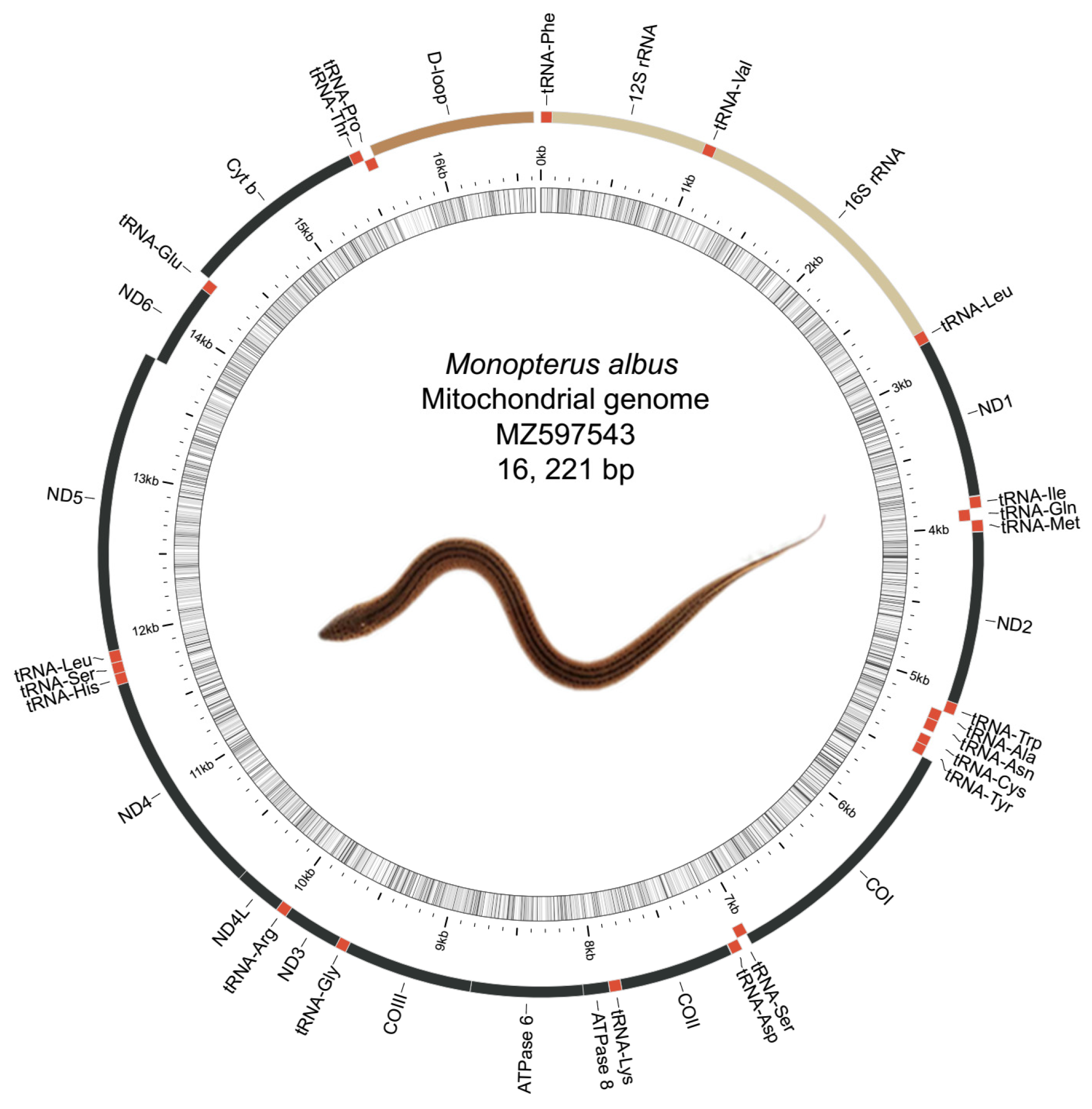
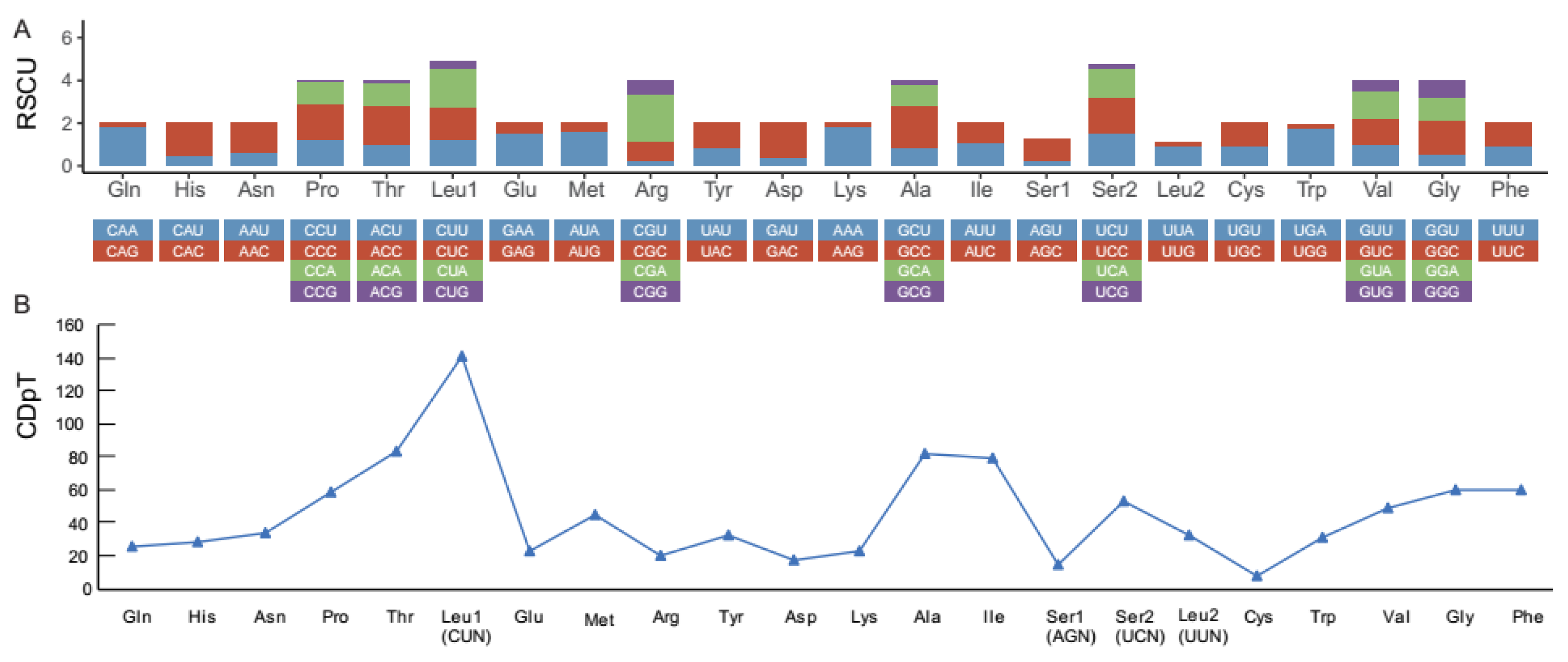
3.1. Mitochondrial Genome Assembly and Annotation
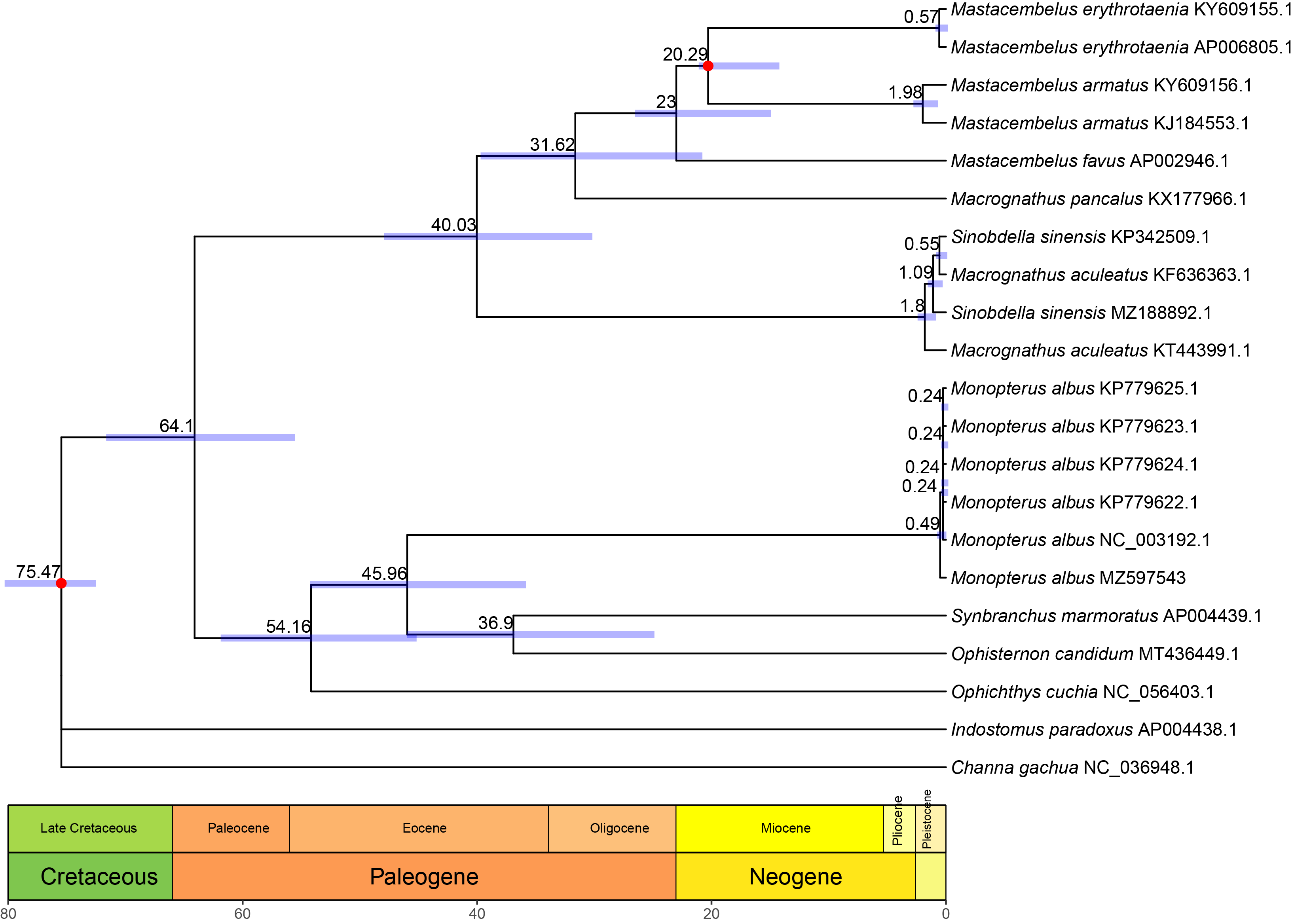
3.2. Phylogenetic Analyses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nelson, J.S.; Grande, T.C.; Wilson, M.V. Fishes of the World; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Yang, D.; Chen, F.; Ruan, G. Aquaculture of the Paddy Eel, Monopterus albus; John Wiley & Sons Ltd.: Hoboken, NJ, USA, 2018; pp. 283–296. [Google Scholar]
- Khanh, N.; Ngan, H. Current practices of rice field eel Monopterus albus (Zuiew, 1973) culture in Vietnam. Aquac. Asia Mag. 2010, 15, 26–29. [Google Scholar]
- Herawati, V.E.; Nugroho, R.A.; Pinandoyo; Hutabarat, J.; Prayitno, B.; Karnaradjasa, O. The growth performance and nutrient quality of asian swamp eel Monopterus albus in central java indonesia in a freshwater aquaculture system with different feeds. J. Aquat. Food Prod. Technol. 2018, 27, 658–666. [Google Scholar] [CrossRef]
- Yang, D.; Chen, F.; Ruan, G.; Li, T. Comparative study on fecundity of different strains of Monopterus albus. J. Hydroecol. 2009, 2, 133–135. [Google Scholar]
- Chen, F.; Yang, D.; Su, Y. Comparison of growth performance of three different body color ricefield eel (Monopterus albus). J. Yangtze Univ. 2009, 6, 33–34, 38. [Google Scholar]
- Cai, X.; Yu, S.M.; Mipam, T.; Zhang, X.Y.; Yue, B.S. Phylogenetic lineages of Monopterus albus (Synbranchiformes: Synbranchidae) in China inferred from mitochondrial control region. J. Zool. Syst. Evol. Res. 2013, 51, 38–44. [Google Scholar] [CrossRef]
- Liang, H.W.; Guo, S.S.; Li, Z.; Luo, X.Z.; Zou, G.W. Assessment of genetic diversity and population structure of swamp eel Monopterus albus in China. Biochem. Syst. Ecol. 2016, 68, 81–87. [Google Scholar] [CrossRef]
