Abstract
Campylobacter jejuni (C. jejuni) is responsible for 80% of human campylobacteriosis and is the leading cause of gastroenteritis globally. The relevant public health risks of C. jejuni are caused by particular virulence genes encompassing its virulome. We analyzed 40,371 publicly available genomes of C. jejuni deposited in the NCBI Pathogen Detection Database, combining their epidemiologic metadata with an in silico bioinformatics analysis to increase our current comprehension of their virulome from a global perspective. The collection presented a virulome composed of 126 identified virulence factors that were grouped in three clusters representing the accessory, the softcore, and the essential core genes according to their prevalence within the genomes. The multilocus sequence type distribution in the genomes was also investigated. An unexpected low prevalence of the full-length flagellin flaA and flaB locus of C. jejuni genomes was revealed, and an essential core virulence gene repertoire prevalent in more than 99.99% of genomes was identified. Altogether, this is a pioneer study regarding Campylobacter jejuni that has compiled a significant amount of data about the Multilocus Sequence Type and virulence factors concerning their global prevalence and distribution over this database.
1. Introduction
Campylobacter is a Gram-negative bacterium that is able to infect several wild and domesticated animals, contaminated water, raw milk, and animal-based food worldwide [1,2]. Among the species, Campylobacter jejuni (C. jejuni) is responsible for 80% of human campylobacteriosis and is the leading cause of gastroenteritis globally [3,4]. In addition, it may cause bacteremia, sepsis, meningitis, urinary tract infections, and hemolytic-uremic syndrome. In some cases, the patient may develop complications after infection such as inflammatory bowel diseases (IBD) and Barrett’s esophagus [5]. Post-infectious sequelae are emerging with severe illnesses such as Guillain–Barré syndrome (GBS) [6], a rare autoimmune inflammatory disease of the Peripheral Nervous System, and Miller Fisher syndrome, which is also a rare disease [7].
The relevant public health risks of C. jejuni are caused by particular virulence genes encompassing its virulome. Unlike the classical virulence factors such as the T3SS and lipopolysaccharide (LPS) observed in other enteropathogens, C. jejuni lacks or does not rely extensively on many classical virulence factors such as enterotoxigenic Escherichia coli and Salmonella spp. [8,9,10]. It has the particular growth necessities apparent in a fragile organism; however, it has a complex array of fitness and virulence factors [11] that aid the bacterium to respond to the defense mounted by the host (adhesion, invasion, and temporary survival inside the human intestinal epithelial cells (IECs) in vitro). Collective components such as the helical shape, amphitrichous flagella, capsular polysaccharide (CPS), Lipooligosaccharide (LOS), O-linked glycosylation of flagellins, and genotypic and phenotypic variation owing to an augmented number of phase-variable loci are some particularities that promote the successful enteric lifestyle of C. jejuni [9]. Some mechanisms of pathogenesis over C. jejuni, such as the discovery of the novel autotransporter CapC and other virulence factors, have constantly been elucidated [12,13].
Campylobacter jejuni is efficient at generating an extensive intraspecies heterogeneity due to its phase variation ability. These properties require sophisticated molecular typing methods to differentiate species and subtypes and elucidate infection sources. Multi Locus Sequence Typing (MLST) has proven to be the molecular method of choice and has provided strong evidence that genetically closely related strains of C. jejuni can colonize different hosts [14]. Closely related strains usually belong to large clonal complexes (CC), which share multiple MLST alleles (STs). Moreover, the MLST has provided unified, comprehensive, and portable Campylobacter isolate characterization, with curated databases of genotypes available (pubMLST.org/Campylobacter) [15]. The genetic diversity of these species is reflected in the more than 11,800 STs registered on the PubMLST database. Notably, more than 8000 C. jejuni sequence types were registered by 2020 [16]. Certainly, the determination of C. jejuni STs is a valuable resource to the scientific community as a whole for the genotyping investigation of this critical and yet incompletely understood pathogen.
There are web servers and resources that offer free comprehensive analysis pipelines, such as the NCBI Pathogen Detection platform (https://www.ncbi.nlm.nih.gov/pathogens/, accessed on 24 September 2021), the PubMLST (https://pubmlst.org/organisms, accessed on 24 September 2021) and Enterobase (http://enterobase.warwick.ac.uk/, accessed on 24 September 2021) provided that there are no privacy or ethical issues in submitting data to an online repository. The PubMLST seems to be more diverse than the NCBI, with fewer redundant assemblies. In addition, the contributions of USA and UK agencies to genome deposits in the NCBI database are much more significant. The PubMLST also contains the integrated software BIGSdb (https://github.com/kjolley/BIGSdb, accessed on 24 September 2021) that allows a calling engine for a gene-by-gene approach in the same browser, facilitating analysis in real-time. On the other hand, there are already several species-specific databases in the NCBI Pathogen Detection platform against which the isolates of interest can be compared. However, this website does not offer the MLST typing or the virulence genes repertoire for the Campylobacter species.
Studies involving whole-genome sequencing (WGS) have facilitated the biological understanding of gastrointestinal pathogens and several existing microorganisms. The advances of WGS generated by high throughput sequencing platforms has allowed the mass submission of genomes to public repositories in the last years [17,18]. Consequently, in silico studies from bacterial genomes have increased in the last few years, providing several possibilities in gene analysis [19,20,21,22]. The software VirulenceFinder—VFDB (http://cge.cbs.dtu.dk/services/VirulenceFinder/, accessed on 29 June 2021) [23,24] uses WGS data for the in silico characterization of several pathogenic bacteria and enables researchers and clinical health personnel to quickly extract and interpret virulence-relevant information from the genome data [21]. Considering the remaining insights that can still be understood about C. jejuni virulence, we designed this study to analyze lots of WGS by an in silico bioinformatics analysis combining epidemiologic metadata to increase our current comprehension of the C. jejuni virulome from a global perspective.
2. Materials and Methods
2.1. Data Collection
We downloaded the entire collection of 61,711 Campylobacter genomes assembled until 4th May 2021 from the Pathogen Detection Web Browser at the National Biotechnology Information Center (NCBI) website (https://www.ncbi.nlm.nih.gov/pathogens/, accessed on 4 May 2021). The software KmerFinder v3.2 [25,26,27] was used to identify the C. jejuni species within all the Campylobacter genomes. The software was installed and run as per the standard instructions informed by the server of the software (https://bitbucket.org/genomicepidemiology/kmerfinder/src/master/, accessed on 20 July 2021).
2.2. Multilocus Sequence Type (MLST) Identification
The STs were determined from contigs of each genome using the software Seemann T. mlst v2.19.0 according to the instructions provided by the server of software (https://github.com/tseemann/mlst, accessed on 20 July 2021). The analysis for the C. jejuni allele scheme was set from the open-access PubMLST.org website database hosted by the University of Oxford [28].
