Abstract
Understanding the gene mechanisms controlling days to heading (DH) is important in rice breeding for adaption in the target environment. Using a recombinant inbred line population derived from the cross between two japonica rice cultivars, Koshihikari and Baegilmi, we identified three consistent quantitative trait loci (QTLs) for DH for two years, qDH3, qDH6, and qDH7 on chromosomes 3, 6, and 7, respectively. While Baegilmi contributed the allele for early heading at qDH6 and qDH7 with the additive effect of five days each, Koshihikari contributed the allele for early heading at qDH3 with the additive effect of three days. Notably, pyramiding two or more alleles for early heading at these QTLs accelerated heading effectively. Sequencing of Hd16, Hd1, and Ghd7, the previously known heading date genes underlying qDH3, qDH6, and qDH7, respectively, revealed that Baegilmi and Koshihikari carry different alleles at the three genes. Molecular markers were developed to screen the allelic compositions of the three genes among 295 Korean commercial rice cultivars. The results showed that few cultivars carry alleles for early heading at the three genes, highlighting that DH can be further accelerated and fine-tuned in breeding programs by combining the desirable alleles of Hd16, Hd1, and Ghd7.
1. Introduction
The transition from vegetative to reproductive stage is a critical developmental event in plants for ensuring offspring survival under favorable environments [1]. Understanding the genetic basis of flowering time control is especially important in crop species for breeding cultivars adapted well in the target environment. In rice, flowering (also referred to as heading) time is regulated by the complex genetic mechanisms involving hundreds of quantitative trait loci (QTLs) and at least 14 cloned genes [2,3].
Various environmental cues are integrated to modulate the expression of genes encoding florigen, which is synthesized in leaves and transferred to the apical meristem to initiate flower development [4]. In rice, florigen is encoded by Heading Date 3a (Hd3a) and RICE FLOWERING LOCUS T 1 (RFT1), the orthologs of Arabidopsis FT [5,6]. Hd3a expression is mainly regulated by Hd1, the ortholog of Arabidopsis CONSTANS (CO), which upregulates Hd3a under short day and downregulates Hd3a under long day [7]. Hd1 expression is upregulated under both short and long days by OsGI, the ortholog of Arabidopsis GIGANTEA (GI) [8]. Unlike bifunctional Hd1, EARLY HEADING DATE 1 (Ehd1) can accelerate heading under both short and long days by upregulating Hd3a and RFT1 under short and long days, respectively [9]. Ehd1 expression is negatively regulated by Grain number, plant height, and heading date 7 (Ghd7) under long day [10]. While the OsGI-Hd1-Hd3a pathway in rice is orthologous to the GI-CO-FT pathway in Arabidopsis, the Ghd7-Ehd1-Hd3a/RFT1 pathway is unique in rice without clear orthologs in Arabidopsis [1]. Recent studies revealed more complex rice-specific gene networks regulating the Ghd7-Ehd1-Hd3a/RFT1 pathway (e.g., Hd16 and Hd17 upregulating Ghd7 under long day) reviewed in [2,3].
To fine-tune days to heading (DH) in breeding programs and maximize yield and grain quality under the target environment, it is essential to characterize the effects of major genes controlling DH, their allelic variation, epistasis, and interaction with environmental factors including daylength and temperature. In Korea, developing early heading rice cultivars is especially important for boosting farmers’ income by enabling diverse double cropping patterns in the rice paddies, e.g., late-planting of rice after harvesting winter crops such as cabbage, barley, and wheat, or early-planting of rice followed by cash crops such as garlic and onion [11,12]. The utilization of early heading rice cultivars can be also useful for reducing cropping duration in order to minimize damages from erratic weather events. Although over 80 rice cultivars classified as the early heading group have been released in Korea, they occupy less than 10% of the rice cultivation area in Korea as many rice growers generally prefer mid-late heading cultivars because of their higher yield and grain quality compared to the early heading cultivars [13].
Baegilmi is an extremely early heading rice cultivar recently released in Korea exhibiting high yield in the mid-north plain (milled rice yield 5.01 MT/ha) and north-east coastal (5.27 MT/ha) regions in Korea [14]. However, the genes conferring early heading in Baegilmi have remained unknown. In this study, we used a recombinant inbred line (RIL) population derived from the cross between Koshihikari and Baegilmi to identify the chromosomal regions harboring genes controlling DH. Candidate genes underlying the major DH QTLs were sequenced in Koshihikari and Baegilmi and their allelic compositions were screened among commercial rice cultivars. Molecular breeding strategy using the allelic variations in major DH genes was discussed to facilitate the fine-tuning of DH in breeding programs.
