Screening of Virulence-Related Transcriptional Regulators in Streptococcus suis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. In Vivo-Induced Experiments
2.3. Generation of Isogenic Gene Deletion Mutants and Complementation Strains
2.4. Bacterial Growth Curves
2.5. Transmission Electron Microscopy
2.6. Experimental Infections of C57BL/6 Mice
2.7. In Vitro Bacterial Survival in the Presence of Mouse Whole Blood
2.8. Experimental Infections In Vivo
2.9. RNA-Seq Analysis
2.10. Quantitative RT-PCR (qRT-PCR)
2.11. Statistical Analysis
3. Results
3.1. Screening for Transcription Regulators by In vivo Induction
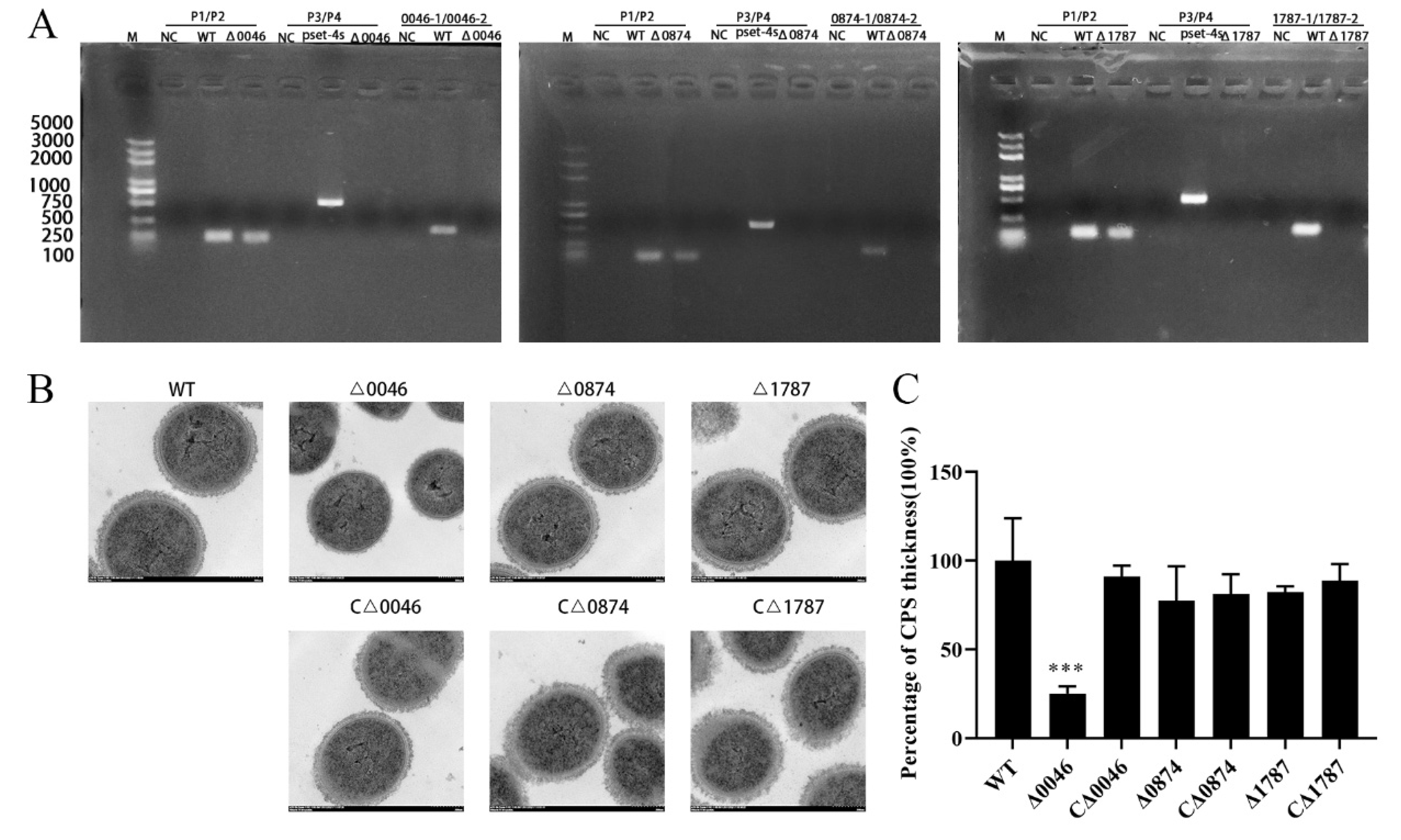
3.2. Construction and Characterization of Mutants and Complementary Strains
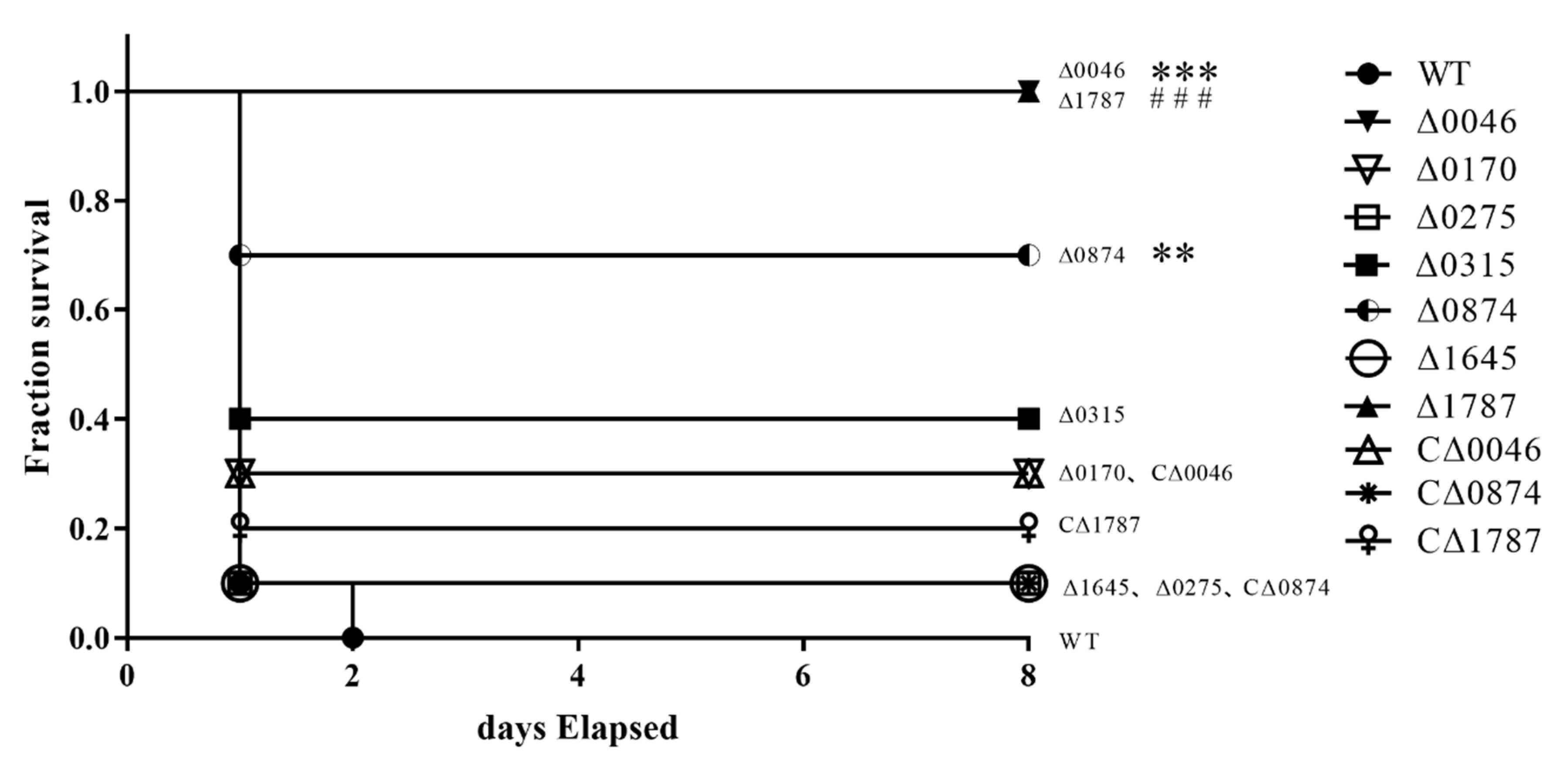
3.3. ComR, sitR and sxvR Deletions Reduce S.suis Virulence in Mice
3.4. The Domains of comR, sitR, and sxvR Indicate That These Three Proteins Are Transcription Regulators
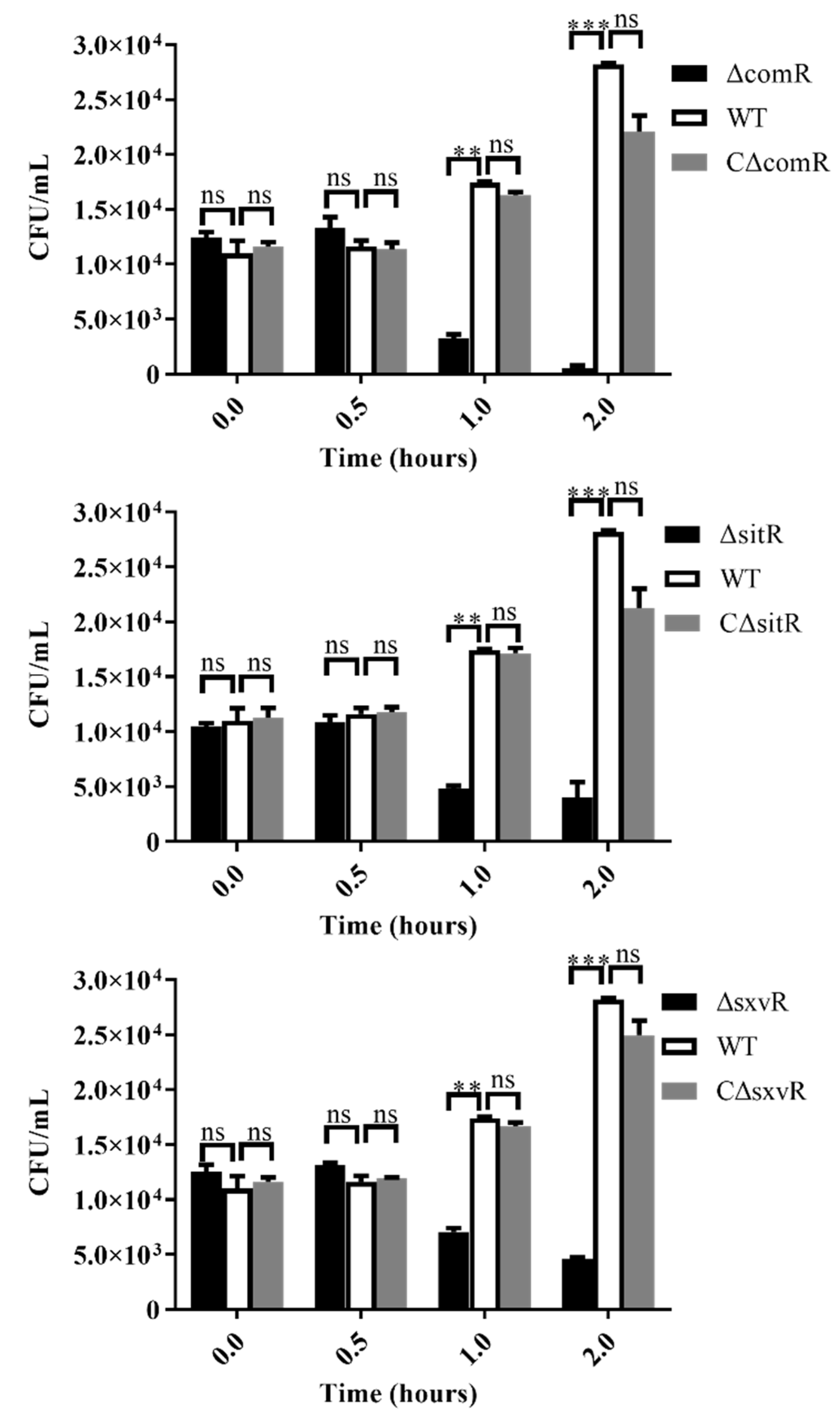
3.5. Survival Ability of Mutants in Whole Blood is Diminished
3.6. ΔcomR, ΔsitR, and ΔsxvR Mutants Exhibit Decreased Pro-Inflammatory Ability and Reduced Bacterial Loads in Mice
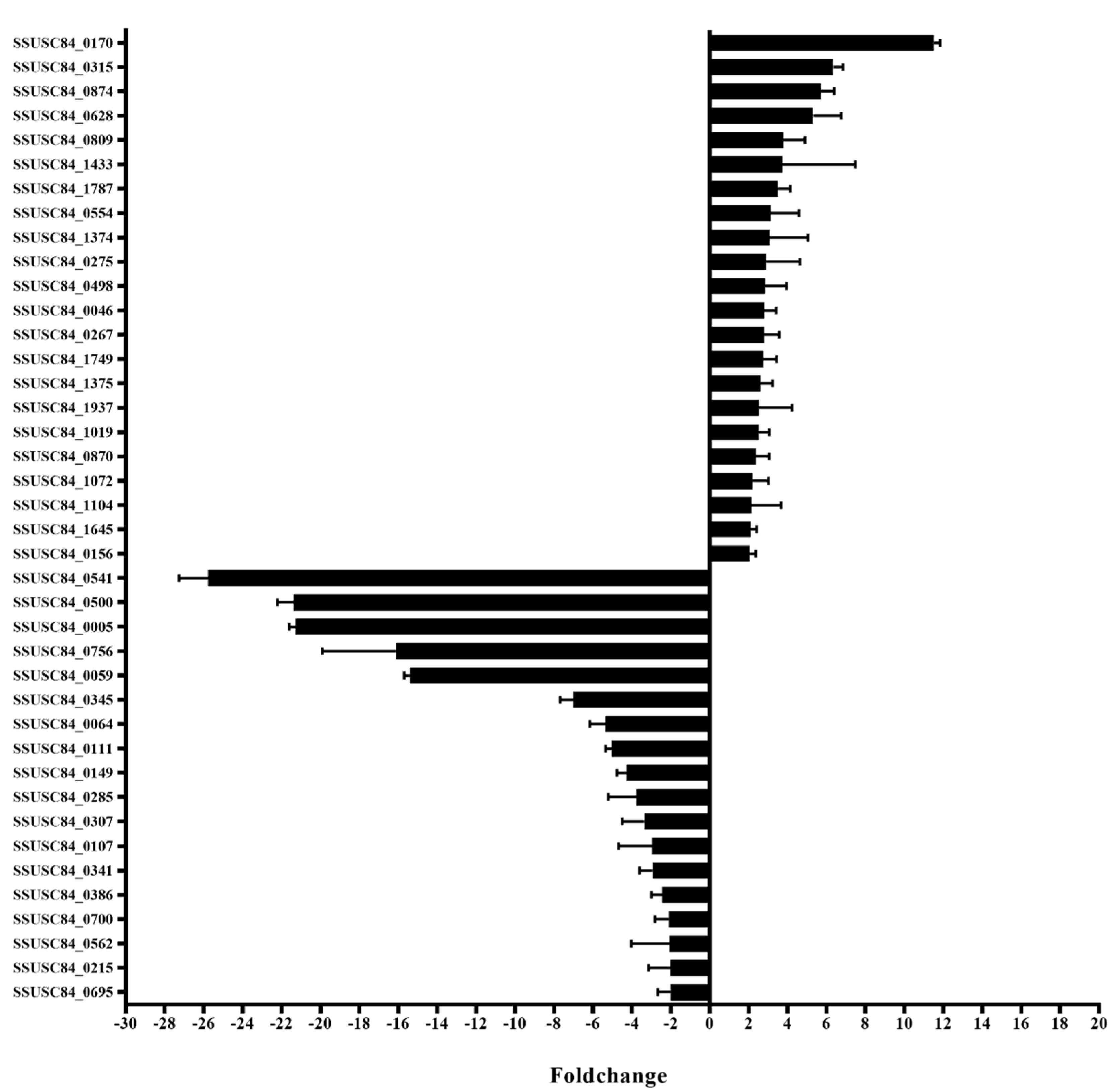
3.7. Differentially Expressed Genes (DEGs) in Mutant Strains