2.3. In Silico Virulome Determination
The bacterial virulome identification was performed according to the instructions provided in the Seemann T. ABRicate software (https://github.com/tseemann/abricate v0.9.9., accessed on 5 August 2021) The virulome was determined by genome hits against the 2597 genes of the virulence database (VFDB) [23,24] using the minimum gene length coverage of 60%. This coverage threshold was set to ensure that genes lying on the edge of a contig or spread over two contigs were not missed due to possible non-perfect assembly [29]. Due to highly variable sequences interspersed among conserved genes and the existence of phase-variable genes such as motA and FlgR [9,30,31], no thresholds were established for the minimum per base identity to avoid missing any gene.
2.4. Statistical Analyses
Statistical analyses were performed using R Studio (http://www.r-project.org, accessed on 12 August 2021) by the Complex Heat map package [32] to cluster and construct abundance heat maps. The prevalence of the virulence genes was calculated by the ratio between each gene and the total number of genes from all the genomes assessed in each geographical region set for this study. The average of the frequencies and the standard deviation were calculated using the formula functions in Microsoft Excel.
3. Results
3.1. Metadata Overview
Through the KmerFinder software, a taxonomic sequence classifier that assigns taxonomic labels to DNA sequences, we identified and collected all the genomes of C. jejuni. Out of the 61,711 Campylobacter genomes, a collection of 40,371 C. jejuni genomes were finally settled for the virulome investigation. The Campylobacter species identified in each genome and the origin of the genomes regarding the geographic location can be seen in Table S1.
3.2. Multilocus Sequence Type (MLST)
The sequence types (STs) were investigated in all the Campylobacter jejuni genomes, although a few could not be determined (Table S1). Over the identified STs, we compiled their global distribution over the five main continents (Figure S1). North America’s genomes presented 745 different STs, followed by Europe (714), Asia (74), South America (64), Oceania (58), and Africa (44). Due to the vast diversity of STs, we ranked the ten most commonly detected STs within this collection of Campylobacter jejuni recovered worldwide. Notably, ST50 or ST353 led the STs in all continents except in Asia, where ST2988 was the most common ST identified within the genomes (Table 1 and Supplementary Figure S1).

Table 1.
Top ten commonly detected Campylobacter jejuni sequence types (STs) recovered from the whole genome sequence in the NCBI Pathogen Detection Database (n = 40,371) from each continent.
3.3. Campylobacter Jejuni Virulome
The Campylobacter jejuni collection presented a virulome composed of 126 identified genes out of the 2597 virulence genes settled in the virulence finder database—VFDB (Table S1). Our clustering analysis revealed that the Campylobacter jejuni virulomes were grouped in three vertical clusters according to their average prevalence and three horizontal clusters according to their prevalence profiles in each geographic location (Figure 1). The virulence genes from the North America, Oceania, and Europe genomes showed similar prevalence profiles and were grouped in Cluster-C. The virulence genes from the African and South American genomes were grouped in Cluster-B. Finally, the virulence genes from the Asian genomes showed a unique prevalence profile and were grouped in Cluster-A.
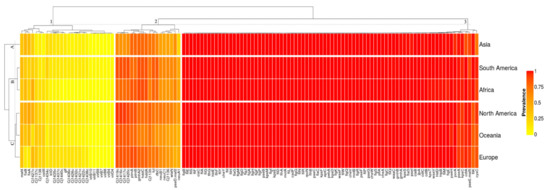
Figure 1.
Color heatmap with the 126 virulence genes comprising the C. jejuni virulome of the 40,371 whole genome sequences collected from the NCBI Pathogen Detection Database. The prevalence ranges from 0 to 1, in which 0 (yellow) is the least prevalent, and 1 (red) is the most prevalent.
Cluster-1 grouped the accessory genes of the virulome with prevalence values ranging from an absence of the genes up to 48.48% (Supplementary Table S1 and Figure S1). The average prevalence in the cluster was 13.17 ± 2.67%. The virulence factors involved the bacterial capsule biosynthesis and transport, especially by sugar and aminotransferase enzymes (Cj1421c, Cj1422c, Cj1426c, Cj1432c, Cj1434c, Cj1435c, Cj1436c, Cj1437c, Cj1438c, Cj1440c, glf, kfiD), the Type IV secretion system (virB10, virB11, virB4, virB7, virB8, virB9, virD4), the immune evasion—LOS (Cj1137c, Cj1138, cstIII), the O-linked flagellar glycosylation (maf4), the capsule biosynthesis and transport (cj1427c), and the motility and export apparatus (flaA, flaB). The sugar transferase Cj1434c displayed the highest prevalence (25.13%) in the genomes from South America. The African genomes displayed the lowest average prevalence of the cluster (9.21%). The plasmid-mediated genes virB-D that encode the Type IV secretion system in C. jejuni were absent in the genomes from Africa and Oceania, were very low in prevalence in Europe (0.78%) and Asia (0.60%), and were low in prevalence in North America (3.88%) and South America (4.56%). The gene cstIII was also low in prevalence in the genomes from Asia (4.79%). In Oceania, the genomes displayed the lowest prevalence (18.13%) of the sugar-nucleotide epimerase/dehydratase cj1427c. The flagellin flgA/flaB genes displayed the lowest prevalence in Europe (12.47/12.13%).
Cluster-2 grouped the softcore genes of the virulome with prevalence ranging from 62.38 up to 77.44%. The average prevalence within this cluster was 71.08 ± 5.68% (Supplementary Table S1 and Figure S1). The virulence factors involved the immune evasion—LOS (gmhA2, Cj1135, neuA1, neuB1, neuC1, Cj1136, wlaN), the capsule biosynthesis and transport (Cj1419c, Cj1416c, Cj1417c, Cj1420c, hddC, hddA, fcl, rfbC), and the motility and export apparatus (ptmA, ptmB, pseD/maf2). The gene neuA1 had a higher prevalence (above 42.11%) in the genomes from North America, Europe, Oceania, South America, and Africa than Asia (23.35%). The genes Cj1419c, Cj1416c, Cj1417c, Cj1420c displayed the highest prevalence in all continents (above 83.04%) except in Asia (below 71.86%). The genes gmhA2, hddC, and hddA displayed a lower prevalence in Oceania (below 67.25%) than the other continents (above 77.54%). The genes neuB1 and neuC1 displayed a higher prevalence (above 71.04%) in North America than in the other continents (below 64.40%). The genes wlaN and pseD/maf2 displayed the lowest prevalence (46.11/44.14%) in the genomes from Asia and Europe, respectively.
Cluster-3 grouped the core genes of the virulome with prevalence ranging from 71.86 up to 100%. The average prevalence within this cluster was 99.44 ± 0.26% (Supplementary Table S1 and Figure S1). This cluster grouped the largest repertoire of genes with the smallest standard deviation in their average prevalence. The cluster was mainly formed by the virulence genes responsible for the motility and export apparatus (55 out of 82) as the flagellar basal body rod and hook protein (flg), the flagellar biosynthesis and assembly protein (flh), the flagellar motor protein (operon fli), the chemotaxis protein (che), and the flagellar motor protein (mot). A group of 32 genes within this virulence factor (cadF, fliL, flgG, motA, flhA, pseB, kpsE, kpsM, flgI, fliD, flgA, fliI, flgF, flgP, flaG, flgH, cheY, flgD, flgJ, flgM, flhF, flhG, hldE, fliY, fliG, fliA, ciaC, fliF, fliH, fliS, fliE, flgB) encompassed the essential core of the virulome in which the average prevalence was above 99.9%. The flagellar basal body rod protein FlgB was detected in all the 40,371 (100%) C. jejuni genomes investigated. The genes fliK, pseE/maf5, kpsC, cdtA, pseA, and cysC displayed the lowest average prevalence within the cluster (below 96.52%).