- Arisuryanti, T. Molecular Genetic and Taxonomic Studies of the Swamp Eel(Monopterus albus Zuiew 1793). Ph.D. Thesis, Charles Darwin University, Northern Territory, Australia, 2016. [Google Scholar]
- Matsumoto, S.; Kon, T.; Yamaguchi, M.; Takeshima, H.; Yamazaki, Y.; Mukai, T.; Kuriiwa, K.; Kohda, M.; Nishida, M. Cryptic diversification of the swamp eel monopterus albus in east and southeast asia, with special reference to the ryukyuan populations. Ichthyol. Res. 2010, 57, 71–77. [Google Scholar] [CrossRef]
- Miah, M.; Ali, H.; Jannat, E.; Naser, M.N.; Ahmed, M.K. Rearing and production performance of freshwater mud eel, Monopterus cuchia in different culture regimes. Adv. Zool. Bot. 2015, 3, 34–36. [Google Scholar] [CrossRef] [Green Version]
- Britz, R.; Dahanukar, N.; Standing, A.; Philip, S.; Kumar, B.; Raghavan, R. Osteology of ‘Monopterus’ roseni with the description of rakthamichthys, new genus, and comments on the generic assignment of the amphipnous group species (Teleostei: Synbranchiformes). Ichthyol. Explor. Freshw. 2021, 30, 221–236. [Google Scholar]
- Kawahara, R.; Miya, M.; Mabuchi, K.; Lavoué, S.; Inoue, J.G.; Satoh, T.P.; Kawaguchi, A.; Nishida, M. Interrelationships of the 11 gasterosteiform families (sticklebacks, pipefishes, and their relatives): A new perspective based on whole mitogenome sequences from 75 higher teleosts. Mol. Phylogenetics Evol. 2008, 46, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Betancur, R.R.; Wiley, E.O.; Arratia, G.; Acero, A.; Bailly, N.; Miya, M.; Lecointre, G.; Orti, G. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017, 17, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, K.J.; Ruber, L.; Bills, R.; Day, J.J. Mastacembelid eels support lake tanganyika as an evolutionary hotspot of diversification. BMC Evol. Biol. 2010, 10, 188. [Google Scholar] [CrossRef] [Green Version]