2. Materials and Methods
2.1. Plant Materials and Phenotype Evaluation
Two japonica rice cultivars, Koshihikari with high eating quality [15] and Baegilmi with early maturity [14], were used in this study. Days to heading (DH) and grain filling rates of the two cultivars were evaluated at the experimental field of the National Institute of Crop Science (NICS), Suwon, Korea (37°27′ N 126°99′ E) in 2014. The seeds of each cultivar were sown on 25 April and transplanted on 25 May under a randomized complete block design (RCBD) with three replications. Each plot comprised of eight 4.5 m rows, with 30 hills per row and three plants per hill. The hills within a row were spaced by 15 cm and the rows were spaced by 30 cm. DH was determined by counting the number of days from sowing to heading when the panicles emerged in 40% of the plants in a plot. To evaluate the rate of grain filling of Baegilmi and Koshihikari, changes in grain weight of the two cultivars were monitored by measuring 1000 grain weight every three to four days during 19–50 days after heading.
To map QTLs for DH, a RIL population (n = 142) was constructed from the cross between Koshihikari and Baegilmi by the single seed descent method. The RIL population (F6 and F7 generation in 2016 and 2017, respectively) and its parents were grown at the experimental field of NICS, Wanju, Korea (35°84′ N 127°05′ E) in 2016 and 2017. The seeds were sown on 9 May and 10 May in 2016 and 2017, respectively, and the seedlings were transplanted four weeks after sowing. Each RIL was transplanted in a 4.5 m row with the individual plants spaced by 15 cm (30 plants per each line) and the rows spaced by 30 cm. DH of each RIL was determined by counting the number of days from sowing to heading when the panicles emerged in 40% of the plants in a row.
Days to heading of 295 commercial rice cultivars released by NICS, Rural Development Administration, Korea, were evaluated under the optimum and early planting conditions at the experimental field of NICS, Wanju, Korea. Sowing and transplanting dates for optimum planting were 9 May and 1 Jun, respectively in 2018, and those for early planting were 10 Apr and 9 May, respectively in 2019. Planting density and DH evaluation were as described above for the RIL population.
2.2. Sequencing Library Construction and Genotyping
Genomic DNA was extracted from the fresh young leaves of the Koshihikari × Baegilmi RILs using the CTAB (cetyl trimethylammonium bromide) method [16] with minor modifications. The quality and quantity of the extracted DNA were checked using the DeNovix DS-11 spectrophotometer (DeNovix, Wilmington, DE, USA) and the Quant-iTTM dsDNA assay kit (Thermo Fisher Scientific, Waltham, MA, USA). The genotyping-by-sequencing (GBS) library was constructed according to [17]. Briefly, each DNA sample was digested with the ApeKI restriction enzyme (New England Biolabs, Ipswitch, MA, USA), ligated with barcode adapters, pooled, and purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany). The pooled libraries were amplified by PCR with an adapter-specific primer set, analyzed for the target length of 170–350 bp using BioAnalyzer 2100 (Agilent, Santa Clara, CA, USA), and sequenced using the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). The sequencing reads were mapped to the Nipponbare IRGSP-1.0 reference using the BWA aligner [18] and single nucleotide polymorphism (SNPs) were extracted in the format of a VCF (variant calling format) file. The SNP filtering and genotype calling of each RIL were carried out using TASSEL-GBS v2 [19] with the filtering conditions (mapping quality ≥30, base quality ≥20, and coverage ≥7) as described in [20]. Genotype calling for each SNP was conducted by defining a homozygous genotype when the same sequence rate is over 0.90 and a heterozygous genotype when the alternative sequence rate is 0.25–0.27.
2.3. Linkage Mapping and Statistical Analysis
Of 893 SNPs segregating in the RIL population, 128 high quality SNPs were selected for the linkage map construction after excluding SNPs with missing rate over 10%, significant segregation distortion (Chi-square test p–value < 0.05), or overlapping genetic positions. The composite interval mapping was implemented using QTL IciMapping version 4.1 [21] with the threshold LOD (logarithm of the odds) score of 3.0. The same program was also used to estimate the additive effects and the phenotypic variation explained by each QTL at the peak LOD. The mean comparisons of DH between the RILs with different allelic combinations of the identified QTLs were conducted by the Duncan’s multiple range test using SAS version 9.4 (Cary, NC, USA). Three-way factorial ANOVAs were conducted to study the main effects of the three DH QTLs and their two-way and three-way interactions using SAS version 9.4.
2.4. Sequencing and Marker Analysis for Hd16, Hd1, and Ghd7
The coding regions of Hd16, Hd1, and Ghd7, the candidate genes underlying the three DH QTLs from the Koshihikari × Baegilmi RIL population, were sequenced to search polymorphisms between Koshihikari and Baegilmi. The tenth exon of Hd16 was amplified by polymerase chain reaction (PCR) using the primers and conditions described in Supplementary Table S1. The two exons of Hd1 and the two exons of Ghd7 were amplified by PCR as described in [22]. Sequencing of the PCR products was performed by Macrogen (Daejeon, Korea). To search polymorphism in the promoter region of Ghd7, the 2 kb upstream region of Ghd7 from Koshihikari and Baegilmi was sequenced using the customized sequencing service at Macrogen (Daejeon, Korea). Molecular markers to differentiate the three main polymorphisms in Hd16, Hd1, and Ghd7 between Koshihikari and Baegilmi were developed (Supplementary Table S1) and used to screen 295 Korean rice cultivars released in 1979–2017 by NICS, Rural Development Administration.