4. Discussion
Campylobacter is the leading cause of bacterial foodborne gastroenteritis in the world [4]. Within this genus, C. jejuni has always been the most relevant clinical [11,33,34], infectious [9], and epidemic zoonotic worldwide [35]. Here we combined more than forty thousand publicly available Campylobacter jejuni genomes of strains isolated from more than forty countries of five continents in the world. The synthesized data will help the scientific community advance more in understanding this species pathogenesis. As far as we know, this is a pioneer study regarding Campylobacter jejuni that has compiled a significant amount of data about the Multilocus Sequence type and the virulence factors concerning their global prevalence and distribution over this database.
Multilocus sequence typing (MLST) is a gold standard informative tool that has successfully characterized the population structure of Campylobacter, identifying lineages such as sequence types (STs) and clonal complexes (CCs) [36,37]. The need for a standard global microbial identification and the power of the MLST explain the many databases and schemes created. The PubMLST (https://pubmlst.org) is the reference database hosting more than 77,000 C. jejuni isolates worldwide. Most of the metadata deposited in this database contain only the STs scheme, although the upload of whole-genome sequences has reached more than 40,000 C. jejuni isolates. Even though most of the C. jejuni ST has already been identified in the PubMLST, our results link them to metadata such as the collection date, the location, the isolation source type of the strains, and the virulence factor composing the C. jejuni virulome of the samples derived from NCBI (Supplementary Table S1). Moreover, the STs displayed in a word cloud diagram quickly highlight their prevalence and distribution worldwide (Supplementary Figure S1). Notably, the wide diversity of C. jejuni STs in the world is probably due to the plasticity in their genetic relationship once the seven schemed housekeeping genes have shown weak clonality among the C. jejuni population [15].
We ranked the top ten identified STs from our genome collection by geographical location (Table 1). The ST50 was the most commonly reported ST in Europe and in Oceania. It was the second most common ST in North America and the sixth most common ST in South America. This sequence type has been recently characterized in C. jejuni genomes from North America, Europe, and Australia, highlighting its importance as a global pathogen [16]. The ST50 distribution presented here corroborates with Wallace et al. [16], who showed that the ST50 is among the top ten commonly reported STs within the more than 77,000 sequences deposited in the PubMLST in all continents except Africa. Interestingly, none of the host generalist sequence types ST21, ST45, and ST48 were reported top in Africa, Asia, and South America, although the chicken specialist ST353 [38,39,40] was most frequently identified in Africa and South America. It is also important to comment that the most hyper-aerotolerant, multi-stress resistant, and strong biofilm-producing C. jejuni isolates belonged to the host generalist clonal complexes ST21, ST45, ST48, and ST206 [41]. In summary, the frequency and distribution of the STs displayed here only represent the genomes collected from the Pathogen Detection Database for this study. To better understand the prevalence and distribution of the C. jejuni STs around the globe, we recommend associating our data with the additional information from the PubMLST database.
A large number of the C. jejuni Capsular Polysaccharide (CPS) genes are involved in capsule biosynthesis and transport. Capsule-related genes were first reported at the beginning of the 2000s in the reports of Karlyshev et al. [42] on the surface of C. jejuni, found on the outermost layer composed of repeated units of several sugars [43,44]. We identified a group of CPS genes in Cluster-1 (Cj1421c, Cj1422c, Cj1426c, Cj1432c, Cj1434c, Cj1435c, Cj1436c, Cj1437c, Cj1438c, Cj1440c, glf, kfiD) with an average prevalence lower than 15% in the C. jejuni genomes worldwide. The remaining CPS genes were grouped in Cluster-2 and 3 and displayed an average prevalence above 70%. The reason for this discrepancy was well understood by the complete genome sequence of the strain NCTC11168 that clarifies the multiple mechanisms of structural variation of CPS, including the exchange of entire clusters by horizontal transfer, gene duplication, deletion, fusion, and contingency gene variation [42]. CPS is also the major sero-determinant of the Penner typing scheme [45], and the high variability of the CPS genes in the C. jejuni genome results in a variety of Penner serotypes [46]. Besides the high genetic rearrangements both within and outside the CPS gene cluster, the locus showed a unique mosaic structure maintained in hypervirulent C. jejuni clones and is a key virulence factor for the induction of systemic infection and abortion in pregnant animals [47]. Lastly, the mutants ΔkpsS-C-M were shown to be deficient in capsule production, resulting in the loss of its ability to induce bacteremia and liver infection in the murine model for the infectivity of C. jejuni [47,48]. The high prevalence of the locus kpsC-T-S-D-F-E-M in our collection of genomes corroborates the importance of these genes for the vitality of C. jejuni and provides evidence that they are potential candidates for the development of vaccines against C. jejuni worldwide.
The virulence genes virB-D are plasmid-mediated, and their prevalence in the C. jejuni genomes is associated with the presence of the plasmid pVir. The plasmid contains components of a type IV secretion system (T4SS) important for several major bacterial pathogens [49]. However, its involvement in pathogenicity which increases bloody diarrhea in C. jejuni enteritis is controversial [50,51,52,53]. The very low prevalence of vir genes in humans and broilers found by Iglesias-Torrens et al. [54] also suggests that the pVir plasmid is not required for C. jejuni to either colonize birds or infect humans. Our results provide a solid demonstration that the plasmid pVir and its virulence vir genes have a very low prevalence (below 5%) in C. jejuni globally, corroborating the hypothesis that the T4SS encoded by the pVir plasmid may not be relevant for the virulence of C. jejuni [52,55].
In 1993, the polymerase chain reaction (PCR) was developed to amplify the C. jejuni flaA gene for restriction fragment length polymorphism (RFLP) analysis for genotyping purposes [56]. Later, in 1997, the demonstration of a recombination within and between flagellin loci of natural strains suggests that flagellin gene typing (restriction fragment length polymorphism analysis of PCR-amplified flagellin genes) cannot be considered a stable method for the long-term monitoring of pathogenic Campylobacter populations [57]. However, in the same year, a short variable region (SVR) was identified from the sequences of flaA and was shown to be conserved among outbreak-related strains [30]. Recent studies have stated that the direct sequencing of the SVR-flaA gene is genetically stable over long periods and is helpful for Campylobacter typing. It has a similar or higher discriminatory power than MLST, especially in short-term and localized epidemiological investigations [58,59,60,61]. Punctual studies from multiple places worldwide have reported a high prevalence of flagellin A and B in C. jejuni [62,63,64,65,66,67,68,69,70,71].
