Analysis of Gene Expression of miRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
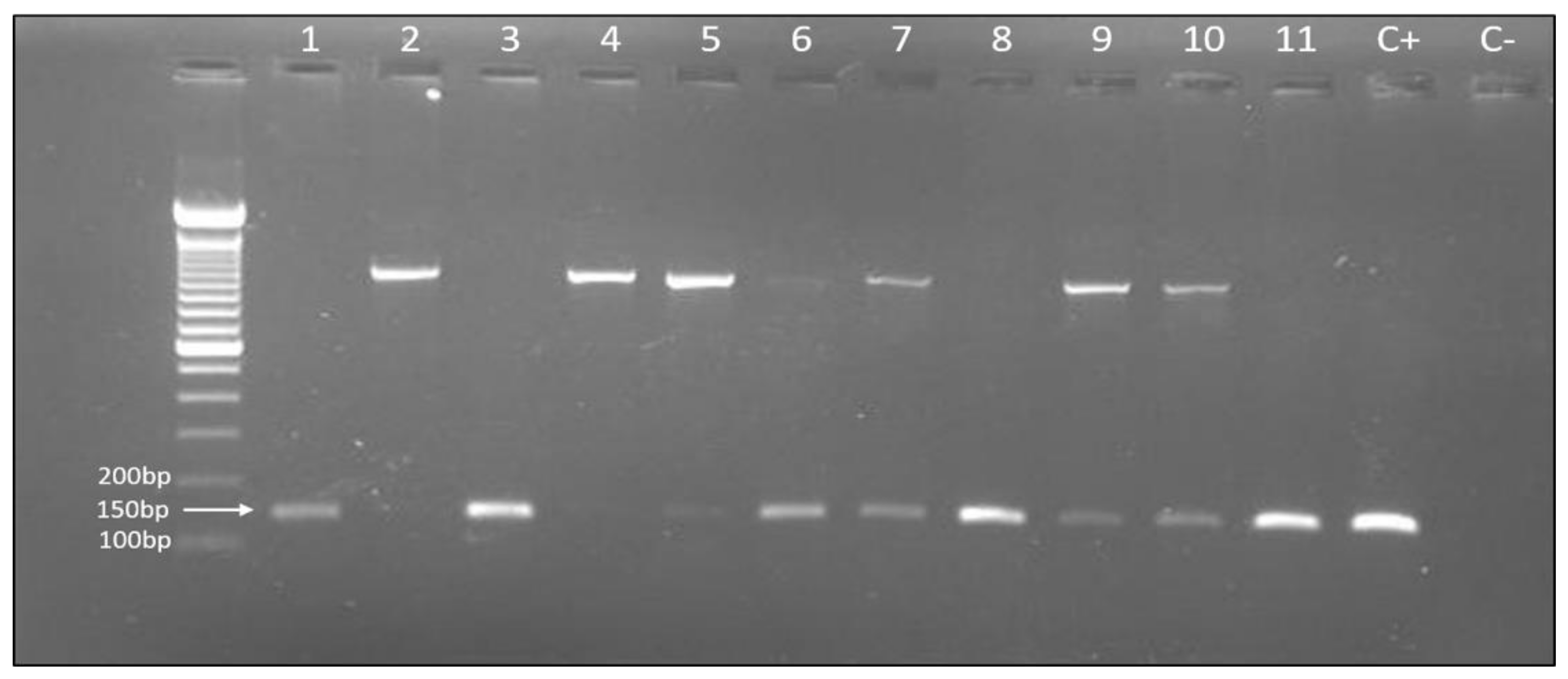
2.2. DNA Extraction and Helicobacter Pylori Detection
2.3. RNA Extraction, cDNA Synthesis, and Real-Time Quantitative PCR (qPCR) Gene Expression Analysis
2.4. Bioinformatics Analysis
2.4.1. Identification of miRNAs and Target Genes in Apoptosis
2.4.2. Screening for Differentially Expressed Genes (DEGs)
2.5. Statistical Analysis
3. Results
3.1. Helicobacter Pylori Detection
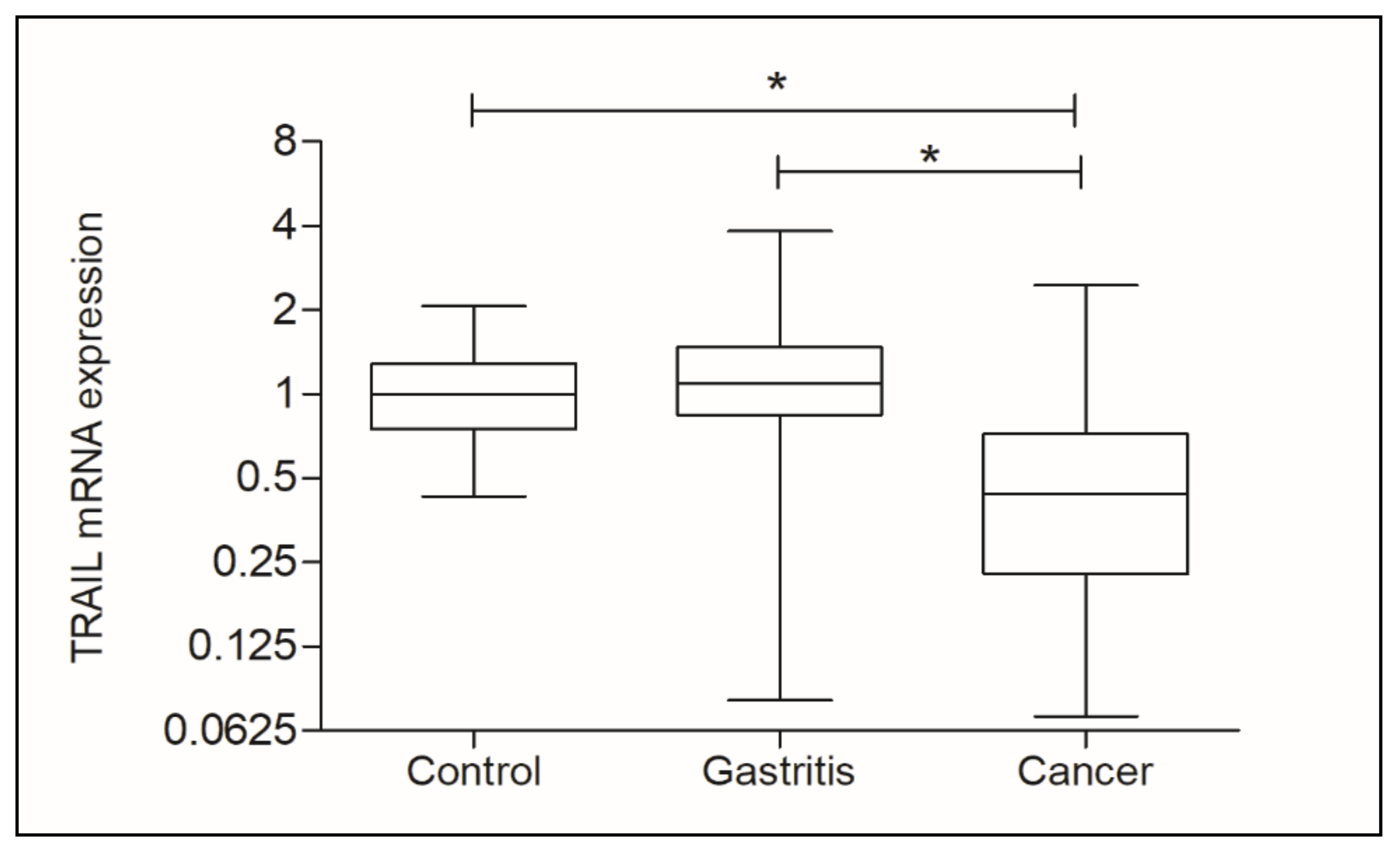
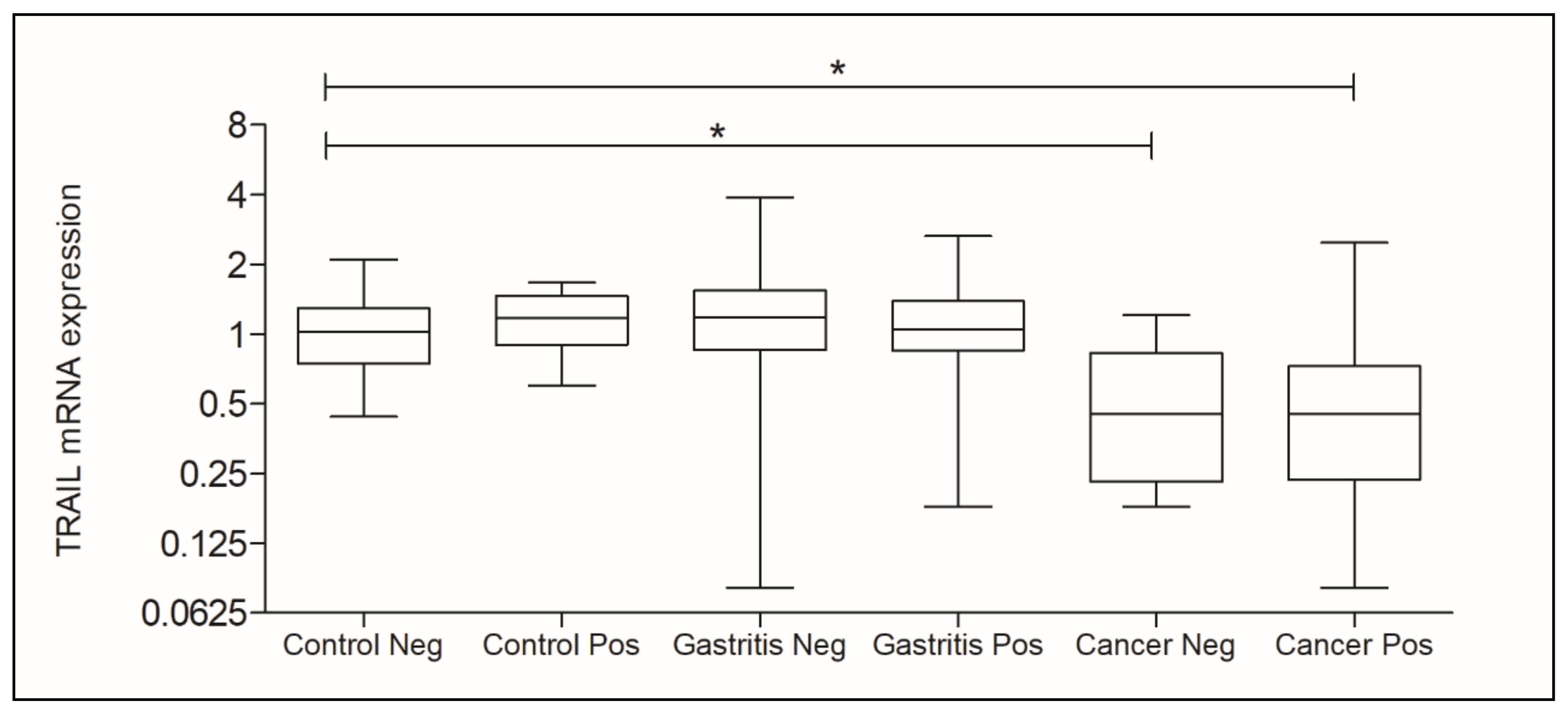
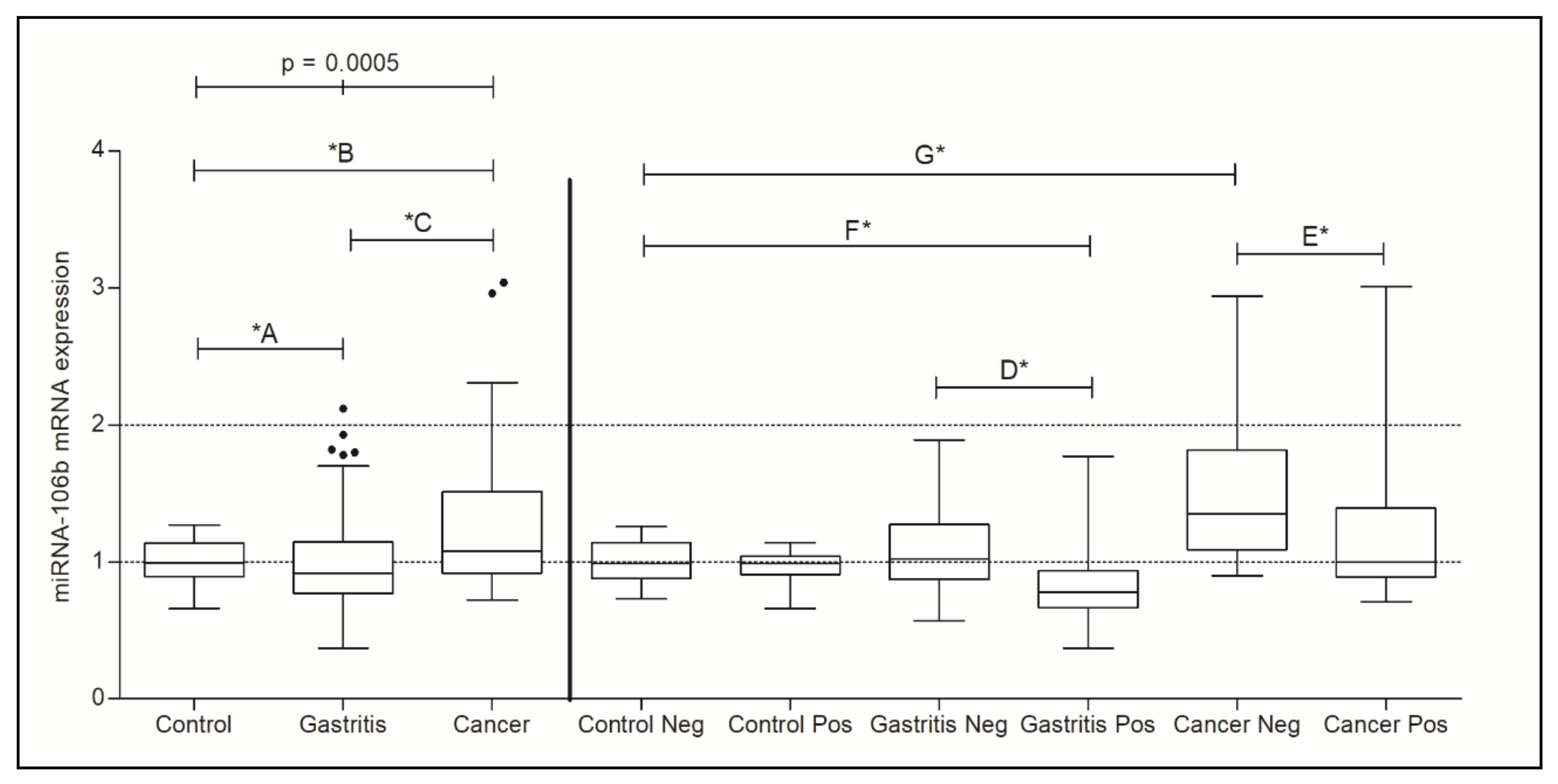
3.2. Analysis of TRAIL and microRNA-106b-5p Gene Expression
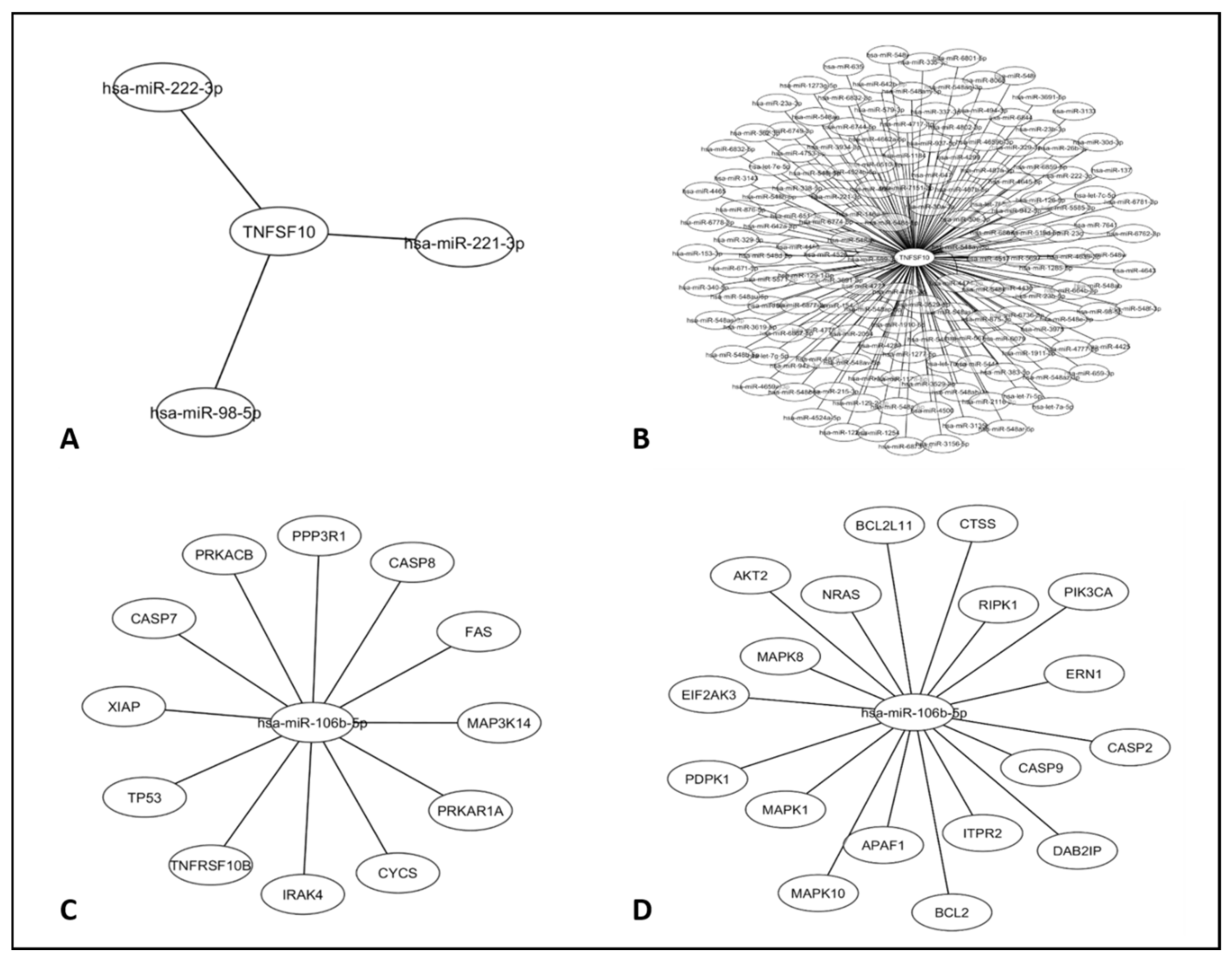
3.3. Bioinformatics Analysis and Interaction Network in the Apoptosis Pathway Considering TRAIL and miR-106b-5p Genes
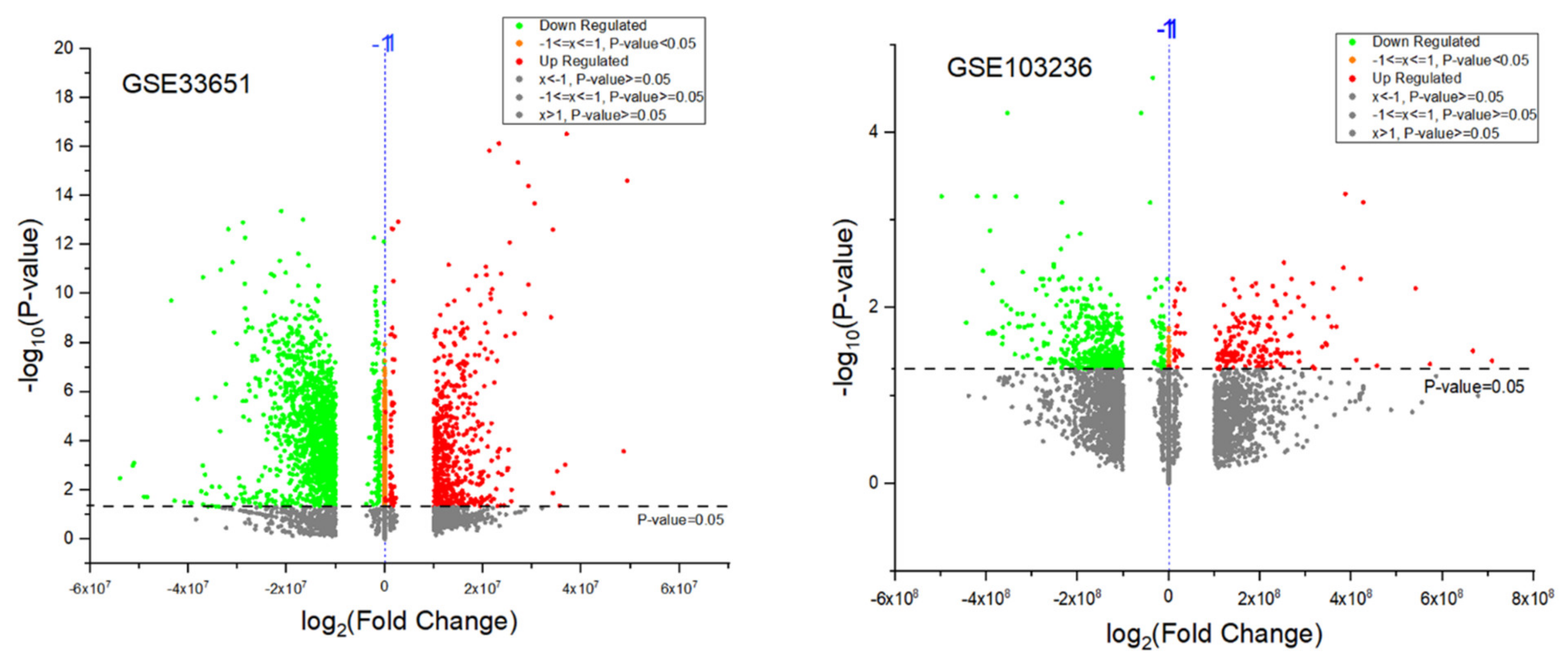
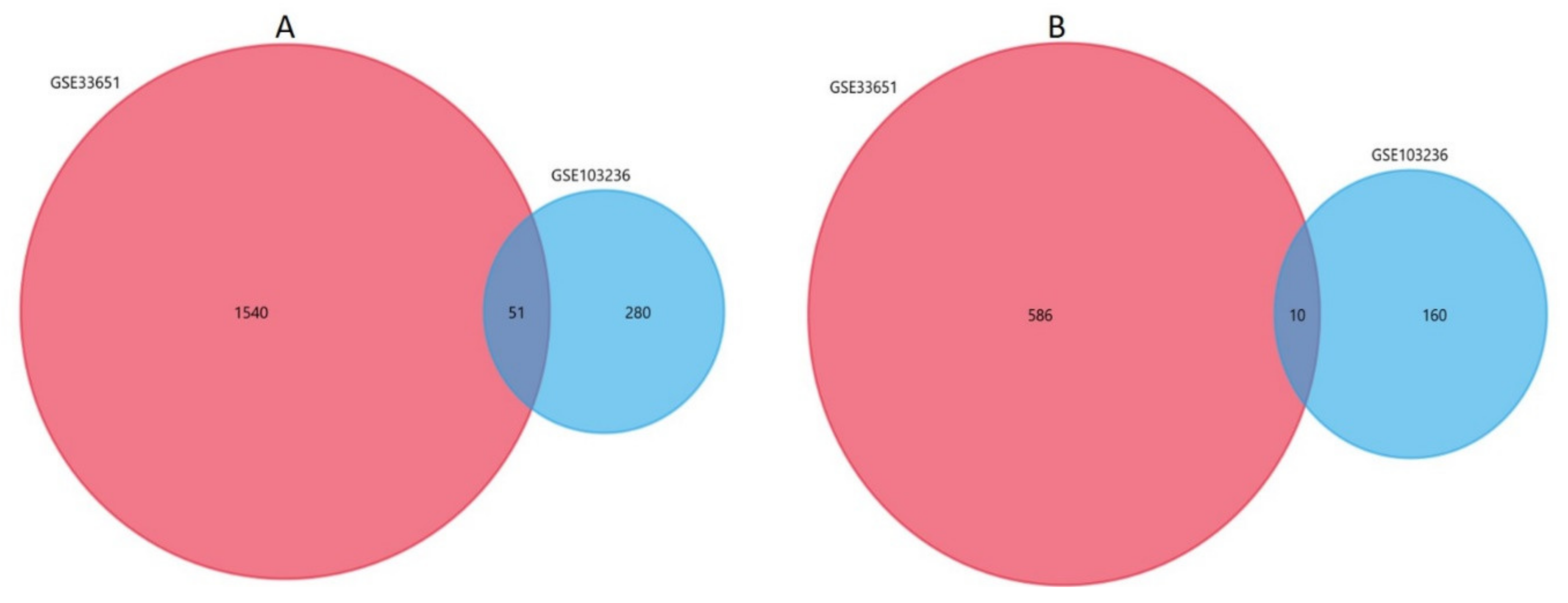
3.4. Identification of Differentially Expressed Genes (DEGs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bozkurt, O.; Firat, S.T.; Dogan, E.; Cosar, R.; Inanc, M.; Ozkan, M. The prognostic value of the change in neutrophil-to-lymphocyte ratio during first-line palliative chemotherapy in patients with metastatic gastric cancer: A retrospective study. J. BUON 2019, 24, 1992–1999. [Google Scholar] [PubMed]
- Tai, Q.; Shao, H.; Liu, Y.; Li, E.; Zhao, R. A comparative analysis on clinical efficacy of FOLFOX6 regimen and DCF regimen as neoadjuvant chemotherapy combined with radical gastrectomy in treating advanced gastric cancer. J. BUON 2019, 24, 2006–2012. [Google Scholar] [PubMed]
- Huang, Y.; Zhang, J.; Hou, L.; Wang, G.; Liu, H.; Zhang, R.; Chen, X.; Zhu, J. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J. Exp. Clin. Cancer Res. 2017, 36, 194. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Li, Z.L.; Qiu, S.F. IFN-gamma Induces Gastric Cancer Cell Proliferation and Metastasis Through Upregulation of Integrin beta3-Mediated NF-kappaB Signaling. Transl. Oncol. 2018, 11, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; El Hajj, N.; Sittler, S.; Lammert, N.; Barnes, R.; Meloni-Ehrig, A. Gastric cancer: Classification, histology and application of molecular pathology. J. Gastrointest. Oncol. 2012, 3, 251–261. [Google Scholar]