3. Results
3.1. Days to Heading and Grain Filling Rate of Baegilmi in Comparison with Koshihikari
The DH of Baegilmi under the optimum planting (i.e., sowing and transplanting on 25 April and 25 May, respectively, in Suwon, Korea) was 82 days, which was 21 days earlier than that of Koshihikari (Figure 1a). To compare the rate of grain filling of Baegilmi and Koshihikari, the grain weight change was monitored by measuring 1000 grain weight (TGW) every 3–4 days from 19 days after heading (DAH) to 50 DAH (Figure 1b). While the TGW of Baegilmi increased faster than that of Koshihikari during the early grain filling stage, reaching 19.7 g at 26 DAH (14.5 g in Koshihikari), the TGW of Koshihikari increased faster in the late grain filling stage, reaching 23.1g at 40 DAH which was similar to Baegilmi (23.2 g). A similar pattern was observed when the changes in grain weight were monitored as the ratio of grain weight to the final grain weight according to the cumulative temperature after heading (Figure 1c). Our results indicated that Baegilmi undergoes rapid grain weight increase in the early stage of grain filling as well as early vegetative-to-reproductive transition compared to Koshihikari.
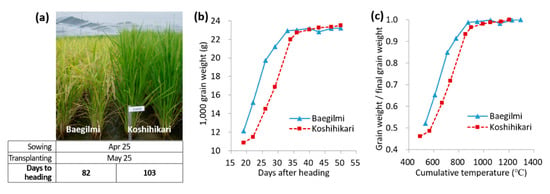
Figure 1.
Maturity of Baegilmi in comparison with Koshihikari. (a) Representative phenotype of Baegilmi and Koshihikari in the field at 112 days after sowing (15 August). Days to heading was determined as the number of days from sowing to heading; (b) Changes in grain weight during 19–50 days after heading; (c) Changes in the proportion of grain weight to the final grain weight according to the cumulative temperature after heading.
3.2. Mapping of qDH3, qDH6, and qDH7 for Days to Heading
The Koshihikari × Baegilmi RIL population (n = 142) was used to map QTLs for DH. A GBS experiment detected 893 SNPs segregating in the mapping population. After removing SNPs with low genotyping quality and overlapping genetic positions, a linkage map spanning a total length of 1293 cM was constructed with the 128 selected SNPs (Supplementary Figure S1). The average number of markers per chromosome was 10.7, ranging from six on chromosome 3 to 17 on chromosome 6. The average interval between two adjacent markers was 11.1 cM.
The Koshihikari × Baegilmi RILs showed continuous DH variation (64–105 days in 2016 and 64–107 days in 2017) with positively skewed distribution (Figure 2a,b). Baegilmi headed 19 and 18 days earlier than Koshihikari in 2016 and 2017, respectively. We detected three major QTLs for DH designated qDH3, qDH6, and qDH7 on chromosomes 3, 6, and 7, respectively, in both 2016 and 2017 (Figure 3a, Table 1). At qDH3, Koshihikari contributed the allele for early heading with the additive effects of −3.1–−3.4 days and the LOD scores of 5.1–7.5, explaining 8.4–10.3% of the DH variation. On the other hand, Baegilmi contributed the allele for early heading at qDH6 and qDH7 (additive effects of 5.2–5.4 days and 5.0–5.3 days, respectively), which showed higher LOD scores (12.6–15.1 and 12.7–13.7, respectively) explaining higher levels of the DH variation (24.7–26.2% and 22.8–25.2%, respectively).
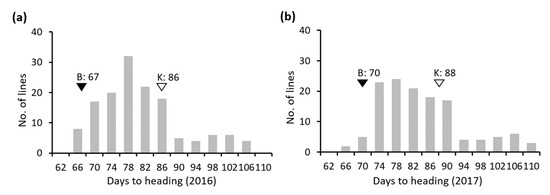
Figure 2.
Frequency distribution of days to heading in the Koshihikari × Baegilmi RIL population evaluated in 2016 (a) and 2017 (b). The values of Baegilmi (B) and Koshihikari (K) are indicated above filled and unfilled triangles, respectively.
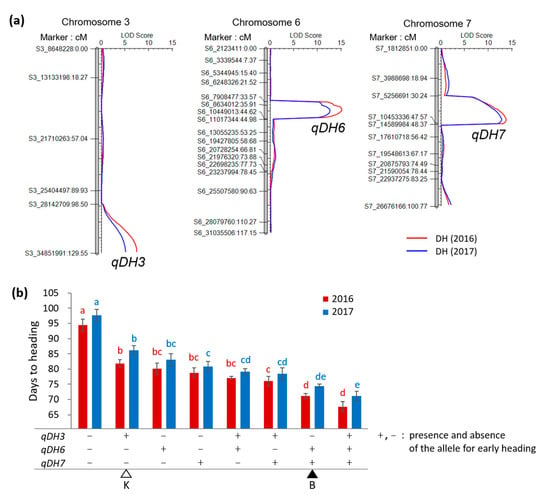
Figure 3.