3.8. Functional Analysis of Virulence-Associated Genes (VAGs) and DEGs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zheng, F.; Ji, H.; Cao, M.; Wang, C.; Feng, Y.; Li, M.; Pan, X.; Wang, J.; Qin, Y.; Hu, F.; et al. Contribution of the Rgg transcription regulator to metabolism and virulence of Streptococcus suis serotype 2. Infect. Immun. 2011, 79, 1319–1328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lin, L.; Fu, L.; Han, L.; Zhang, A. Neutrophil extracellular Traps play an important role in clearance of Streptococcus suis in vivo. Microbiol. Immunol. 2016, 60, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Filippitzi, M.E.; Goumperis, T.; Robinson, T.; Saegerman, C. Microbiological zoonotic emerging risks, transmitted between livestock animals and humans (2007–2015). Transbound. Emerg. Dis. 2017, 64, 1059–1070. [Google Scholar] [CrossRef]
- Dutkiewicz, J.; Zajac, V.; Sroka, J.; Wasinski, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wojcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II-Pathogenesis. Ann. Agric. Environ. Med. 2018, 25, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Tien le, H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA-DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Fittipaldi, N.; Segura, M.; Grenier, D.; Gottschalk, M. Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 2012, 7, 259–279. [Google Scholar] [CrossRef]
- Zhao, J.; Pan, S.; Lin, L.; Fu, L.; Yang, C.; Xu, Z.; Wei, Y.; Jin, M.; Zhang, A. Streptococcus suis serotype 2 strains can induce the formation of neutrophil extracellular traps and evade trapping. FEMS Microbiol. Lett. 2015, 362, fnv022. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis virulence factors: Are they all really critical? Trends Microbiol. 2017, 25, 585–599. [Google Scholar] [CrossRef]