- Gall, A.; Gaudet, R.G.; Gray-Owen, S.D.; Salama, N.R. TIFA Signaling in Gastric Epithelial Cells Initiates the cag Type 4 Secretion System-Dependent Innate Immune Response to Helicobacter pylori Infection. MBio 2017, 8, e01168-17. [Google Scholar] [CrossRef] [PubMed]
- Gobert, A.P.; Verriere, T.; Asim, M.; Barry, D.P.; Piazuelo, M.B.; de Sablet, T.; Delgado, A.G.; Bravo, L.E.; Correa, P.; Peek, R.M., Jr.; et al. Heme oxygenase-1 dysregulates macrophage polarization and the immune response to Helicobacter pylori. J. Immunol. 2014, 193, 3013–3022. [Google Scholar] [CrossRef]
- Yang, Y.; Du, J.; Liu, F.; Wang, X.; Li, X.; Li, Y. Role of caspase-3/E-cadherin in helicobacter pylori-induced apoptosis of gastric epithelial cells. Oncotarget 2017, 8, 59204–59216. [Google Scholar] [CrossRef]
- Yang, Y.; Deng, C.S.; Peng, J.Z.; Wong, B.C.; Lam, S.K.; Xia, H.H. Effect of Helicobacter pylori on apoptosis and apoptosis related genes in gastric cancer cells. Mol. Pathol. 2003, 56, 19–24. [Google Scholar] [CrossRef][Green Version]
- Yang, Y.; Huang, X.; Chen, S.; Ma, G.; Zhu, M.; Yan, F.; Yu, J. Resveratrol induced apoptosis in human gastric carcinoma SGC-7901 cells via activation of mitochondrial pathway. Asia Pac. J. Clin. Oncol. 2018, 14, e317–e324. [Google Scholar] [CrossRef]
- Jones, N.L.; Day, A.S.; Jennings, H.A.; Sherman, P.M. Helicobacter pylori induces gastric epithelial cell apoptosis in association with increased Fas receptor expression. Infect. Immun. 1999, 67, 4237–4242. [Google Scholar] [CrossRef] [PubMed]
- Grivicich, I.; Regner, A.; da Rocha, A.B. Morte celular por apoptose. Rev. Bras. Cancerol. 2007, 53, 335–343. [Google Scholar]