QTLs for days to heading from the Koshihikari × Baegilmi RIL population. (a) Mapping of qDH3, qDH6, and qDH7 on chromosomes 3, 6, and 7, respectively. Days to heading of the RILs observed in 2016 and 2017 were used for mapping. The number after the letter ‘S’ in the marker name indicates the chromosome number followed by the physical position according to the IRGSP-1.0 reference; (b) Days to heading of the RILs with different allelic combinations of qDH3, qDH6, and qDH7. The markers S3_34851991, S6_8634012, and S7_10453336 were used to represent qDH3, qDH6, and qDH7, respectively. + and − indicate the presence and absence of the allele for early heading, respectively. The genotypes of Baegilmi (B) and Koshihikari (K) are indicated by filled and unfilled triangles, respectively. Different letters above the bars indicate significant difference according to the Duncan’s multiple range test at p < 0.05. Error bars indicate standard errors.

Table 1.
Quantitative trait loci (QTLs) for days to heading from the Koshihikari × Baegilmi recombinant inbred line (RIL) population.
To analyze the main effects of the three QTLs and their two-way and three-way interactions, 2 × 2 × 2 factorial ANOVAs were conducted for DH of the Koshihikari × Baegilmi RILs (Table 2). The main effects of qDH3, qDH6, and qDH7 were highly significant (3.8 × 10−15 < p < 2.6 × 10−5) in both 2016 and 2017. On average, the RILs carrying the Koshihikari alleles at qDH3 headed 5.1–5.2 days earlier than those carrying the Baegilmi alleles. On the other hand, the RILs carrying the Baegilmi alleles at qDH6 or qDH7 headed 9.5–9.9 or 10.7–11.4 days earlier than those with the Koshihikari alleles, respectively. The RILs with different allele combinations of qDH3, qDH6, and qDH7 showed that accumulating two or more alleles for early heading of theses QTLs can shorten DH effectively, with the pyramiding effect of qDH6 and qDH7 being greater than that of qDH3 (Figure 3b).

Table 2.
Three-way ANOVAs of qDH3, qDH6, and qDH7 for days to heading.
The two-way and three-way interactions among qDH3, qDH6, and qDH7 were marginally significant (0.02 < p < 0.05) and explained little phenotypic variation (1.3–1.7%) (Table 2). The qDH3 × qDH6 interaction was significant only in 2016, where the effect of qDH6 accelerating heading was greater under the absence of the allele for early heading at qDH3 (Figure 4a). Similarly, the effect of qDH7 accelerating heading was greater under the absence of the allele for early heading at qDH3, and this was significant in both 2016 and 2017 (Figure 4b,c). The qDH3 × qDH6 × qDH7 interaction was significant only in 2016. Under the absence of the allele for early heading at qDH3, the qDH6 × qDH7 interaction pattern was similar to the qDH3 × qDH6 and qDH3 × qDH7 interactions (Figure 4d), i.e., the effect of one QTL accelerating heading was greater under the absence of the allele for early heading at the other QTL. However, under the presence of the allele for early heading at qDH3, the qDH6 × qDH7 interaction pattern was reversed (Figure 4e), i.e., the effect of one QTL accelerating heading was greater under the presence of the allele for early heading at the other QTL.
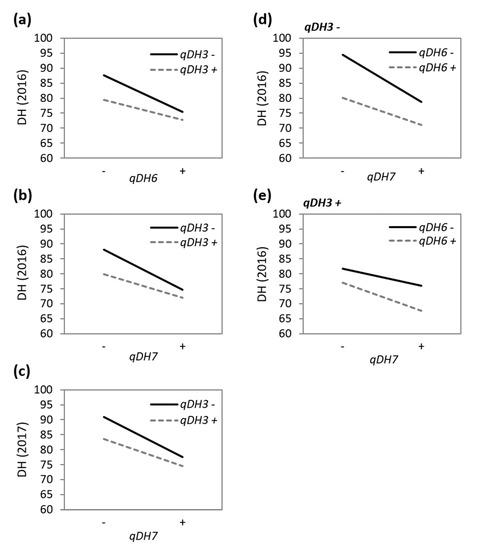
Figure 4.
Interactions among the three QTLs for days to heading. (a) qDH3 × qDH6 interaction in 2016; (b) qDH3 × qDH7 interaction in 2016; (c) qDH3 × qDH7 interaction in 2017; (d) qDH6 × qDH7 interaction under the absence of the allele for early heading at qDH3 in 2016; (e) qDH6 × qDH7 under the presence of the allele for early heading at qDH3 in 2016. Only the significant (p < 0.05) interactions are plotted (see Table 2).