- Zhang, Y.; Zong, B.; Wang, X.; Zhu, Y.; Hu, L.; Li, P.; Zhang, A.; Chen, H.; Liu, M.; Tan, C. Fisetin Lowers Streptococcus suis serotype 2 pathogenicity in mice by inhibiting the hemolytic activity of suilysin. Front. Microbiol. 2018, 9, 1723. [Google Scholar] [CrossRef]
- Gottschalk, M.G.; Lacouture, S.; Dubreuil, J.D. Characterization of Streptococcus suis capsular type 2 haemolysin. Microbiology 1995, 141, 189–195. [Google Scholar] [CrossRef]
- de Buhr, N.; Neumann, A.; Jerjomiceva, N.; von Kockritz-Blickwede, M.; Baums, C.G. Streptococcus suis DNase SsnA contributes to degradation of neutrophil extracellular traps (NETs) and evasion of NET-mediated antimicrobial activity. Microbiology 2014, 160, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Yu, J.; Xu, Z.; Liu, L.; Song, Y.; Sun, X.; Zhang, A.; Jin, M. HP1330 contributes to Streptococcus suis virulence by inducing toll-like receptor 2- and ERK1/2-dependent pro-inflammatory responses and influencing in vivo S. suis loads. Front. Immunol. 2017, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef] [PubMed]
- Houde, M.; Gottschalk, M.; Gagnon, F.; Van Calsteren, M.R.; Segura, M. Streptococcus suis capsular polysaccharide inhibits phagocytosis through destabilization of lipid microdomains and prevents lactosylceramide-dependent recognition. Infect. Immun. 2012, 80, 506–517. [Google Scholar] [CrossRef]
