RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription
Abstract
1. Introduction
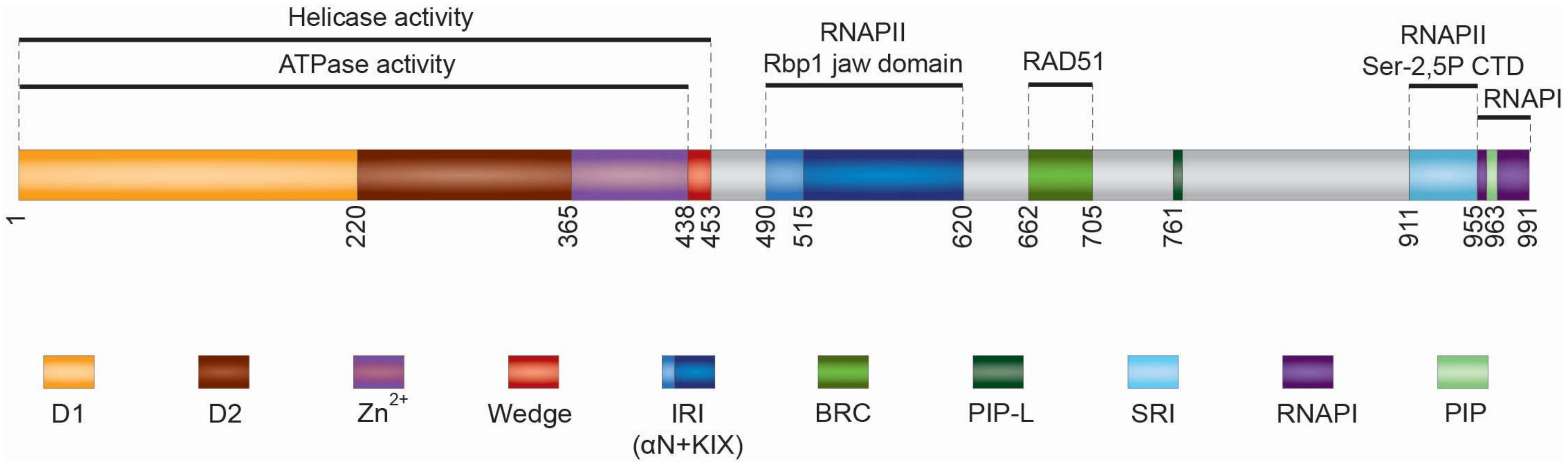
2. Structure and Biochemical Properties of RECQ5
3. RECQ5 Regulates Homologous Recombination
4. RECQ5 Regulates Transcription Elongation and Prevents Transcription Stress
5. RECQ5 Promotes Resolution of Transcription-Replication Conflicts
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Fairman-Williams, M.E.; Guenther, U.P.; Jankowsky, E. SF1 and SF2 helicases: Family matters. Curr. Opin. Struct. Biol. 2010, 20, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Chu, W.K.; Hickson, I.D. RecQ helicases: Multifunctional genome caretakers. Nat. Rev. Cancer 2009, 9, 644–654. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, K.A.; Gangloff, S.; Rothstein, R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010, 44, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Hartung, F.; Puchta, H. The RecQ gene family in plants. J. Plant. Physiol. 2006, 163, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Hickson, I.D. RecQ helicases: Caretakers of the genome. Nat. Rev. Cancer 2003, 3, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Urban, V.; Dobrovolna, J.; Janscak, P. Distinct functions of human RecQ helicases during DNA replication. Biophys. Chem. 2017, 225, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Croteau, D.L.; Popuri, V.; Opresko, P.L.; Bohr, V.A. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014, 83, 519–552. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M., Jr.; Bohr, V.A. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007, 35, 7527–7544. [Google Scholar] [CrossRef] [PubMed]
- Kitao, S.; Ohsugi, I.; Ichikawa, K.; Goto, M.; Furuichi, Y.; Shimamoto, A. Cloning of two new human helicase genes of the RecQ family: Biological significance of multiple species in higher eukaryotes. Genomics 1998, 54, 443–452. [Google Scholar] [CrossRef]
- Sekelsky, J.J.; Brodsky, M.H.; Rubin, G.M.; Hawley, R.S. Drosophila and human RecQ5 exist in different isoforms generated by alternative splicing. Nucleic Acids Res. 1999, 27, 3762–3769. [Google Scholar] [CrossRef] [PubMed]
- Shimamoto, A.; Nishikawa, K.; Kitao, S.; Furuichi, Y. Human RecQ5beta, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3alpha and 3beta. Nucleic Acids Res. 2000, 28, 1647–1655. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, Y.; Lu, X.; Barnes, E.; Yan, M.; Lou, H.; Luo, G. Recql5 and Blm RecQ DNA helicases have nonredundant roles in suppressing crossovers. Mol. Cell Biol. 2005, 25, 3431–3442. [Google Scholar] [CrossRef]
- Hu, Y.; Raynard, S.; Sehorn, M.G.; Lu, X.; Bussen, W.; Zheng, L.; Stark, J.M.; Barnes, E.L.; Chi, P.; Janscak, P.; et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007, 21, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Tang, L.; Cai, M.; Chen, H.; Wong, J.; Zhang, P. RECQL5 plays an essential role in maintaining genome stability and viability of triple-Negative breast cancer cells. Cancer Med. 2019, 8, 4743–4752. [Google Scholar] [CrossRef] [PubMed]