Conversely, our results demonstrate that the flagellin genes flaA and flaB are the unique genes within the motility and export apparatus virulence factor group that displayed an average prevalence below 35% and a particularly low average prevalence worldwide. These prevalence averages were even lower in genomes from Europe that displayed values below 12.5%. These results corroborate other studies that have investigated the virulence genes of C. jejuni from contigs of whole-genome sequences. In a recent study involving 81 C. jejuni and C. coli strains from Chile, flagellin genes flaA and flaB had a low prevalence within the genomes (9 out of 81) [72]. In Ireland, 50% of the C. jejuni sequenced strains from clinical and poultry sources presented the genes flaA and flaB. Curiously, in this study, a higher recovery rate of flaA and flaB previously amplified by a conventional PCR was observed via WGS screening, showing the better resolution of this technique for virulence detection [73]. In a genome-wide association study with C.jejuni isolates from Poland, the authors displayed similar results, reporting that the majority of the virulence genes, with rare exceptions such as flaA, flaB, or flaE, were evenly distributed throughout the genomes of all the STs under study [74]. Finally, in a multi-omics approach study that revealed the potential core vaccine targets for C. jejuni, flaA and flaB were not found within the core or essential core virulome of 173 strains [75].
We verified the presence of the flaA/B gene in full annotations of 233 complete genome sequences of C. jejuni deposited in the RefSeq database of the NCBI (data not shown). This investigation was necessary to rule out the possibility that the low prevalence of the flaA/B gene was caused by (1) the gene lying on the edge of contigs, (2) spread over contigs discarded by low quality, or (3) final draft genomes assembled with low completeness/quality. Out of the 233 genomes, the flaA and flaB genes were annotated in 17 (7.29%) and 4 (1.71%), respectively. We believe some well-known facts can explain these results. First, the intragenomic recombination in the flagellin locus of C. jejuni resulted either in the deletion or repositioning (into flaB) of sequences initially located in the flaA gene [76]. Second, it has been suggested that flaB may serve as a gene donor, of which sequences can be introduced through homologous recombination into the flaA gene, either to compensate for deleterious mutations or possibly to increase the immunogenic repertoire of a given C. jejuni strain [57,76,77]. Third, in the flagellin locus of C. jejuni, sequences essential for the transport and assembly of flagellin monomers into flagellin filaments are located in the conserved regions. In contrast, the internal region lacks functional information and is more highly variable and susceptible to homologous recombination [57,78]. The low prevalence of the full-length flagellin flaA and flaB locus of C. jejuni demonstrated here supports the argument against the long-term stability of flagellin gene typing, highlighting the potential low reliability of these methods for investigation into closely related Campylobacter strains [79].
It is also worth noting the low prevalence of the gene wlaN in Cluster-2. The wlaN gene is responsible for the production of Beta-1,3 galactosyltransferase which is involved in cell wall synthesis, and in the cytotoxin production of Campylobacter and the cytolethal distending toxin genes cdtA, cdtB, and cdtC [80]. The product of the wlaN gene also shows ganglioside mimicking structures that are putative to be involved in the development of Guillain–Barré syndrome after C. jejuni infection [81,82]. The prevalence of the wlaN gene is controversial within the literature. A low prevalence was reported in the Campylobacter strains from Japan [83], Bangladesh [62], Ireland [84], and Brazil [85], whereas a study in Korea identified the wlaN gene among 100% of human and 78.6% of animal C. jejuni isolates investigated [86]. The mass screening of C. jejuni genomes presented here showed that the average prevalence of this gene in the world is about 56%. Besides, wlaN is considered a vital trait gene for Campylobacter virulence, and the intermediary prevalence displayed here indicates that their role in the pathogenesis of C. jejuni should be further well-evaluated.
Cluster-3 grouped the core genes of the virulome. A group of 32 genes encompassed an essential core with an average prevalence above 99.99% within the 40,371 C. jejuni genomes studied here. The intense presence of the strict genes (cadF, fliL, flgG, motA, flhA, pseB, kpsE, kpsM, flgI, fliD, flgA, fliI, flgF, flgP, flaG, flgH, cheY, flgD, flgJ, flgM, flhF, flhG, hldE, fliY, fliG, fliA, ciaC, fliF, fliH, fliS, fliE, flgB) in this large number of genomes worldwide develops our the understanding of the pathogenic potential of C. jejuni. The applicability of the identification of the core genes, for example, is essential for studies where the identification of the virulent bacterial proteins can help the design of potential drugs and vaccines, especially in bacteria such C. jejuni that show extensive genetic variations and a smaller core genome than other foodborne pathogens [75]. Another applicability of the identification of core genes is in the fast detection by a PCR-based assay. As we discussed above, PCR assays based on the whole or specific regions of the flagellin genes flaA and flaB to detect the genotype of C. jejuni require a better understanding and confirmation of their functionality. The detection of the gene flgA in 99.9% and flgB in 100% of the 40,371 C. jejuni genomes studied here provide evidence for its possible application on assays such as those discussed above. An insertional mutation flgA caused its complete absence in C. jejuni NCTC11168 [87]. This explains the presence in all genomes and proves that the gene is essential for flagellar biosynthesis and biofilm formation. Of the 126 genes that encompass the core virulome identified in this study, 59 (~47%) are related to motility, export apparatus, and adherence. They play vital roles in the rotation and switching of the bacterial flagella and are conducive to Campylobacter adhesion and biofilm formation [9]. Moreover, it was proven that the flagellar proteins’ export apparatus as a type-III export system in C. jejuni is required for the secretion of invasion antigens (CiaC virulence protein), essential for host invasion.
5. Conclusions
The current study evaluated the prevalence of C. jejuni virulence markers in a large number of genomes from a diverse set of locations never investigated in the Campylobacter field before. Our data showed the virulence of C. jejuni in a new perspective, bringing novel insights to the scientific community. The in silico analysis of WGS revealed a notable diversity in the occurrence of clinically relevant virulence genes, highlighted by the complete or partial lack of the flaA and flaB loci in a high number of genomes from all the continents. This observation requires further investigation to support the explaining causes we presented above. Altogether, we combined valuable epidemiological and genomic data from several virulence profiles in an extensive collection of C. jejuni strains worldwide.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/genes12101538/s1, Figure S1: Word-cloud map displaying the frequency intensity of the MLST from the collected C. jejuni genomes in a global perspective, Table S1: Metadata, virulence frequency, and clustering distribution from the C. jejuni genomes utilized in this study.
Author Contributions
Conceptualization, P.P.; Data curation, P.P. and A.B.P.; Formal analysis, P.P.; Funding acquisition, C.A.C.J.; Investigation, P.P.; Methodology, P.P.; Project administration, C.A.C.J.; Resources, C.A.C.J.; Software, P.P.; Supervision, S.d.S.D. and C.A.C.J.; Visualization, P.P., A.B.P. and A.M.P.d.S.; Writing—original draft, P.P.; Writing—review and editing, P.P., A.B.P., A.M.P.d.S., S.d.S.D. and C.A.C.J. All authors have read and agreed to the published version of the manuscript.