- Benga, L.; Goethe, R.; Rohde, M.; Valentin-Weigand, P. Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell. Microbiol. 2004, 6, 867–881. [Google Scholar] [CrossRef]
- Tan, C.; Liu, M.; Jin, M.; Liu, J.; Chen, Y.; Wu, T.; Fu, T.; Bei, W.; Chen, H. The key virulence-associated genes of Streptococcus suis type 2 are upregulated and differentially expressed in vivo. FEMS Microbiol. Lett. 2008, 278, 108–114. [Google Scholar] [CrossRef]
- Beier, D.; Gross, R. Regulation of bacterial virulence by two-component systems. Curr. Opin. Microbiol. 2006, 9, 143–152. [Google Scholar] [CrossRef]
- Han, H.; Liu, C.; Wang, Q.; Xuan, C.; Zheng, B.; Tang, J.; Yan, J.; Zhang, J.; Li, M.; Cheng, H.; et al. The two-component system Ihk/Irr contributes to the virulence of Streptococcus suis serotype 2 strain 05ZYH33 through alteration of the bacterial cell metabolism. Microbiology 2012, 158, 1852–1866. [Google Scholar] [CrossRef]
- Chen, W.J.; Zhu, T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004, 9, 591–596. [Google Scholar] [CrossRef]
- Camejo, A.; Buchrieser, C.; Couve, E.; Carvalho, F.; Reis, O.; Ferreira, P.; Sousa, S.; Cossart, P.; Cabanes, D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009, 5, e1000449. [Google Scholar] [CrossRef]