3.3. Hd16, Hd1, and Ghd7 Underlying the Days to Heading QTLs
As the three QTLs for DH encompass previously isolated heading date genes—Hd16 (Os03g0793500) at qDH3, Hd1 (Os06g0275000) at qDH6, and Ghd7 (Os07g0261200) at qDH7 [7,10,23]—we sequenced the coding regions of the three genes from Koshihikari and Baegilmi to search sequence polymorphisms.
It was previously shown that relative to the functional Hd16 allele of Nipponbare, Koshihikari carries a G-to-A (alanine-to-threonine) mutation in the 10th exon of Hd16 that is responsible for early heading under long day [23]. Sequence analysis of the 10th exon of Hd16 revealed that the sequence of Baegilmi is identical to Nipponbare (Figure 5a). This was consistent with our QTL analysis where Koshihikari provided the allele for early heading and Baegilmi provided the allele for late heading at qDH3 (Table 1), suggesting Hd16 as a strong candidate gene for qDH3.
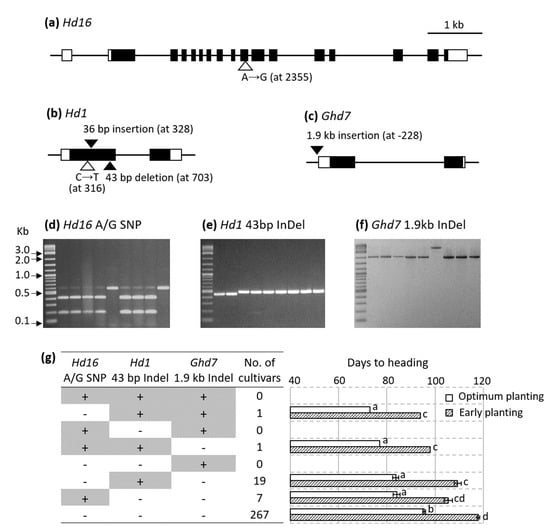
Figure 5.
Sequence polymorphisms of Hd16 (Os03g0793500), Hd1 (Os06g0275000), and Ghd7 (Os07g0261200) in Baegilmi relative to Koshihikari (a–c) and their genotyping among Korean rice cultivars (d–g). Coding region and untranslated region are depicted as filled and unfilled squares, respectively, while introns are depicted as black lines (a–c). Unfilled and filled triangles indicate SNP and insertion/deletion of Baegilmi in comparison with Koshihikari, respectively (a–c). The Hd16 A SNP (Koshihikari allele) and the G SNP (Baegilmi allele) are visualized as intact (579 bp) and digested (393 bp + 186 bp) bands, respectively (d). The Hd1 43 bp deletion (Baegilmi allele) is visualized as a lower band (e). The Ghd7 1.9 kb insertion (Baegilmi allele) is visualized as an upper band (f). + and − indicate the presence and absence of the allele for early heading at each gene among 295 Korean rice cultivars, respectively, and the error bars in the bar graph indicate standard errors (g). Sowing and transplanting dates for optimum planting were 9 May and 1 June, respectively in 2018, and those for early planting were 10 April and 9 May, respectively in 2019, in Wanju, Korea. Different letters next to the bars indicate significant difference (p < 0.05) according to the Duncan’s multiple range test in each year.
Sequence analysis of the Hd1 coding region revealed that Koshihikari carries the functional Hd1 allele identical to Nipponbare as previously reported [22], while Baegilmi carries a non-functional allele with three sequence polymorphisms relative to Koshihikari—a C-to-T SNP, a 36-bp insertion, and a 43-bp deletion in the first exon of Hd1 (Figure 5b). The Hd1 polymorphisms in Baegilmi are identical to those reported in HS66 (GeneBank ID AB041841), the γ adiation mutant of the Japanese cultivar Ginbouzu [7].
While the sequence analysis found no polymorphism in the coding region of Ghd7 between Koshihikari and Baegilmi, we identified a 1901 bp insertion at the −228 bp position from the start codon of Ghd7 in Baegilmi (Figure 5c). The position of the 1901 bp insertion and its sequence were identical to those of the putative retrotransposon inserted in the promoter region of Ghd7 reported in the Japanese cultivar Sorachi (GenBank ID LC472532) [24].