- Malmstrom, M.; Matschiner, M.; Torresen, O.K.; Jakobsen, K.S.; Jentoft, S. Whole genome sequencing data and de novo draft assemblies for 66 teleost species. Sci. Data 2017, 4, 160132. [Google Scholar] [CrossRef] [PubMed]
- Allio, R.; Donega, S.; Galtier, N.; Nabholz, B. Large variation in the ratio of mitochondrial to nuclear mutation rate across animals: Implications for genetic diversity and the use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 2017, 34, 2762–2772. [Google Scholar] [CrossRef] [Green Version]
- Miya, M.; Kawaguchi, A.; Nishida, M. Mitogenomic exploration of higher teleostean phylogenies: A case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol. Biol. Evol. 2001, 18, 1993–2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, P.A.; Archer, F.I.; Foote, A.D.; Vilstrup, J.; Allen, E.E.; Wade, P.; Durban, J.; Parsons, K.; Pitman, R.; Li, L.; et al. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 2010, 20, 908–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.Y.; Hasegawa, H.; Cooley, J.R.; Simon, C.; Yoshimura, J.; Cai, W.Z.; Sota, T.; Li, H. Mitochondrial genomics reveals shared phylogeographic patterns and demographic history among three periodical cicada species groups. Mol. Biol. Evol. 2019, 36, 1187–1200. [Google Scholar] [CrossRef]
- Tian, H.F.; Hu, Q.M.; Li, Z. A high-quality de novo genome assembly of one swamp eel (Monopterus albus) strain with Pacbio and Hi-C sequencing data. G3 2021, 11, jkaa032. [Google Scholar] [CrossRef]