- Milohanic, E.; Glaser, P.; Coppee, J.Y.; Frangeul, L.; Vega, Y.; Vazquez-Boland, J.A.; Kunst, F.; Cossart, P.; Buchrieser, C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 2003, 47, 1613–1625. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.J.; Pridmore, A.C.; Lee, M.E.; Read, R.C. Induction of pro-inflammatory cytokine release by human macrophages during exposure of Streptococcus pneumoniae to penicillin is influenced by minimum inhibitory concentration ratio. Int. J. Antimicrob. Agents 2005, 26, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, D.; Osaki, M.; Sekizaki, T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 2001, 46, 140–148. [Google Scholar] [CrossRef]
- Warrens, A.N.; Jones, M.D.; Lechler, R.I. Splicing by overlap extension by PCR using asymmetric amplification: An improved technique for the generation of hybrid proteins of immunological interest. Gene 1997, 186, 29–35. [Google Scholar] [CrossRef]
- Gao, T.; Tan, M.; Liu, W.; Zhang, C.; Zhang, T.; Zheng, L.; Zhu, J.; Li, L.; Zhou, R. GidA, a tRNA modification enzyme, contributes to the growth, and virulence of Streptococcus suis serotype 2. Front. Cell. Infect. Microbiol. 2016, 6, 44. [Google Scholar] [CrossRef] [PubMed]
- Pian, Y.; Gan, S.; Wang, S.; Guo, J.; Wang, P.; Zheng, Y.; Cai, X.; Jiang, Y.; Yuan, Y. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 2012, 80, 2402–2413. [Google Scholar] [CrossRef]
- Liu, L.; Unit of Animal Infectious Diseases, National Key Laboratory of Agricultural Microbiology, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, China. Unpublished work. 2020.