3.4. Allelic Composition of Hd16, Hd1, and Ghd7 among Commercial Rice Cultivars
To study the allelic composition of the three DH genes in commercial rice cultivars, we screened 295 Korean rice cultivars released in 1979–2017 using the molecular markers designed to genotype the sequence polymorphisms in Hd16 (A/G SNP), Hd1 (43 bp Indel), and Ghd7 (1.9 kb Indel) (Supplementary Table S1, Figure 5d–f). Of the eight (2 × 2 × 2) possible allelic combinations, five were observed among the 295 cultivars (Supplementary Table S2, Figure 5g). The majority (>90%) carried the alleles for late heading for all three polymorphisms, namely, the Hd16 G SNP, the Hd1 43-bp insertion, and the Ghd7 1.9-kb deletion. Among 26 cultivars carrying an allele for early heading in one of three DH genes, 19 carried the Hd1 43-bp deletion and seven carried the Hd16 A SNP. Only two out of the 295 Korean rice cultivars carried alleles for early heading in two of the three DH genes—Jopum carried the alleles for early heading at Hd16 and Hd1, while Baegilmi carried the alleles for early heading at Hd1 and Ghd7. None of the 295 cultivars carried the alleles for early heading at all three genes. The average DH of the five cultivar groups according to the allelic combinations of Hd16 (A/G SNP), Hd1 (43 bp Indel), and Ghd7 (1.9 kb Indel) indicated that pyramiding the alleles for early heading at the three genes would accelerate DH effectively in the genetic background of commercial Korean rice cultivars (Figure 5g).
4. Discussion
Baegilmi is an extremely early heading Korean rice cultivar that can be incorporated in diverse double cropping systems to improve land use efficiency [14]. To genetically dissect the early heading characteristics of Baegilmi, we used a RIL population derived from the cross between Koshihikari and Baegilmi and identified the chromosomal locations harboring the genes controlling DH. We identified three major QTLs for DH, of which the allele for early heading was contributed by Baegilmi at qDH6 and qDH7, and by Koshihikari at qDH3. As pyramiding of the alleles for early heading at each QTL was effective in accelerating heading, the Koshihikari × Baegilmi RIL population provides useful breeding lines with different allelic combinations of the three QTLs conferring varying DH, ranging from 71 days to 98 days under the natural long day condition in Wanju (36° N), Korea (Figure 3b). Therefore, the RIL population would be useful for selecting promising breeding lines potentially inheriting the good eating quality of Koshihikari with various heading dates that can be adapted in different environments and cropping patterns. To support this idea, we are currently evaluating the RILs with different allelic combinations of the three DH QTLs in terms of important agronomic traits such as yield performance and grain quality.
All three DH QTLs identified in this study harbor previously cloned heading date genes, i.e., Hd1 [7], Ghd7 [10], and Hd16 [23] at qDH6, qDH7, and qDH3, respectively. Hd1, the ortholog of Arabidopsis CO, is the first heading date gene cloned in rice [7]. While Hd1 accelerates heading under short day by upregulating Hd3a, it downregulates Hd3a under long day and represses heading [6,7,8,25]. Due to this dual function of Hd1 depending on daylength, nonfunctional hd1 alleles confer early heading phenotype under long day as the loss-of-function of Hd1 de-represses Hd3a and promotes heading [26,27]. The nonfunctional hd1 alleles have played an important role in expanding rice cultivation in high latitude areas with natural long day conditions where early heading is critical for rice plants to mature before the cold winter [27]. Hd1 sequencing revealed that Baegimli carries a nonfunctional hd1 allele due to the 43 bp deletion in exon 1 causing a frameshift (Figure 5b). The 43 bp deletion in Hd1 was first reported in the γ ray mutant cultivar HS66 [7], and has been observed in many rice cultivars grown in high latitude areas such as Jilin (44° N) and Liaoning (41° N) of China [28] and Italy (35–47° N) [27]. Among the 295 Korean rice cultivars screened in this study, 21 including Baegilmi carried the nonfunctional hd1 allele due to the 43 bp deletion and showed earlier heading phenotype compared to those without the 43 bp deletion (Figure 5g). As there are many other nonfunctional hd1 alleles arising from frameshift or premature stop codon at different genic positions [22,26,27,28], further work is underway to define additional allelic variation in Hd1 among the Korean rice cultivars.
Similar to Hd1, Ghd7 encoding a CCT (CO, CO-LIKE, and TIMING OF CAB1) domain protein delays heading under long day by downregulating Hd3a [10]. In addition, similar to nonfunctional hd1, nonfunctional ghd7 alleles such as ghd7-0 (full gene deletion) and ghd7-0a (premature stop codon) accelerate heading under long day and contribute to the adaption of rice in high latitude areas [10,28]. Relative to Koshihikari, Baegilmi has a 1901 bp insertion in the promoter region (−228 bp from the start codon) of Ghd7 (Figure 5c). The 1901 bp insertion is identical to that reported in Ghd7 from a Japanese cultivar Sorachi (GenBank ID LC472533) with 449 bp long terminal repeats at each end and 5 bp (AGGTA) target site duplication (Fujino and Yamanouch 2020). This allele has been mainly found in rice cultivars bred in Hokkaido (42–45° N) of Japan, providing an important genetic source for rice adaption in high latitude [24,29]. Among the 295 Korean rice cultivars screened in this study, Baegilmi was the only cultivar carrying the loss-of-function ghd7 allele due to the 1901 bp retrotransposon insertion (Figure 5g), suggesting that this allele would provide a valuable genetic source to accelerate heading in breeding programs. Interestingly, the same retrotransposon insertion has been also found in exon 2 of Hd1 from a Taiwanese landrace Muteka (GenBank ID KR230393), creating a loss-of-function hd1 allele [30]. This illustrates that transposable elements can play important roles in functionally diversifying the heading date genes.