- Tavera-Tapia, A.; de la Hoya, M.; Calvete, O.; Martin-Gimeno, P.; Fernandez, V.; Macias, J.A.; Alonso, B.; Pombo, L.; de Diego, C.; Alonso, R.; et al. RECQL5: Another DNA helicase potentially involved in hereditary breast cancer susceptibility. Hum. Mutat. 2019, 40, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Dou, S.X.; Zhang, X.D.; Wang, P.Y.; Kanagaraj, R.; Liu, J.L.; Janscak, P.; Hu, J.S.; Xi, X.G. The zinc-Binding motif of human RECQ5beta suppresses the intrinsic strand-Annealing activity of its DExH helicase domain and is essential for the helicase activity of the enzyme. Biochem. J. 2008, 412, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.A.; Aitkenhead, H.; Savitsky, P.; Gileadi, O. Insights into the RecQ helicase mechanism revealed by the structure of the helicase domain of human RECQL5. Nucleic Acids Res. 2017, 45, 4231–4243. [Google Scholar] [CrossRef]
- Schwendener, S.; Raynard, S.; Paliwal, S.; Cheng, A.; Kanagaraj, R.; Shevelev, I.; Stark, J.M.; Sung, P.; Janscak, P. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J. Biol. Chem. 2010, 285, 15739–15745. [Google Scholar] [CrossRef]
- Islam, M.N.; Paquet, N.; Fox, D.; Dray, E.; Zheng, X.F.; Klein, H.; Sung, P.; Wang, W. A variant of the breast cancer type 2 susceptibility protein (BRC) repeat is essential for the RECQL5 helicase to interact with RAD51 recombinase for genome stabilization. J. Biol. Chem. 2012, 287, 23808–23818. [Google Scholar] [CrossRef]
- Aygun, O.; Svejstrup, J.; Liu, Y. A RECQ5-RNA polymerase II association identified by targeted proteomic analysis of human chromatin. Proc. Natl. Acad. Sci. USA 2008, 105, 8580–8584. [Google Scholar] [CrossRef]
- Aygun, O.; Xu, X.; Liu, Y.; Takahashi, H.; Kong, S.E.; Conaway, R.C.; Conaway, J.W.; Svejstrup, J.Q. Direct inhibition of RNA polymerase II transcription by RECQL5. J. Biol. Chem. 2009, 284, 23197–23203. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.N.; Fox, D.; Guo, R.; Enomoto, T.; Wang, W. RecQL5 promotes genome stabilization through two parallel mechanisms—Interacting with RNA polymerase II and acting as a helicase. Mol. Cell Biol. 2010, 30, 2460–2472. [Google Scholar] [CrossRef] [PubMed]
- Kanagaraj, R.; Huehn, D.; MacKellar, A.; Menigatti, M.; Zheng, L.; Urban, V.; Shevelev, I.; Greenleaf, A.L.; Janscak, P. RECQ5 helicase associates with the C-Terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res. 2010, 38, 8131–8140. [Google Scholar] [CrossRef] [PubMed]
- Kassube, S.A.; Jinek, M.; Fang, J.; Tsutakawa, S.; Nogales, E. Structural mimicry in transcription regulation of human RNA polymerase II by the DNA helicase RECQL5. Nat. Struct. Mol. Biol. 2013, 20, 892–899. [Google Scholar] [CrossRef]
- Urban, V.; Dobrovolna, J.; Huhn, D.; Fryzelkova, J.; Bartek, J.; Janscak, P. RECQ5 helicase promotes resolution of conflicts between replication and transcription in human cells. J. Cell Biol. 2016, 214, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Kanagaraj, R.; Saydam, N.; Garcia, P.L.; Zheng, L.; Janscak, P. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006, 34, 5217–5231. [Google Scholar] [CrossRef]
- Li, M.; Xu, X.; Chang, C.W.; Zheng, L.; Shen, B.; Liu, Y. SUMO2 conjugation of PCNA facilitates chromatin remodeling to resolve transcription-Replication conflicts. Nat. Commun. 2018, 9, 2706. [Google Scholar] [CrossRef]
- Garcia, P.L.; Liu, Y.; Jiricny, J.; West, S.C.; Janscak, P. Human RECQ5beta, a protein with DNA helicase and strand-Annealing activities in a single polypeptide. EMBO J. 2004, 23, 2882–2891. [Google Scholar] [CrossRef]
- Thakur, J.K.; Yadav, A.; Yadav, G. Molecular recognition by the KIX domain and its role in gene regulation. Nucleic Acids Res. 2014, 42, 2112–2125. [Google Scholar] [CrossRef]