- Barragan, M.J.; Blazquez, B.; Zamarro, M.T.; Mancheno, J.M.; Garcia, J.L.; Diaz, E.; Carmona, M. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 2005, 280, 10683–10694. [Google Scholar] [CrossRef]
- Zhang, C.; Sun, W.; Tan, M.; Dong, M.; Liu, W.; Gao, T.; Li, L.; Xu, Z.; Zhou, R. The eukaryote-like serine/threonine kinase STK regulates the growth and metabolism of zoonotic Streptococcus suis. Front. Cell. Infect. Microbiol. 2017, 7, 66. [Google Scholar] [CrossRef]
- Zaccaria, E.; van Baarlen, P.; de Greeff, A.; Morrison, D.A.; Smith, H.; Wells, J.M. Control of competence for DNA transformation in Streptococcus suis by genetically transferable pherotypes. PLoS ONE 2014, 9, e99394. [Google Scholar] [CrossRef]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the eye of the cytokine storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef]
- Dominguez-Punaro, M.C.; Segura, M.; Plante, M.M.; Lacouture, S.; Rivest, S.; Gottschalk, M. Streptococcus suis serotype 2, an important swine and human pathogen, induces strong systemic and cerebral inflammatory responses in a mouse model of infection. J. Immunol. 2007, 179, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Auger, J.P.; Fittipaldi, N.; Benoit-Biancamano, M.O.; Segura, M.; Gottschalk, M. virulence studies of different sequence types and geographical origins of Streptococcus suis serotype 2 in a mouse model of infection. Pathogens 2016, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Mashburn-Warren, L.; Morrison, D.A.; Federle, M.J. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 2010, 78, 589–606. [Google Scholar] [CrossRef] [PubMed]
- Zaccaria, E.; Wels, M.; van Baarlen, P.; Wells, J.M. Temporal regulation of the transformasome and competence development in Streptococcus suis. Front. Microbiol. 2016, 7, 1922. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Junges, R.; Amdal, H.A.; Chen, T.; Morrison, D.A.; Petersen, F.C. A positive feedback loop mediated by Sigma X enhances expression of the streptococcal regulator ComR. Sci. Rep. 2017, 7, 5984. [Google Scholar] [CrossRef]
- Zhong, X.; Zhang, Y.; Zhu, Y.; Dong, W.; Ma, J.; Pan, Z.; Roy, S.; Lu, C.; Yao, H. The two-component signaling system VraSRSS is critical for multidrug resistance and full virulence in Streptococcus suis serotype 2. Infect. Immun. 2018, 86, e00096-18. [Google Scholar] [CrossRef]
- Smith, H.E.; Damman, M.; van der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756. [Google Scholar] [CrossRef]
- Li, M.; Cai, R.J.; Li, C.L.; Song, S.; Li, Y.; Jiang, Z.Y.; Yang, D.X. Deletion of ssnA attenuates the pathogenicity of Streptococcus suis and confers protection against Serovar 2 strain challenge. PLoS ONE 2017, 12, e0169791. [Google Scholar] [CrossRef]
- Li, M.; Wang, C.; Feng, Y.; Pan, X.; Cheng, G.; Wang, J.; Ge, J.; Zheng, F.; Cao, M.; Dong, Y.; et al. SalK/SalR, a two-component signal transduction system, is essential for full virulence of highly invasive Streptococcus suis serotype 2. PLoS ONE 2008, 3, e2080. [Google Scholar] [CrossRef]