- Xu, L.; Zhang, Y.; Qu, X.; Che, X.; Guo, T.; Li, C.; Ma, R.; Fan, Y.; Ma, Y.; Hou, K.; et al. DR5-Cbl-b/c-Cbl-TRAF2 complex inhibits TRAIL-induced apoptosis by promoting TRAF2-mediated polyubiquitination of caspase-8 in gastric cancer cells. Mol. Oncol. 2017, 11, 1733–1751. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.S.; Wang, X.W.; He, W.; Ma, X.T.; Wang, H.Y.; Han, M.; Li, B.H. TRAIL inhibits platelet-induced colorectal cancer cell invasion. J. Int. Med. Res. 2019, 47, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Qu, X.; Zhang, Y.; Hu, X.; Yang, X.; Hou, K.; Teng, Y.; Zhang, J.; Sada, K.; Liu, Y. Oxaliplatin enhances TRAIL-induced apoptosis in gastric cancer cells by CBL-regulated death receptor redistribution in lipid rafts. FEBS Lett. 2009, 583, 943–948. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.C.; Han, S.I. MDL-12330A potentiates TRAIL-induced apoptosis in gastric cancer cells through CHOP-mediated DR5 upregulation. Korean J. Physiol. Pharmacol. 2017, 21, 397–405. [Google Scholar] [CrossRef]
- Fuentes, R.G.; Toume, K.; Arai, M.A.; Sadhu, S.K.; Ahmed, F.; Ishibashi, M. Scopadulciol, Isolated from Scoparia dulcis, Induces beta-Catenin Degradation and Overcomes Tumor Necrosis Factor-Related Apoptosis Ligand Resistance in AGS Human Gastric Adenocarcinoma Cells. J. Nat. Prod. 2015, 78, 864–872. [Google Scholar] [CrossRef]
- Xu, L.; Qu, X.; Luo, Y.; Zhang, Y.; Liu, J.; Qu, J.; Zhang, L.; Liu, Y. Epirubicin enhances TRAIL-induced apoptosis in gastric cancer cells by promoting death receptor clustering in lipid rafts. Mol. Med. Rep. 2011, 4, 407–411. [Google Scholar] [CrossRef]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Fan, L.; Zhu, C.; Qiu, R.; Zan, P.; Zheng, Z.; Xu, T.; Li, G. MicroRNA-661 Enhances TRAIL or STS Induced Osteosarcoma Cell Apoptosis by Modulating the Expression of Cytochrome c1. Cell. Physiol. Biochem. 2017, 41, 1935–1946. [Google Scholar] [CrossRef]
- Huang, G.; Chen, X.; Cai, Y.; Wang, X.; Xing, C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol. Rep. 2017, 37, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.X.; Zhang, B.L.; Yang, M.Y.; Liu, H.; Xiao, C.H.; Zhang, S.G.; Liu, R. MicroRNA-106b-5p promotes hepatocellular carcinoma development via modulating FOG2. OncoTargets Ther. 2019, 12, 5639–5647. [Google Scholar] [CrossRef]