Funding
The research was funded by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) Brazil—grant number E-26/203.049/2017 and E-26/202.227/2018, the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)—grant number [311422/2016-0], and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Brazil—Finance Code001.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors are thankful for the financial support provided by the Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) Brazil—, the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) Brazil.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Young, K.T.; Davis, L.M.; DiRita, V.J. Campylobacter jejuni: Molecular Biology and Pathogenesis. Nat. Rev. Microbiol. 2007, 5, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Epping, L.; Walther, B.; Piro, R.M.; Knüver, M.-T.; Huber, C.; Thürmer, A.; Flieger, A.; Fruth, A.; Janecko, N.; Wieler, L.H.; et al. Genome-Wide Insights into Population Structure and Host Specificity of Campylobacter jejuni. Sci. Rep. 2021, 11, 10358. [Google Scholar] [CrossRef] [PubMed]
- Bolton, D.J. Campylobacter Virulence and Survival Factors. Food Microbiol. 2015, 48, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Kirk, M.D.; Pires, S.M.; Black, R.E.; Caipo, M.; Crump, J.A.; Devleesschauwer, B.; Döpfer, D.; Fazil, A.; Fischer-Walker, C.L.; Hald, T.; et al. World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis. PLoS Med. 2015, 12, e1001921. [Google Scholar] [CrossRef]
- Nielsen, H.L.; Dalager-Pedersen, M.; Nielsen, H. Risk of Inflammatory Bowel Disease after Campylobacter jejuni and Campylobacter concisus Infection: A Population-Based Cohort Study. Scand. J. Gastroenterol. 2019, 54, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, Y.; Rojas, M.; Pacheco, Y.; Acosta-Ampudia, Y.; Ramírez-Santana, C.; Monsalve, D.M.; Gershwin, M.E.; Anaya, J.-M. Guillain–Barré Syndrome, Transverse Myelitis and Infectious Diseases. Cell. Mol. Immunol. 2018, 15, 547–562. [Google Scholar] [CrossRef]
- Leonhard, S.E.; Mandarakas, M.R.; Gondim, F.A.A.; Bateman, K.; Ferreira, M.L.B.; Cornblath, D.R.; van Doorn, P.A.; Dourado, M.E.; Hughes, R.A.C.; Islam, B.; et al. Diagnosis and Management of Guillain–Barré Syndrome in Ten Steps. Nat. Rev. Neurol. 2019, 15, 671–683. [Google Scholar] [CrossRef]
- Gaytán, M.O.; Martínez-Santos, V.I.; Soto, E.; González-Pedrajo, B. Type Three Secretion System in Attaching and Effacing Pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 129. [Google Scholar] [CrossRef]
- Burnham, P.M.; Hendrixson, D.R. Campylobacter jejuni: Collective Components Promoting a Successful Enteric Lifestyle. Nat. Rev. Microbiol. 2018, 16, 551–565. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Ferrari, R.G.; Conte-Junior, C.A. Virulence Factors in Salmonella Typhimurium: The Sagacity of a Bacterium. Curr. Microbiol. 2019, 76, 762–773. [Google Scholar] [CrossRef]
- Elmi, A.; Nasher, F.; Dorrell, N.; Wren, B.; Gundogdu, O. Revisiting Campylobacter jejuni Virulence and Fitness Factors: Role in Sensing, Adapting, and Competing. Front. Cell. Infect. Microbiol. 2021, 10, 607704. [Google Scholar] [CrossRef] [PubMed]
- Mehat, J.W.; Park, S.F.; van Vliet, A.H.M.; La Ragione, R.M. CapC, a Novel Autotransporter and Virulence Factor of Campylobacter jejuni. Appl. Environ. Microbiol. 2018, 84, e01032-18. [Google Scholar] [CrossRef] [PubMed]
- Mehat, J.W.; La Ragione, R.M.; van Vliet, A.H.M. Campylobacter jejuni and Campylobacter coli Autotransporter Genes Exhibit Lineage-Associated Distribution and Decay. BMC Genom. 2020, 21, 314. [Google Scholar] [CrossRef] [PubMed]
- Gripp, E.; Hlahla, D.; Didelot, X.; Kops, F.; Maurischat, S.; Tedin, K.; Alter, T.; Ellerbroek, L.; Schreiber, K.; Schomburg, D.; et al. Closely Related Campylobacter jejuni Strains from Different Sources Reveal a Generalist Rather than a Specialist Lifestyle. BMC Genom. 2011, 12, 584. [Google Scholar] [CrossRef]
- Colles, F.M.; Maiden, M.C.J. Campylobacter Sequence Typing Databases: Applications and Future Prospects. Microbiology 2012, 158, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.L.; Cribb, D.M.; Bulach, D.M.; Ingle, D.J.; Joensen, K.G.; Nielsen, E.M.; Leekitcharoenphon, P.; Stingl, K.; Kirk, M.D. Campylobacter jejuni ST50, a Pathogen of Global Importance: A Comparative Genomic Analysis of Isolates from Australia, Europe and North America. Zoonoses Public Health 2021, 68, 638–649. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2017, 45, D37–D42. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Ferrari, R.G.; Panzenhagen, P.; Rodrigues, G.L.; Conte-Junior, C.A. Virulence Genes Identification and Characterization Revealed the Presence of the Yersinia High Pathogenicity Island (HPI) in Salmonella from Brazil. Gene 2021, 787, 145646. [Google Scholar] [CrossRef]
- De Fátima RauberWürfel, S.; Jorge, S.; de Oliveira, N.R.; Kremer, F.S.; Sanchez, C.D.; Campos, V.F.; da Silva Pinto, L.; da Silva, W.P.; Dellagostin, O.A. Campylobacter jejuni Isolated from Poultry Meat in Brazil: In Silico Analysis and Genomic Features of Two Strains with Different Phenotypes of Antimicrobial Susceptibility. Mol. Biol. Rep. 2020, 47, 671–681. [Google Scholar] [CrossRef]
- MalbergTetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico Genotyping of Escherichia coli Isolates for Extraintestinal Virulence Genes by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef]
- Rodrigues, G.L.; Panzenhagen, P.; Ferrari, R.G.; dos Santos, A.; Paschoalin, V.M.F.; Conte-Junior, C.A. Frequency of Antimicrobial Resistance Genes in Salmonella From Brazil by in Silico Whole-Genome Sequencing Analysis: An Overview of the Last Four Decades. Front. Microbiol. 2020, 11, 1864. [Google Scholar] [CrossRef]