- Tan, M.F.; Gao, T.; Liu, W.Q.; Zhang, C.Y.; Yang, X.; Zhu, J.W.; Teng, M.Y.; Li, L.; Zhou, R. MsmK, an ATPase, contributes to utilization of multiple carbohydrates and host colonization of Streptococcus suis. PLoS ONE 2015, 10, e0130792. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Y.; Zhao, Y.; Zhao, H.; Bai, J.; Chen, J.; Zhou, Y.; Wang, C.; Li, Y. Sub-MIC tylosin inhibits Streptococcus suis biofilm formation and results in differential protein expression. Front. Microbiol. 2016, 7, 384. [Google Scholar] [CrossRef] [PubMed]
- Laurenceau, R.; Krasteva, P.V.; Diallo, A.; Ouarti, S.; Duchateau, M.; Malosse, C.; Chamot-Rooke, J.; Fronzes, R. Conserved Streptococcus pneumoniae spirosomes suggest a single type of transformation pilus in competence. PLoS Pathog. 2015, 11, e1004835. [Google Scholar] [CrossRef]
- Yu, Y.; Qian, Y.; Du, D.; Xu, C.; Dai, C.; Li, Q.; Liu, H.; Shao, J.; Wu, Z.; Zhang, W. SBP2 plays an important role in the virulence changes of different artificial mutants of Streptococcus suis. Mol. Biosyst. 2016, 12, 1948–1962. [Google Scholar] [CrossRef] [PubMed]
- Fulde, M.; Willenborg, J.; de Greeff, A.; Benga, L.; Smith, H.E.; Valentin-Weigand, P.; Goethe, R. ArgR is an essential local transcriptional regulator of the arcABC operon in Streptococcus suis and is crucial for biological fitness in an acidic environment. Microbiology 2011, 157, 572–582. [Google Scholar] [CrossRef] [PubMed]
- Haikarainen, T.; Thanassoulas, A.; Stavros, P.; Nounesis, G.; Haataja, S.; Papageorgiou, A.C. Structural and thermodynamic characterization of metal ion binding in Streptococcus suis Dpr. J. Mol. Biol. 2011, 405, 448–460. [Google Scholar] [CrossRef]
- Norris, M.H.; Rahman Khan, M.S.; Schweizer, H.P.; Tuanyok, A. An avirulent Burkholderia pseudomallei purM strain with atypical type B LPS: Expansion of the toolkit for biosafe studies of melioidosis. BMC Microbiol. 2017, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- de Buhr, N.; Stehr, M.; Neumann, A.; Naim, H.Y.; Valentin-Weigand, P.; von Kockritz-Blickwede, M.; Baums, C.G. Identification of a novel DNase of Streptococcus suis (EndAsuis) important for neutrophil extracellular trap degradation during exponential growth. Microbiology 2015, 161, 838–850. [Google Scholar] [CrossRef]
- Zheng, C.; Ren, S.; Xu, J.; Zhao, X.; Shi, G.; Wu, J.; Li, J.; Chen, H.; Bei, W. Contribution of NADH oxidase to oxidative stress tolerance and virulence of Streptococcus suis serotype 2. Virulence 2017, 8, 53–65. [Google Scholar] [CrossRef]

| Pathway | ID | Input Number | Background Number | p-Value |
|---|---|---|---|---|
| ΔcomR vs. WT | ||||
| ABC transporters | sss02010 | 28 | 75 | 5.76 × 10−13 |
| Tyrosine metabolism | sss00350 | 3 | 5 | 0.009213 |
| Biotin metabolism | sss00780 | 3 | 5 | 0.009213 |
| Fatty acid metabolism | sss01212 | 4 | 12 | 0.012373 |
| Fatty acid biosynthesis | sss00061 | 4 | 13 | 0.015451 |
| Arginine and proline metabolism | sss00330 | 3 | 12 | 0.055231 |
| ΔsitR vs. WT | ||||
| Purine metabolism | sss00230 | 8 | 51 | 1.487 × 10−6 |
| Biosynthesis of secondary metabolites | sss01110 | 7 | 152 | 0.0087595 |
| One carbon pool by folate | sss00670 | 2 | 10 | 0.014002 |
| ΔsxvR vs. WT | ||||
| Purine metabolism | sss00230 | 6 | 51 | 0.0006233 |