- Xu, K.; Xiong, W.; Zhao, S.; Wang, B. MicroRNA-106b serves as a prognostic biomarker and is associated with cell proliferation, migration, and invasion in osteosarcoma. Oncol. Lett. 2019, 18, 3342–3348. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef]
- Wang, M.; Yang, Y.O.; Jin, Q.; Shang, L.; Zhang, J. Function of miR-25 in the invasion and metastasis of esophageal squamous carcinoma cells and bioinformatical analysis of the miR-106b-25 cluster. Exp. Ther. Med. 2018, 15, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Stolte, M.; Meining, A. The updated Sydney system: Classification and grading of gastritis as the basis of diagnosis and treatment. Can. J. Gastroenterol. 2001, 15, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.; Orcini, W.; Peruquetti, R.; Cardoso Smith, M.; Payão, S.; Rasmussen, L. Prevalence of Helicobacter pylori cag A and sab A Genotypes in Patients with Gastric Disease. Adv. Microbiol. 2019, 9, 239–247. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Warren, J.R.; Marshall, B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1983, 1, 1273–1275. [Google Scholar]
- Schulz, C.; Schutte, K.; Mayerle, J.; Malfertheiner, P. The role of the gastric bacterial microbiome in gastric cancer: Helicobacter pylori and beyond. Therap. Adv. Gastroenterol. 2019, 12, 1756284819894062. [Google Scholar] [CrossRef]
- Camilo, V.; Sugiyama, T.; Touati, E. Pathogenesis of Helicobacter pylori infection. Helicobacter 2017, 22, e12405. [Google Scholar] [CrossRef] [PubMed]
- Venerito, M.; Vasapolli, R.; Rokkas, T.; Delchier, J.C.; Malfertheiner, P. Helicobacter pylori, gastric cancer and other gastrointestinal malignancies. Helicobacter 2017, 22, e12413. [Google Scholar] [CrossRef] [PubMed]
- Kivrak Salim, D.; Sahin, M.; Koksoy, S.; Adanir, H.; Suleymanlar, I. Local Immune Response in Helicobacter pylori Infection. Medicine 2016, 95, e3713. [Google Scholar] [CrossRef] [PubMed]