- Kizer, K.O.; Phatnani, H.P.; Shibata, Y.; Hall, H.; Greenleaf, A.L.; Strahl, B.D. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell Biol. 2005, 25, 3305–3316. [Google Scholar] [CrossRef]
- Li, M.; Phatnani, H.P.; Guan, Z.; Sage, H.; Greenleaf, A.L.; Zhou, P. Solution structure of the Set2-Rpb1 interacting domain of human Set2 and its interaction with the hyperphosphorylated C-Terminal domain of Rpb1. Proc. Natl. Acad. Sci. USA 2005, 102, 17636–17641. [Google Scholar] [CrossRef] [PubMed]
- Vojnic, E.; Simon, B.; Strahl, B.D.; Sattler, M.; Cramer, P. Structure and carboxyl-Terminal domain (CTD) binding of the Set2 SRI domain that couples histone H3 Lys36 methylation to transcription. J. Biol. Chem. 2006, 281, 13–15. [Google Scholar] [CrossRef] [PubMed]
- Phatnani, H.P.; Greenleaf, A.L. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006, 20, 2922–2936. [Google Scholar] [CrossRef]
- Li, M.; Xu, X.; Liu, Y. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol. Cell Biol. 2011, 31, 2090–2099. [Google Scholar] [CrossRef] [PubMed]
- Di Marco, S.; Hasanova, Z.; Kanagaraj, R.; Chappidi, N.; Altmannova, V.; Menon, S.; Sedlackova, H.; Langhoff, J.; Surendranath, K.; Huhn, D.; et al. RECQ5 Helicase Cooperates with MUS81 Endonuclease in Processing Stalled Replication Forks at Common Fragile Sites during Mitosis. Mol. Cell 2017, 66, 658.e8–671.e8. [Google Scholar] [CrossRef] [PubMed]
- Heyer, W.D.; Ehmsen, K.T.; Liu, J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010, 44, 113–139. [Google Scholar] [CrossRef] [PubMed]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef]
- Mitchel, K.; Zhang, H.; Welz-Voegele, C.; Jinks-Robertson, S. Molecular structures of crossover and noncrossover intermediates during gap repair in yeast: Implications for recombination. Mol. Cell 2010, 38, 211–222. [Google Scholar] [CrossRef]
- Zheng, L.; Kanagaraj, R.; Mihaljevic, B.; Schwendener, S.; Sartori, A.A.; Gerrits, B.; Shevelev, I.; Janscak, P. MRE11 complex links RECQ5 helicase to sites of DNA damage. Nucleic Acids Res. 2009, 37, 2645–2657. [Google Scholar] [CrossRef]
- Paliwal, S.; Kanagaraj, R.; Sturzenegger, A.; Burdova, K.; Janscak, P. Human RECQ5 helicase promotes repair of DNA double-Strand breaks by synthesis-Dependent strand annealing. Nucleic Acids Res. 2014, 42, 2380–2390. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Seki, M.; Narita, Y.; Nakagawa, T.; Yoshimura, A.; Otsuki, M.; Kawabe, Y.; Tada, S.; Yagi, H.; Ishii, Y.; et al. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol. Cell Biol. 2003, 23, 3527–3535. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Izumikawa, K.; Yanagida, M.; Hayano, T.; Tachikawa, H.; Komatsu, W.; Shimamoto, A.; Futami, K.; Furuichi, Y.; Shinkawa, T.; Yamauchi, Y.; et al. Association of human DNA helicase RecQ5beta with RNA polymerase II and its possible role in transcription. Biochem. J. 2008, 413, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Saponaro, M.; Kantidakis, T.; Mitter, R.; Kelly, G.P.; Heron, M.; Williams, H.; Soding, J.; Stewart, A.; Svejstrup, J.Q. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 2014, 157, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Helmrich, A.; Ballarino, M.; Tora, L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell 2011, 44, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Muse, T.; Aguilera, A. Transcription-Replication conflicts: How they occur and how they are resolved. Nat. Rev. Mol. Cell Biol. 2016, 17, 553–563. [Google Scholar] [CrossRef]