- Chen, D.X.; Chu, W.Y.; He, Y.; Liang, X.F.; Mai, K.S. Characteristics and phylogenetic studies of complete mitochondrial DNA based on the ricefield eel (Monopterus albus) from four different areas. Mitochondrial DNA A 2016, 27, 2419–2420. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Hunt, M.; De Silva, N.; Otto, T.D.; Parkhill, J.; Keane, J.A.; Harris, S.R. Circlator: Automated circularization of genome assemblies using long sequencing reads. Genome Biol. 2015, 16, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Novoplasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed] [Green Version]
- Iwasaki, W.; Fukunaga, T.; Isagozawa, R.; Yamada, K.; Maeda, Y.; Satoh, T.P.; Sado, T.; Mabuchi, K.; Takeshima, H.; Miya, M.; et al. Mitofish and mitoannotator: A mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol. Biol. Evol. 2013, 30, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlic, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. Phylosuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. Partitionfinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. Dnasp 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.40. Available online: http://mesquiteproject.org (accessed on 20 December 2018).
- Chu, T.J.; Jin, Z.; Liu, K. The mitochondrial genome of Sinobdella sinensis (Synbranchiformes: Mastacembelidae) from China’s Qiantang river. Mitochondrial DNA B 2021, 6, 2741–2742. [Google Scholar] [CrossRef] [PubMed]
- Mello, B. Estimating timetrees with mega and the timetree resource. Mol. Biol. Evol. 2018, 35, 2334–2342. [Google Scholar] [CrossRef] [Green Version]
- Mello, B.; Tao, Q.Q.; Barba-Montoya, J.; Kumar, S. Molecular dating for phylogenies containing a mix of populations and species by using bayesian and reltime approaches. Mol. Ecol. Resour. 2021, 21, 122–136. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. Timetree: A resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.C.; Lam, T.T.Y.; Zhu, H.C.; Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.H.; Li, M.; Xu, S.N.; Liu, L.; Chen, Z.Z.; Zou, K.S. Complete mitogenomes of three carangidae (Perciformes) fishes: Genome description and phylogenetic considerations. Int. J. Mol. Sci. 2020, 21, 4685. [Google Scholar] [CrossRef]