- Suerbaum, S.; Michetti, P. Helicobacter pylori infection. N. Engl. J. Med. 2002, 347, 1175–1186. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, B.; Chen, D.; Setroikromo, R.; Haisma, H.J.; Quax, W.J. Histone Deacetylase Inhibitors Sensitize TRAIL-Induced Apoptosis in Colon Cancer Cells. Cancers 2019, 11, 645. [Google Scholar] [CrossRef] [PubMed]
- Jo, E.B.; Lee, Y.S.; Lee, H.; Park, J.B.; Park, H.; Choi, Y.L.; Hong, D.; Kim, S.J. Combination therapy with c-met inhibitor and TRAIL enhances apoptosis in dedifferentiated liposarcoma patient-derived cells. BMC Cancer 2019, 19, 496. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Cui, Y.; Liu, M.; Tan, Z.; Jiang, Y. Downregulation of miR-125b promotes resistance of glioma cells to TRAIL through overexpression of Tafazzin which is a mitochondrial protein. Aging 2019, 11, 2670–2680. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, C.C.; Pan, Z.G.; Zhou, C.W. Irigenin sensitizes TRAIL-induced apoptosis via enhancing pro-apoptotic molecules in gastric cancer cells. Biochem. Biophys. Res. Commun. 2018, 496, 998–1005. [Google Scholar] [CrossRef]
- Thorburn, A. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) pathway signaling. J. Thorac. Oncol. 2007, 2, 461–465. [Google Scholar] [CrossRef]
- Rasheduzzaman, M.; Moon, J.H.; Lee, J.H.; Nazim, U.M.; Park, S.Y. Telmisartan generates ROS-dependent upregulation of death receptor 5 to sensitize TRAIL in lung cancer via inhibition of autophagy flux. Int. J. Biochem. Cell Biol. 2018, 102, 20–30. [Google Scholar] [CrossRef]
- Fan, Y.; Sheng, W.; Meng, Y.; Cao, Y.; Li, R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif. Cells Nanomed. Biotechnol. 2020, 48, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Fodde, R. The APC gene in colorectal cancer. Eur. J. Cancer 2002, 38, 867–871. [Google Scholar] [CrossRef]
- Fujimoto, T.; Tomizawa, M.; Yokosuka, O. SiRNA of frizzled-9 suppresses proliferation and motility of hepatoma cells. Int. J. Oncol. 2009, 35, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Qi, D.; Jiang, J.; Zhang, J.; Yu, C. Expression pattern of p-Smad2/Smad4 as a predictor of survival in invasive breast ductal carcinoma. Oncol. Lett. 2020, 19, 1789–1798. [Google Scholar] [CrossRef]