- Macheret, M.; Halazonetis, T.D. Intragenic origins due to short G1 phases underlie oncogene-Induced DNA replication stress. Nature 2018, 555, 112–116. [Google Scholar] [CrossRef]
- Hamperl, S.; Cimprich, K.A. The contribution of co-Transcriptional RNA: DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst.) 2014, 19, 84–94. [Google Scholar] [CrossRef]
- Hamperl, S.; Bocek, M.J.; Saldivar, J.C.; Swigut, T.; Cimprich, K.A. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell 2017, 170, 774.e19–786.e19. [Google Scholar] [CrossRef]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.W.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef]
- Bhowmick, R.; Minocherhomji, S.; Hickson, I.D. RAD52 Facilitates Mitotic DNA Synthesis Following Replication Stress. Mol. Cell 2016, 64, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-Strand break repair-Independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Chappidi, N.; Nascakova, Z.; Boleslavska, B.; Zellweger, R.; Isik, E.; Andrs, M.; Menon, S.; Dobrovolna, J.; Balbo Pogliano, C.; Matos, J.; et al. Fork Cleavage-Religation Cycle and Active Transcription Mediate Replication Restart after Fork Stalling at Co-Transcriptional R-Loops. Mol. Cell 2020, 77, 528–541. [Google Scholar] [CrossRef] [PubMed]
- Zellweger, R.; Dalcher, D.; Mutreja, K.; Berti, M.; Schmid, J.A.; Herrador, R.; Vindigni, A.; Lopes, M. Rad51-Mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 2015, 208, 563–579. [Google Scholar] [CrossRef]
- Neelsen, K.J.; Lopes, M. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015, 16, 207–220. [Google Scholar] [CrossRef]
- Mourgues, S.; Gautier, V.; Lagarou, A.; Bordier, C.; Mourcet, A.; Slingerland, J.; Kaddoum, L.; Coin, F.; Vermeulen, W.; Gonzales de Peredo, A.; et al. ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc. Natl. Acad. Sci. USA 2013, 110, 17927–17932. [Google Scholar] [CrossRef]
- Helmrich, A.; Ballarino, M.; Nudler, E.; Tora, L. Transcription-Replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013, 20, 412–418. [Google Scholar] [CrossRef]
- Li, M.; Pokharel, S.; Wang, J.T.; Xu, X.; Liu, Y. RECQ5-Dependent SUMOylation of DNA topoisomerase I prevents transcription-Associated genome instability. Nat. Commun. 2015, 6, 6720. [Google Scholar] [CrossRef]
- Wasserman, M.R.; Schauer, G.D.; O’Donnell, M.E.; Liu, S. Replication Fork Activation Is Enabled by a Single-Stranded DNA Gate in CMG Helicase. Cell 2019, 178, 600–611. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrs, M.; Hasanova, Z.; Oravetzova, A.; Dobrovolna, J.; Janscak, P. RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription. Genes 2020, 11, 232. https://doi.org/10.3390/genes11020232
Andrs M, Hasanova Z, Oravetzova A, Dobrovolna J, Janscak P. RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription. Genes. 2020; 11(2):232. https://doi.org/10.3390/genes11020232
Chicago/Turabian StyleAndrs, Martin, Zdenka Hasanova, Anna Oravetzova, Jana Dobrovolna, and Pavel Janscak. 2020. "RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription" Genes 11, no. 2: 232. https://doi.org/10.3390/genes11020232
APA StyleAndrs, M., Hasanova, Z., Oravetzova, A., Dobrovolna, J., & Janscak, P. (2020). RECQ5: A Mysterious Helicase at the Interface of DNA Replication and Transcription. Genes, 11(2), 232. https://doi.org/10.3390/genes11020232