- Britz, R.; Sudasinghe, H.; Sykes, D.; Ranasinghe, R.H.T. Ophichthys desilvai, a poorly known synbranchid eel from Sri Lanka (Teleostei: Synbranchidae). Ichthyol. Explor. Freshw. 2020, 30, 1–16. [Google Scholar]
- Perdices, A.; Doadrio, I.; Bermingham, E. Evolutionary history of the synbranchid eels (Teleostei: Synbranchidae) in Central America and the Caribbean islands inferred from their molecular phylogeny. Mol. Phylogenetics Evol. 2005, 37, 460–473. [Google Scholar] [CrossRef]
- Xue, L.Z.; Gao, Y.; Wu, M.Y.; Tian, T.; Fan, H.P.; Huang, Y.J.; Huang, Z.; Li, D.P.; Xu, L.H. Telomere-to-telomere assembly of a fish Y chromosome reveals the origin of a young sex chromosome pair. Genome Biol. 2021, 22, 203. [Google Scholar] [CrossRef] [PubMed]
- Supiwong, W.; Pinthong, K.; Seetapan, K.; Saenjundaeng, P.; Bertollo, L.A.C.; de Oliveira, E.A.; Yano, C.F.; Liehr, T.; Phimphan, S.; Tanomtong, A.; et al. Karyotype diversity and evolutionary trends in the Asian swamp eel Monopterus albus (Synbranchiformes, Synbranchidae): A case of chromosomal speciation? BMC Evol. Biol. 2019, 19, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nirchio, M.; Mariguela, T.C.; Ferreira, I.A.; Foresti, F.; Oliveira, C. Karyotype and nuleolus organizer regions of ophisternon aenigmaticum (Teleostei: Synbranchiformes: Synbranchidae) from Venezuela. Interciencia 2011, 36, 229–233. [Google Scholar]
- Wilkens, H.; Strecker, U. Regressive and constructive traits in astyanax surface and cave fish. In Evolution in the Dark; Springer: Berlin/Heidelberg, Germany, 2017; pp. 79–189. [Google Scholar]