| One carbon pool by folate | sss00670 | 2 | 10 | 0.0216202 |
| Biosynthesis of secondary metabolites | sss01110 | 7 | 152 | 0.030136 |
| Alanine, aspartate and glutamate metabolism | sss00250 | 2 | 15 | 0.0419204 |
| Gene_id | Genename | Foldchange (log2) | Padj | Description |
|---|---|---|---|---|
| ΔcomR vs. WT | ||||
| SSUSC84_0504 | cps2E | −0.7278 | 6.7712 × 10−47 | putative galactosyl transferase |
| SSUSC84_0505 | cps2F | −0.75585 | 4.5179 × 10−50 | putative rhamnosyl transferase |
| SSUSC84_0506 | cps2G | −0.9129 | 7.2375 × 10−72 | putative glycosyl transferase |
| SSUSC84_0507 | cps2H | −0.96001 | 1.2376 × 10−79 | putative membrane protein |
| SSUSC84_0509 | cps2J | −1.1937 | 4.035 × 10−115 | putative glycosyltransferase |
| SSUSC84_0510 | cps2K | −1.0932 | 1.3498 × 10−96 | putative glycosyl transferase |
| SSUSC84_1270 | ftsX | −0.8355 | 1.9701 × 10−38 | putative cell division protein |
| SSUSC84_0648 | nox | −0.99753 | 1.7347 × 10−09 | NADH oxidase |
| SSUSC84_1782 | ssnA | −1.4199 | 1.0774 × 10−85 | surface-anchored DNA nuclease |
| SSUSC84_1234 | sao | −1.5613 | 1.3274 × 10−210 | putative surface-anchored protein |
| SSUSC84_0520 | neuA | −1.7595 | 1.6735 × 10−254 | N-acylneuraminate cytidylyltransferase |
| SSUSC84_0517 | neuB | −1.5933 | 3.3455 × 10−195 | putative N-acetylneuraminic acid synthase |
| SSUSC84_0518 | neuC | −1.6449 | 2.7075 × 10−214 | putative UDP-N-acetylglucosamine 2-epimerase |
| SSUSC84_1502 | ofs | −2.3267 | 0 | serum opacity factor |
| SSUSC84_1907 | SBP2 | −2.0237 | 3.44 × 10−236 | putative accessory pilus subunit |
| SSUSC84_0849 | SalR | −0.59813 | 0.00049797 | response regulator protein |
| SSUSC84_1566 | csrR | −0.59471 | 1.5809 × 10−17 | response regulator protein |
| ΔsitR vs. WT | ||||
| SSUSC84_1927 | argR | −0.61878 | 0.00013602 | putative arginine repressor |
| SSUSC84_1526 | dpr | −0.74885 | 0.020193 | Dps-like peroxide resistance protein Dpr |
| SSUSC84_0031 | purD | −0.68965 | 0.00043377 | phosphoribosylamine-glycine ligase |
| ΔsxvR vs. WT | ||||
| SSUSC84_0648 | nox | −0.91682 | 2.2094 × 10−7 | NADH oxidase |
| SSUSC84_1782 | ssnA | −0.71857 | 3.5286 × 10−25 | surface-anchored DNA nuclease |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Zhang, Q.; Xu, Z.; Chen, B.; Zhang, A.; Sun, X.; Jin, M. Screening of Virulence-Related Transcriptional Regulators in Streptococcus suis. Genes 2020, 11, 972. https://doi.org/10.3390/genes11090972
Liu L, Zhang Q, Xu Z, Chen B, Zhang A, Sun X, Jin M. Screening of Virulence-Related Transcriptional Regulators in Streptococcus suis. Genes. 2020; 11(9):972. https://doi.org/10.3390/genes11090972
Chicago/Turabian StyleLiu, Liang, Qiang Zhang, Zhongmin Xu, Bo Chen, Anding Zhang, Xiaomei Sun, and Meilin Jin. 2020. "Screening of Virulence-Related Transcriptional Regulators in Streptococcus suis" Genes 11, no. 9: 972. https://doi.org/10.3390/genes11090972
APA StyleLiu, L., Zhang, Q., Xu, Z., Chen, B., Zhang, A., Sun, X., & Jin, M. (2020). Screening of Virulence-Related Transcriptional Regulators in Streptococcus suis. Genes, 11(9), 972. https://doi.org/10.3390/genes11090972