- Chen, L. VFDB: A Reference Database for Bacterial Virulence Factors. Nucleic Acids Res. 2004, 33, D325–D328. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Saputra, D.; Sicheritz-Ponten, T.; Lund, O.; Svendsen, C.A.; Frimodt-Møller, N.; Aarestrup, F.M. Rapid Whole-Genome Sequencing for Detection and Characterization of Microorganisms Directly from Clinical Samples. J. Clin. Microbiol. 2014, 52, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Larsen, M.V.; Cosentino, S.; Lukjancenko, O.; Saputra, D.; Rasmussen, S.; Hasman, H.; Sicheritz-Pontén, T.; Aarestrup, F.M.; Ussery, D.W.; Lund, O. Benchmarking of Methods for Genomic Taxonomy. J. Clin. Microbiol. 2014, 52, 1529–1539. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Aarestrup, F.M.; Lund, O. Rapid and Precise Alignment of Raw Reads against Redundant Databases with KMA. BMC Bioinform. 2018, 19, 307. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-Access Bacterial Population Genomics: BIGSdb Software, the PubMLST.Org Website and Their Applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Clausen, P.T.L.C.; Zankari, E.; Aarestrup, F.M.; Lund, O. Benchmarking of Methods for Identification of Antimicrobial Resistance Genes in Bacterial Whole Genome Data. J. Antimicrob. Chemother. 2016, 71, 2484–2488. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Helsel, L.O.; Fields, P.I.; Hiett, K.L. Discrimination of Campylobacter jejuni Isolates by Fla Gene Sequencing. J. Clin. Microbiol. 1997, 35, 2810–2814. [Google Scholar] [CrossRef]
- Cody, A.J.; Maiden, M.J.C.; Dingle, K.E. Genetic Diversity and Stability of the PorA Allele as a Genetic Marker in Human Campylobacter Infection. Microbiology 2009, 155, 4145–4154. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Skirrow, M.B. Campylobacter Enteritis: A “New” Disease. Br. Med. J. 1977, 2, 9–11. [Google Scholar] [CrossRef]
- Skirrow, M.B. John McFadyean and the Centenary of the First Isolation of Campylobacter Species. Clin. Infect. Dis. 2006, 43, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Mourkas, E.; Taylor, A.J.; Méric, G.; Bayliss, S.C.; Pascoe, B.; Mageiros, L.; Calland, J.K.; Hitchings, M.D.; Ridley, A.; Vidal, A.; et al. Agricultural Intensification and the Evolution of Host Specialism in the Enteric Pathogen Campylobacter jejuni. Proc. Natl. Acad. Sci. USA 2020, 117, 11018–11028. [Google Scholar] [CrossRef]
- Maiden, M.C.J.; van Rensburg, M.J.J.; Bray, J.E.; Earle, S.G.; Ford, S.A.; Jolley, K.A.; McCarthy, N.D. MLST Revisited: The Gene-by-Gene Approach to Bacterial Genomics. Nat. Rev. Microbiol. 2013, 11, 728–736. [Google Scholar] [CrossRef]
- Dingle, K.E.; Colles, F.M.; Wareing, D.R.A.; Ure, R.; Fox, A.J.; Bolton, F.E.; Bootsma, H.J.; Willems, R.J.L.; Urwin, R.; Maiden, M.C.J. Multilocus Sequence Typing System for Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, S.K.; Cheng, L.; Méric, G.; Haan, C.P.A.; Llarena, A.; Marttinen, P.; Vidal, A.; Ridley, A.; Clifton-Hadley, F.; Connor, T.R.; et al. Cryptic Ecology among Host Generalist Campylobacter jejuni in Domestic Animals. Mol. Ecol. 2014, 23, 2442–2451. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Colles, F.; Richardson, J.; Cody, A.J.; Elson, R.; Lawson, A.; Brick, G.; Meldrum, R.; Little, C.L.; Owen, R.J.; et al. Host Association of Campylobacter Genotypes Transcends Geographic Variation. Appl. Environ. Microbiol. 2010, 76, 5269–5277. [Google Scholar] [CrossRef]
- Dearlove, B.L. Rapid Host Switching in Generalist Campylobacter Strains Erodes the Signal for Tracing Human Infections. ISME J. 2016, 10, 721–729. [Google Scholar] [CrossRef]
- Mouftah, S.F.; Cobo-Díaz, J.F.; Álvarez-Ordóñez, A.; Mousa, A.; Calland, J.K.; Pascoe, B.; Sheppard, S.K.; Elhadidy, M. Stress Resistance Associated with Multi-Host Transmission and Enhanced Biofilm Formation at 42 °C among Hyper-Aerotolerant Generalist Campylobacter jejuni. Food Microbiol. 2021, 95, 103706. [Google Scholar] [CrossRef] [PubMed]
- Karlyshev, A.V.; Champion, O.L.; Churcher, C.; Brisson, J.-R.; Jarrell, H.C.; Gilbert, M.; Brochu, D.; St Michael, F.; Li, J.; Wakarchuk, W.W.; et al. Analysis of Campylobacter jejuni Capsular Loci Reveals Multiple Mechanisms for the Generation of Structural Diversity and the Ability to Form Complex Heptoses: C. jejuni CPS Biosynthesis. Mol. Microbiol. 2004, 55, 90–103. [Google Scholar] [CrossRef] [PubMed]
- McNally, D.J.; Hui, J.P.M.; Aubry, A.J.; Mui, K.K.K.; Guerry, P.; Brisson, J.-R.; Logan, S.M.; Soo, E.C. Functional Characterization of the Flagellar Glycosylation Locus in Campylobacter jejuni 81–176 Using a Focused Metabolomics Approach. J. Biol. Chem. 2006, 281, 18489–18498. [Google Scholar] [CrossRef]
- Dzieciatkowska, M.; Brochu, D.; van Belkum, A.; Heikema, A.P.; Yuki, N.; Houliston, R.S.; Richards, J.C.; Gilbert, M.; Li, J. Mass Spectrometric Analysis of Intact Lipooligosaccharide: Direct Evidence for O-Acetylated Sialic Acids and Discovery of O-Linked Glycine Expressed by Campylobacter jejuni. Biochemistry 2007, 46, 14704–14714. [Google Scholar] [CrossRef]
- Karlyshev, A.V.; Linton, D.; Gregson, N.A.; Lastovica, A.J.; Wren, B.W. Genetic and Biochemical Evidence of a Campylobacter jejuni Capsular Polysaccharide That Accounts for Penner Serotype Specificity: Genetics and Biochemistry of C. jejuni LPS Biosynthesis. Mol. Microbiol. 2002, 35, 529–541. [Google Scholar] [CrossRef] [PubMed]
- Bacon, D.J.; Szymanski, C.M.; Burr, D.H.; Silver, R.P.; Alm, R.A.; Guerry, P. A Phase-Variable Capsule Is Involved in Virulence of Campylobacter jejuni 81-176: C. jejuni Capsular Phase Variation. Mol. Microbiol. 2001, 40, 769–777. [Google Scholar] [CrossRef] [PubMed]