Hd16 encoding casein kinase I delays heading under long day by activating Ghd7 through phosphorylation, and the missense mutation in exon 10 of Hd16 from Koshihikari decreases the kinase activity of Hd16, thus accelerating heading under long day [23]. The screening of over 300 worldwide rice cultivars and 30 wild rice accessions revealed that the Koshihikari-type Hd16 allele is found only in Japanese japonica cultivars [23]. Among the 295 Korean rice cultivars, only eight (i.e., Manan, Boseok, Jinkwang, Pungmi, Pungmi 1, Cheonga, Jungsaenggold, and Jopum) carried the Koshihikari-type Hd16 and showed earlier heading phenotype compared to those carrying the Nipponbare-type Hd16 (Figure 5g). These cultivars would provide useful genetic sources for developing early heading rice cultivars.
We previously reported that Baegilmi was developed by chemically mutagenizing Koshihikari [14]. However, it is extremely unlikely that the EMS mutagenesis would have created the above-mentioned sequence polymorphisms in Hd1, Ghd7, and Hd16 that had been reported previously. This indicates that in the initial stages of breeding Baegilmi, the Koshihikari seed stock used for mutagenesis might have been contaminated during seed handling (e.g., threshing) with a different germplasm which may carry all or some of the previously known alleles, i.e., the HS66-type Hd1, the Sorachi-type Ghd7, and the Nipponbare-type Hd16. We currently do not know the exact source of the potential contamination. Although lacking a clear parentage, the practical value of Baegilmi as a cultivar is unaffected because Baegilmi carries a unique allelic composition of the three heading date genes and exhibits the second earliest heading date among the 295 Korean rice cultivars released in 1979–2017 [31]. Our study also suggests that developing cultivars with earlier heading would be possible by pyramiding the alleles for early heading at Hd1, Ghd7, and Hd16, as none of the 295 cultivars screened in this study carried the alleles for early heading at all three genes.
5. Conclusions
Baegilmi is a japonica rice cultivar with extremely early heading that is useful for diversifying cropping patterns in Korea. From the Koshihikari × Baegilmi RIL population, we detected three QTLs for days to heading, qDH3, qDH6, and qDH7. Different allelic combinations of the three QTLs in the RIL population provided breeding lines that can be useful for developing high quality japonica rice cultivars inheriting Koshihikari’s high eating quality with varying DH. Screening Korean rice cultivars for the allelic compositions of Hd16 (A/G SNP), Hd1 (43 bp Indel), and Ghd7 (1.9 kb Indel) underlying qDH3, qDH6, and qDH7, respectively, showed that few cultivars carry one or more alleles for early heading at the three genes, suggesting that DH can be further accelerated and fine-tuned in breeding programs by using the allelic variations in Hd16, Hd1, and Ghd7.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/9/957/s1, Supplementary Figure S1: Linkage map from the Koshihikari × Baegilmi RIL population (n = 142) using 128 SNP markers, Supplementary Table S1: Molecular markers designed to genotype sequence polymorphisms in Hd16, Hd1, and Ghd7, Supplementary Table S2: Allelic distributions of the polymorphisms in Hd16, Hd1, and Ghd7 in 295 Korean commercial rice cultivars.
Author Contributions
Conceptualization by Y.M. and J.-U.J.; phenotype evaluation and data curation by J.-M.J., S.-K.H., J.K., and C.L.; genotyping analysis by G.P.L.; statistical analysis by Y.M. and J.-U.J.; writing—original draft preparation by Y.M.; writing—review and editing by J.-U.J.; project administration by Y.M. and J.-U.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Rural Development Administration (RDA), Republic of Korea, grant number PJ01357205.