- Manicum, T.; Ni, F.; Ye, Y.; Fan, X.; Chen, B.C. Prognostic values of E2F mRNA expression in human gastric cancer. Biosci. Rep. 2018, 38, BSR20181264. [Google Scholar] [CrossRef]
- Marcolino, T.F.; Pimenta, C.A.M.; Artigiani Neto, R.; Castelo, P.; Silva, M.S.; Forones, N.M.; Oshima, C.T.F. p53, Cyclin-D1, beta-catenin, APC and c-myc in Tumor Tissue from Colorectal and Gastric Cancer Patients with Suspected Lynch Syndrome by the Bethesda Criteria. Asian Pac. J. Cancer Prev. 2020, 21, 343–348. [Google Scholar] [CrossRef]
- Min, K.W.; Kim, D.H.; Do, S.I.; Chae, S.W.; Kim, K.; Sohn, J.H.; Lee, H.J.; Do, I.G.; Pyo, J.S.; Kim, Y.; et al. Expression Pattern of Smad4/GATA3 as a Predictor of Survival in Invasive Ductal Carcinoma of the Breast. Pathobiology 2017, 84, 130–138. [Google Scholar] [CrossRef]
- Qu, Y.; Zhang, H.; Duan, J.; Liu, R.; Deng, T.; Bai, M.; Huang, D.; Li, H.; Ning, T.; Zhang, L.; et al. MiR-17-5p regulates cell proliferation and migration by targeting transforming growth factor-beta receptor 2 in gastric cancer. Oncotarget 2016, 7, 33286–33296. [Google Scholar] [CrossRef]
- Sun, D.; Chen, L.; Lv, H.; Gao, Y.; Liu, X.; Zhang, X. Circ_0058124 Upregulates MAPK1 Expression to Promote Proliferation, Metastasis and Metabolic Abilities in Thyroid Cancer Through Sponging miR-940. OncoTargets Ther. 2020, 13, 1569–1581. [Google Scholar] [CrossRef]
- Yin, W.; Chen, J.; Wang, G.; Zhang, D. MicroRNA106b functions as an oncogene and regulates tumor viability and metastasis by targeting LARP4B in prostate cancer. Mol. Med. Rep. 2019, 20, 951–958. [Google Scholar] [CrossRef]
- Li, Y.; Tian, J.; Guo, Z.J.; Zhang, Z.B.; Xiao, C.Y.; Wang, X.C. Expression of microRNAs-106b in nonsmall cell lung cancer. J. Cancer Res. Ther. 2018, 14, S295–S298. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Shi, L.; Chen, W.; Fang, P.; Li, J.; Jin, L.; Pan, Z.; Pan, C. MiR-106b inhibitors sensitize TRAIL-induced apoptosis in hepatocellular carcinoma through increase of death receptor 4. Oncotarget 2017, 8, 41921–41931. [Google Scholar] [CrossRef] [PubMed]