- Sahin, O.; Terhorst, S.A.; Burrough, E.R.; Shen, Z.; Wu, Z.; Dai, L.; Tang, Y.; Plummer, P.J.; Ji, J.; Yaeger, M.J.; et al. Key Role of Capsular Polysaccharide in the Induction of Systemic Infection and Abortion by Hypervirulent Campylobacter jejuni. Infect. Immun. 2017, 85, e00001-17. [Google Scholar] [CrossRef]
- Stahl, M.; Ries, J.; Vermeulen, J.; Yang, H.; Sham, H.P.; Crowley, S.M.; Badayeva, Y.; Turvey, S.E.; Gaynor, E.C.; Li, X.; et al. A Novel Mouse Model of Campylobacter jejuni Gastroenteritis Reveals Key Pro-Inflammatory and Tissue Protective Roles for Toll-like Receptor Signaling during Infection. PLoS Pathog. 2014, 10, e1004264. [Google Scholar] [CrossRef]
- Christie, P.J. Type IV Secretion: Intercellular Transfer of Macromolecules by Systems Ancestrally Related to Conjugation Machines. Mol. Microbiol. 2001, 40, 294–305. [Google Scholar] [CrossRef]
- Tracz, D.M.; Keelan, M.; Ahmed-Bentley, J.; Gibreel, A.; Kowalewska-Grochowska, K.; Taylor, D.E. PVir and Bloody Diarrhea in Campylobacter jejuni Enteritis. Emerg. Infect. Dis. 2005, 11, 839–843. [Google Scholar] [CrossRef]
- Bacon, D.J.; Alm, R.A.; Burr, D.H.; Hu, L.; Kopecko, D.J.; Ewing, C.P.; Trust, T.J.; Guerry, P. Involvement of a Plasmid in Virulence of Campylobacter jejuni 81-176. Infect. Immun. 2000, 68, 4384–4390. [Google Scholar] [CrossRef]
- Louwen, R.P.L.; van Belkum, A.; Wagenaar, J.A.; Doorduyn, Y.; Achterberg, R.; Endtz, H.P. Lack of Association between the Presence of the PVir Plasmid and Bloody Diarrhea in Campylobacter jejuni Enteritis. J. Clin. Microbiol. 2006, 44, 1867–1868. [Google Scholar] [CrossRef] [PubMed]
- Marasini, D.; Fakhr, M. Exploring PFGE for Detecting Large Plasmids in Campylobacter jejuni and Campylobacter coli Isolated from Various Retail Meats. Pathogens 2014, 3, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Torrens, Y.; Miró, E.; Guirado, P.; Llovet, T.; Muñoz, C.; Cerdà-Cuéllar, M.; Madrid, C.; Balsalobre, C.; Navarro, F. Population Structure, Antimicrobial Resistance, and Virulence-Associated Genes in Campylobacter jejuni Isolated from Three Ecological Niches: Gastroenteritis Patients, Broilers, and Wild Birds. Front. Microbiol. 2018, 9, 1676. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Ott, R.; Pohl, S.; Burghard, S.; Weig, M.; Groß, U. Identification and Characterization of a Major Subgroup of Conjugative Campylobacter jejuni Plasmids. J. Infect. 2005, 50, 12–21. [Google Scholar] [CrossRef]
- Nachamkin, I.; Bohachick, K.; Patton, C.M. Flagellin Gene Typing of Campylobacter jejuni by Restriction Fragment Length Polymorphism Analysis. J. Clin. Microbiol. 1993, 31, 1531–1536. [Google Scholar] [CrossRef]
- Harrington, C.S.; Thomson-Carter, F.M.; Carter, P.E. Evidence for Recombination in the Flagellin Locus of Campylobacter jejuni: Implications for the Flagellin Gene Typing Scheme. J. Clin. Microbiol. 1997, 35, 2386–2392. [Google Scholar] [CrossRef]
- Wassenaar, T.M.; Bleumink-Pluym, N.M.; van der Zeijst, B.A. Inactivation of Campylobacter jejuni Flagellin Genes by Homologous Recombination Demonstrates That flaA but Not flaB Is Required for Invasion. EMBO J. 1991, 10, 2055–2061. [Google Scholar] [CrossRef]
- Wirz, S.E.; Overesch, G.; Kuhnert, P.; Korczak, B.M. Genotype and Antibiotic Resistance Analyses of Campylobacter Isolates from Ceca and Carcasses of Slaughtered Broiler Flocks. Appl. Environ. Microbiol. 2010, 76, 6377–6386. [Google Scholar] [CrossRef]
- Magnússon, S.H.; Guðmundsdóttir, S.; Reynisson, E.; Rúnarsson, Á.R.; Harðardóttir, H.; Gunnarson, E.; Georgsson, F.; Reiersen, J.; Marteinsson, V.T. Comparison of Campylobacter jejuni Isolates from Human, Food, Veterinary and Environmental Sources in Iceland Using PFGE, MLST and Fla-SVR Sequencing. J. Appl. Microbiol. 2011, 111, 971–981. [Google Scholar] [CrossRef]
- Gomes, C.N.; Souza, R.A.; Passaglia, J.; Duque, S.S.; Medeiros, M.I.C.; Falcão, J.P. Genotyping of Campylobacter coli Strains Isolated in Brazil Suggests Possible Contamination amongst Environmental, Human, Animal and Food Sources. J. Med. Microbiol. 2016, 65, 80–90. [Google Scholar] [CrossRef]
- Talukder, K.A.; Aslam, M.; Islam, Z.; Azmi, I.J.; Dutta, D.K.; Hossain, S.; Nur-E-Kamal, A.; Nair, G.B.; Cravioto, A.; Sack, D.A.; et al. Prevalence of Virulence Genes and Cytolethal Distending Toxin Production in Campylobacter jejuni Isolates from Diarrheal Patients in Bangladesh. J. Clin. Microbiol. 2008, 46, 1485–1488. [Google Scholar] [CrossRef]
- Wieczorek, K.; Szewczyk, R.; Osek, J. Prevalence, Antimicrobial Resistance, and Molecular Characterization of Campylobacter jejuni and C. coli Isolated from Retail Raw Meat in Poland. Vet. Med. 2012, 57, 293–299. [Google Scholar] [CrossRef]
- Casabonne, C.; Gonzalez, A.; Aquili, V.; Subils, T.; Balague, C. Prevalence of Seven Virulence Genes of Campylobacter jejuni Isolated from Patients with Diarrhea in Rosario, Argentina. Int. J. Infect. 2016, 3, e37727. [Google Scholar] [CrossRef]
- Hanning, I.; Biswas, D.; Herrera, P.; Roesler, M.; Ricke, S.C. Prevalence and Characterization of Campylobacter jejuni Isolated from Pasture Flock Poultry. J. Food Sci. 2010, 75, M496–M502. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Jang, S.S.; Choo, E.; Heu, S.; Ryu, S. Prevalence, Genetic Diversity, and Antibiotic Resistance Patterns of Campylobacter jejuni from Retail Raw Chickens in Korea. Int. J. Food Microbiol. 2007, 114, 50–59. [Google Scholar] [CrossRef]
- Melo, R.T.; Grazziotin, A.L.; Júnior, E.C.V.; Prado, R.R.; Mendonça, E.P.; Monteiro, G.P.; Peres, P.A.B.M.; Rossi, D.A. Evolution of Campylobacter jejuni of Poultry Origin in Brazil. Food Microbiol. 2019, 82, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Rossler, E.; Olivero, C.; Soto, L.P.; Frizzo, L.S.; Zimmermann, J.; Rosmini, M.R.; Sequeira, G.J.; Signorini, M.L.; Zbrun, M.V. Prevalence, Genotypic Diversity and Detection of Virulence Genes in Thermotolerant Campylobacter at Different Stages of the Poultry Meat Supply Chain. Int. J. Food Microbiol. 2020, 326, 108641. [Google Scholar] [CrossRef]