| deltaL | bp-RELL | p-KH | p-SH | p-WKH | p-WSH | c-ELW | p-AU | |
|---|---|---|---|---|---|---|---|---|
| Unconstrained | 0 | 0.988 | 0.98 | 1 | 0.98 | 1 | 0.9875 | 0.9927 |
| Indostomus paradoxus grouped with Synbranchidae | 31.339 | −0.011 | −0.02 | +0.093 | −0.02 | −0.0330 | −0.0112 | −0.0118 |
| Indostomus paradoxus grouped with Mastacembelidae | 37.324 | −0.001 | −0.003 | −0.047 | −0.003 | −0.004 | −0.0013 | −0.0035 |
| Monophyly of Sinobdella sinensis and Macrognathus aculeatus | 71.940 | 0 | 0 | −0.002 | 0 | 0 | 0 | −0.0047 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, H.; Hu, Q.; Lu, H.; Li, Z. The Complete Mitochondrial Genome of One Breeding Strain of Asian Swamp Eel (Monopterus albus, Zuiew 1793) Using PacBio and Illumina Sequencing Technologies and Phylogenetic Analysis in Synbranchiformes. Genes 2021, 12, 1567. https://doi.org/10.3390/genes12101567
Tian H, Hu Q, Lu H, Li Z. The Complete Mitochondrial Genome of One Breeding Strain of Asian Swamp Eel (Monopterus albus, Zuiew 1793) Using PacBio and Illumina Sequencing Technologies and Phylogenetic Analysis in Synbranchiformes. Genes. 2021; 12(10):1567. https://doi.org/10.3390/genes12101567
Chicago/Turabian StyleTian, Haifeng, Qiaomu Hu, Hongyi Lu, and Zhong Li. 2021. "The Complete Mitochondrial Genome of One Breeding Strain of Asian Swamp Eel (Monopterus albus, Zuiew 1793) Using PacBio and Illumina Sequencing Technologies and Phylogenetic Analysis in Synbranchiformes" Genes 12, no. 10: 1567. https://doi.org/10.3390/genes12101567
APA StyleTian, H., Hu, Q., Lu, H., & Li, Z. (2021). The Complete Mitochondrial Genome of One Breeding Strain of Asian Swamp Eel (Monopterus albus, Zuiew 1793) Using PacBio and Illumina Sequencing Technologies and Phylogenetic Analysis in Synbranchiformes. Genes, 12(10), 1567. https://doi.org/10.3390/genes12101567