Acknowledgments
We thank Sunghee Kim and Kyoungran Yoo for providing excellent technical supports.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shrestha, R.; Gómez-Ariza, J.; Brambilla, V.; Fornara, F. Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 2014, 114, 1445–1458. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Matsubara, K.; Yano, M. Genetic control of flowering time in rice: Integration of Mendelian genetics and genomics. Theor. Appl. Genet. 2016, 129, 2241–2252. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, K.; Yano, M. Genetic and molecular dissection of flowering time control in rice. In Rice Genomics, Genetics and Breeding; Sasaki, T., Ashikari, M., Eds.; Springer: Singapore, 2018; pp. 177–190. [Google Scholar]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotech. 2015, 32, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Tamaki, S.; Matsuo, S.; Hann, L.W.; Yokoi, S.; Shimamoto, K. Hd3a protein is a mobile flowering signal in rice. Science 2007, 316, 1033–1036. [Google Scholar] [CrossRef] [PubMed]
- Yano, M.; Katayose, Y.; Ashikari, M.; Yamanouchi, U.; Monna, L.; Fuse, T.; Baba, T.; Yamamoto, K.; Umehara, Y.; Nagamura, Y.; et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 2000, 12, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef]
- Doi, K.; Izawa, T.; Fuse, T.; Yamanouchi, U.; Kubo, T.; Shimatani, Z.; Yano, M.; Yoshimura, A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004, 18, 926–936. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef]
- Kim, S.Y.; Seo, J.H.; Bae, H.K.; Hwang, C.D.; Ko, J.M. Rice cultivars adaptable for rice based cropping systems in a paddy field in the Yeongnam plain area of Korea. Korean J. Agri. Sci. 2018, 45, 355–363. [Google Scholar] [CrossRef]
- Kim, Y.D.; Kang, S.G.; Ku, B.I.; Choi, M.K.; Park, H.K.; Park, T.S.; Back, N.H.; Kim, S.J.; Ko, J.K. Selection of suitable rice cultivars for silage barley-rice double cropping in Honam plain area. Korean J. Int. Agric. 2010, 22, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Jeung, J.; Cho, Y.; Kim, B. Development of early maturing rice lines with genes conferring resistance to bacterial blight and rice stripe virus for enhancing the adaptability in plain area. Korean J. Breed. Sci 2015, 47, 118–127. [Google Scholar] [CrossRef]
- Mo, Y.; Jeong, J.-M.; Kim, W.-J.; Kim, B.-K.; Jeung, J.-U. ‘Baegilmi’, an extremely early maturing blast resistant rice with good grain appearance. Korean J. Breed. Sci. 2019, 51, 151–159. [Google Scholar] [CrossRef][Green Version]
- Kobayashi, A.; Hori, K.; Yamamoto, T.; Yano, M. Koshihikari: A premium short-grain rice cultivar—Its expansion and breeding in Japan. Rice 2018, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Jang, Y.J.; Seo, M.; Hersh, C.P.; Rhee, S.J.; Kim, Y.; Lee, G.P. An evolutionarily conserved non-synonymous SNP in a leucine-rich repeat domain determines anthracnose resistance in watermelon. Theor. Appl. Genet. 2019, 132, 473–488. [Google Scholar] [CrossRef]
- Meng, L.; Li, H.; Zhang, L.; Wang, J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop. J. 2015, 3, 269–283. [Google Scholar] [CrossRef]
- Kim, S.R.; Torollo, G.; Yoon, M.R.; Kwak, J.; Lee, C.K.; Prahalada, G.D.; Choi, I.R.; Yeo, U.S.; Jeong, O.Y.; Jena, K.K.; et al. Loss-of-function alleles of Heading date 1 (Hd1) are associated with adaptation of temperate japonica rice plants to the tropical region. Front. Plant Sci. 2018, 9, 1827. [Google Scholar] [CrossRef]
- Hori, K.; Ogiso-Tanaka, E.; Matsubara, K.; Yamanouchi, U.; Ebana, K.; Yano, M. Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 2013, 76, 36–46. [Google Scholar] [CrossRef]
- Fujino, K.; Yamanouchi, U. Genetic effect of a new allele for the flowering time locus Ghd7 in rice. Breed. Sci. 2020, 70, 342–346. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Teshima, K.M.; Yokoi, S.; Innan, H.; Shimamoto, K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl. Acad. Sci. USA 2009, 106, 4555–4560. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ariza, J.; Galbiati, F.; Goretti, D.; Brambilla, V.; Shrestha, R.; Pappolla, A.; Courtois, B.; Fornara, F. Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 2015, 66, 2027–2039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, X.; Yan, W.; Zhang, Z.; Lu, L.; Han, Z.; Zhao, H.; Liu, H.; Song, P.; Hu, Y.; et al. Combinations of the Ghd7, Ghd8 and Hd1 genes largely define the ecogeographical adaptation and yield potential of cultivated rice. New Phytol. 2015, 208, 1056–1066. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Okumoto, Y.; Tsukiyama, T.; Xu, C.; Teraishi, M.; Tanisaka, T. Allelic differentiation at the E1/Ghd7 locus has allowed expansion of rice cultivation area. Plants 2019, 8, 550. [Google Scholar] [CrossRef]
- Wei, F.J.; Tsai, Y.C.; Wu, H.P.; Huang, L.T.; Chen, Y.C.; Chen, Y.F.; Wu, C.C.; Tseng, Y.T.; Hsing, Y.C. Both Hd1 and Ehd1 are important for artificial selection of flowering time in cultivated rice. Plant Sci. 2015, 242, 187–194. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.M.; Park, H.S.; Baek, M.K.; Suh, J.P.; Kim, C.S.; Lee, K.M.; Park, S.G.; Cho, Y.C. Characterization of grain-related traits of 300 Korean rice varieties. In Proceedings of the 2019 Korean Society of Breeding Science (KSBS) & Society for the Advancement of Breeding Research in Asia and Oceania (SABRAO), Gwangju, Korea, 2–5 July 2019. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).