- Ye, F.; Tang, C.; Shi, W.; Qian, J.; Xiao, S.; Gu, M.; Dang, Y.; Liu, J.; Chen, Y.; Shi, R.; et al. A MDM2-dependent positive-feedback loop is involved in inhibition of miR-375 and miR-106b induced by Helicobacter pylori lipopolysaccharide. Int. J. Cancer 2015, 136, 2120–2131. [Google Scholar] [CrossRef] [PubMed]
- Carrasco, G.; Corvalan, A.H. Helicobacter pylori-Induced Chronic Gastritis and Assessing Risks for Gastric Cancer. Gastroenterol. Res. Pract. 2013, 2013, 393015. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Q.; Yang, K.; Brigstock, D.R.; Zhang, L.; Xiu, M.; Sun, L.; Gao, R.P. Rare case of Helicobacter pylori-positive multiorgan IgG4-related disease and gastric cancer. World J. Gastroenterol. 2015, 21, 3429–3434. [Google Scholar] [CrossRef]
- Targosz, A.; Brzozowski, T.; Pierzchalski, P.; Szczyrk, U.; Ptak-Belowska, A.; Konturek, S.J.; Pawlik, W. Helicobacter pylori promotes apoptosis, activates cyclooxygenase (COX)-2 and inhibits heat shock protein HSP70 in gastric cancer epithelial cells. Inflamm. Res. 2012, 61, 955–966. [Google Scholar] [CrossRef]
| Groups | Control | Gastritis | Cancer | Patients Ethnic Origins (%) |
|---|---|---|---|---|
| Male (%) | 22 (36.7) | 53 (40.16) | 30 (57.7) | European (95%) |
| Female (%) | 38 (63.3) | 79 (59.84) | 22 (42.3) | Japanese (2.5%) |
| Total (%) | 60 (100) | 132 (100) | 52 (100) | African (2.5%) |
| Mean Age ± SD | 55 ± 15.6 | 54 ± 16 | 53 ± 10 |
| Control (%) | Gastritis (%) | Cancer (%) | Total (%) | |
|---|---|---|---|---|
| H. pylori + | 11 (18.3) | 54 (40.9) | 38 (73.1) | 103 (42.2) |
| H. pylori − | 49 (81.7) | 78 (59.1) | 14 (26.9) | 141 (57.8) |
| OR (95% CI) | 3.08 (1.45–6.64) | 12.09 (4.73–28.7) | ||
| p | 0.0028 * | <0.0001 * | ||
| Total | 60 (100) | 132 (100) | 52 (100) | 244 (100) |
| Group (RQ Mean) | Compared Groups | p-Value |
|---|---|---|
| Control (0.9950) | Control vs. Gastritis | 0.0305 *A |
| Gastritis (0.9200) | Control vs. Cancer | 0.0307 *B |
| Cancer (1.080) | Gastritis vs. Cancer | 0.0004 *C |