- Abd El-Hamid, M.I.; Abd El-Aziz, N.K.; Samir, M.; El-Naenaeey, E.Y.; Abo Remela, E.M.; Mosbah, R.A.; Bendary, M.M. Genetic Diversity of Campylobacter jejuni Isolated From Avian and Human Sources in Egypt. Front. Microbiol. 2019, 10, 2353. [Google Scholar] [CrossRef]
- Ammar, A.M.; El-Naenaeey, E.-S.Y.; El-Malt, R.M.S.; El-Gedawy, A.A.; Khalifa, E.; Elnahriry, S.S.; Abd El-Hamid, M.I. Prevalence, Antimicrobial Susceptibility, Virulence and Genotyping of Campylobacter jejuni with a Special Reference to the Anti-Virulence Potential of Eugenol and β-Resorcylic Acid on Some Multi-Drug Resistant Isolates in Egypt. Animals 2020, 11, 3. [Google Scholar] [CrossRef]
- Han, X.; Guan, X.; Zeng, H.; Li, J.; Huang, X.; Wen, Y.; Zhao, Q.; Huang, X.; Yan, Q.; Huang, Y.; et al. Prevalence, Antimicrobial Resistance Profiles and Virulence-Associated Genes of Thermophilic Campylobacter spp. Isolated from Ducks in a Chinese Slaughterhouse. Food Control 2019, 104, 157–166. [Google Scholar] [CrossRef]
- Bravo, V.; Katz, A.; Porte, L.; Weitzel, T.; Varela, C.; Gonzalez-Escalona, N.; Blondel, C.J. Genomic Analysis of the Diversity, Antimicrobial Resistance and Virulence Potential of Clinical Campylobacter jejuni and Campylobacter coli Strains from Chile. PLoS Negl. Trop. Dis. 2021, 15, e0009207. [Google Scholar] [CrossRef]
- Truccollo, B.; Whyte, P.; Burgess, C.; Bolton, D. Genetic Characterisation of a Subset of Campylobacter jejuni Isolates from Clinical and Poultry Sources in Ireland. PLoS ONE 2021, 16, e0246843. [Google Scholar] [CrossRef]
- Fiedoruk, K.; Daniluk, T.; Rozkiewicz, D.; Oldak, E.; Prasad, S.; Swiecicka, I. Whole-Genome Comparative Analysis of Campylobacter jejuni Strains Isolated from Patients with Diarrhea in Northeastern Poland. Gut Pathog. 2019, 11, 32. [Google Scholar] [CrossRef]
- Cao, H.; Xu, H.; Ning, C.; Xiang, L.; Ren, Q.; Zhang, T.; Zhang, Y.; Gao, R. Multi-Omics Approach Reveals the Potential Core Vaccine Targets for the Emerging Foodborne Pathogen Campylobacter jejuni. Front. Microbiol. 2021, 12, 665858. [Google Scholar] [CrossRef]
- Alm, R.A.; Guerry, P.; Trust, T.J. Significance of Duplicated Flagellin Genes in Campylobacter. J. Mol. Biol. 1993, 230, 359–363. [Google Scholar] [CrossRef][Green Version]
- Santoyo, G.; Romero, D. Gene Conversion and Concerted Evolution in Bacterial Genomes. FEMS Microbiol. Rev. 2005, 29, 169–183. [Google Scholar] [CrossRef] [PubMed]
- Meinersmann, R.J.; Hiett, K.L. Concerted Evolution of Duplicate Fla Genes in Campylobacter. Microbiology 2000, 146, 2283–2290. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Magana, M.; Chatzipanagiotou, S.; Burriel, A.R.; Ioannidis, A. Inquiring into the Gaps of Campylobacter Surveillance Methods. Vet. Sci. 2017, 4, 36. [Google Scholar] [CrossRef] [PubMed]
- Linton, D.; Gilbert, M.; Hitchen, P.G.; Dell, A.; Morris, H.R.; Wakarchuk, W.W.; Gregson, N.A.; Wren, B.W. Phase Variation of a β-1,3 Galactosyltransferase Involved in Generation of the Ganglioside GM1-like Lipo-Oligosaccharide of Campylobacter jejuni: C. jejuni LOS Phase Variation. Mol. Microbiol. 2002, 37, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Koga, M.; Gilbert, M.; Takahashi, M.; Li, J.; Koike, S.; Hirata, K.; Yuki, N. Comprehensive Analysis of Bacterial Risk Factors for the Development of Guillain-Barré Syndrome after Campylobacter jejuni Enteritis. J. Infect. Dis. 2006, 193, 547–555. [Google Scholar] [CrossRef]
- Guirado, P.; Paytubi, S.; Miró, E.; Iglesias-Torrens, Y.; Navarro, F.; Cerdà-Cuéllar, M.; Stephan-Otto Attolini, C.; Balsalobre, C.; Madrid, C. Differential Distribution of the WlaN and CgtB Genes, Associated with Guillain-Barré Syndrome, in Campylobacter jejuni Isolates from Humans, Broiler Chickens, and Wild Birds. Microorganisms 2020, 8, 325. [Google Scholar] [CrossRef]
- Datta, S.; Niwa, H.; Itoh, K. Prevalence of 11 Pathogenic Genes of Campylobacter jejuni by PCR in Strains Isolated from Humans, Poultry Meat and Broiler and Bovine Faeces. J. Med. Microbiol. 2003, 52, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Koolman, L.; Whyte, P.; Burgess, C.; Bolton, D. Distribution of Virulence-Associated Genes in a Selection of Campylobacter Isolates. Foodborne Pathog. Dis. 2015, 12, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Arguello, Y.M.; Perdoncini, G.; Rodrigues, L.B.; Ruschel dos Santos, L.; Apellanis Borges, K.; QuediFurian, T.; Pippi Salle, C.T.; de Souza Moraes, H.L.; Pereira Gomes, M.J.; Pinheiro do Nascimento, V. Identification of Pathogenic Genes in Campylobacter jejuni Isolated from Broiler Carcasses and Broiler Slaughterhouses. Sci. Rep. 2021, 11, 4588. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Lee, M.Y.; Kim, S.J.; Jeon, S.-E.; Cha, I.; Hong, S.; Chung, G.T.; Huh, M.-J.; Kang, Y.-H.; Yoo, C.-K.; et al. High-Level Ciprofloxacin-Resistant Campylobacter jejuni Isolates Circulating in Humans and Animals in Incheon, Republic of Korea. Zoonoses Public Health 2016, 63, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-S.; Park, C.; Kim, Y.-J. Role of FlgA for Flagellar Biosynthesis and Biofilm Formation of Campylobacter jejuni NCTC11168. J. Microbiol. Biotechnol. 2015, 25, 1871–1879. [Google Scholar] [CrossRef]
- Neal-McKinney, J.M.; Konkel, M.E. The Campylobacter jejuni CiaC Virulence Protein Is Secreted from the Flagellum and Delivered to the Cytosol of Host Cells. Front. Cell. Infect. Microbiol. 2012, 2, 31. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).