| Control Neg. (0.9900) | Control Neg. vs. Control Pos. | 0.4559 |
| Control Pos. (0.9900) | Gastritis Neg. vs. Gastritis Pos. | <0.0001 *D |
| Gastritis Neg. (1.020) | Cancer Neg. vs. Cancer Pos. | 0.0192 *E |
| Gastritis Pos. (0.7800) | Control Neg. vs. Gastritis Neg. | 0.4068 |
| Cancer Neg. (1.350) | Control Neg. vs. Gastritis Pos. | <0.0001 *F |
| Cancer Pos. (1.000) | Control Neg. vs. Cancer Neg. | 0.0001 *G |
| Control Neg. vs. Cancer Pos. | 0.5370 |
| DEGs | Genes |
|---|---|
| Upregulated | MT1E, C1orf132, SST, GPX3, MAL, ATP4B, RNASE1, SCUBE2, C16orf89, CHGA |
| Downregulated | IVNS1ABP, THY1, SMARCA4, HAVCR2, INHBA, GTPBP4, TMEM158, MFSD12, COL18A1, PLA2G7, SSR2, COL1A1, CTHRC1, SERPINH1, IFI30, OLFML2B, SOD2, SPARC, FAM20C, MFSD13A, MYO1B, C1orf112, AGPAT4, KIF26B, S100A10, SULF1, UCK2, CENPF, LOX, PMEPA1, NCR3LG1, CTSA, ANGPT2, CENPN, SPON2, NUF2, APOC1, PGM2L1, FAP, COL12A1, KIF18B, ARHGAP39, CKS1B, IGF2BP3, FRP4, MMP3, LRP8, CLDN4, SLC4A11, LINC01296, EPHB2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pereira, J.; Santos, M.; Delabio, R.; Barbosa, M.; Smith, M.; Payão, S.; Rasmussen, L. Analysis of Gene Expression of miRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer. Genes 2020, 11, 393. https://doi.org/10.3390/genes11040393
Pereira J, Santos M, Delabio R, Barbosa M, Smith M, Payão S, Rasmussen L. Analysis of Gene Expression of miRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer. Genes. 2020; 11(4):393. https://doi.org/10.3390/genes11040393
Chicago/Turabian StylePereira, Jéssica, Mônica Santos, Roger Delabio, Mônica Barbosa, Marília Smith, Spencer Payão, and Lucas Rasmussen. 2020. "Analysis of Gene Expression of miRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer" Genes 11, no. 4: 393. https://doi.org/10.3390/genes11040393
APA StylePereira, J., Santos, M., Delabio, R., Barbosa, M., Smith, M., Payão, S., & Rasmussen, L. (2020). Analysis of Gene Expression of miRNA-106b-5p and TRAIL in the Apoptosis Pathway in Gastric Cancer. Genes, 11(4), 393. https://doi.org/10.3390/genes11040393





