Heteroplasmy and Copy Number in the Common m.3243A>G Mutation—A Post-Mortem Genotype–Phenotype Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient
2.2. Literature Cases
2.3. Muscle Histopathology-Patient
2.4. Restriction Fragment Length Polymorphism (RFLP)-Polymerase Chain Reaction (PCR)-Patient
2.5. Determination of mtDNA Copy Numbers-Patient
2.6. Ethical Statement
2.7. Statistical Analysis
3. Results
3.1. Muscle Histopathology (Patient)
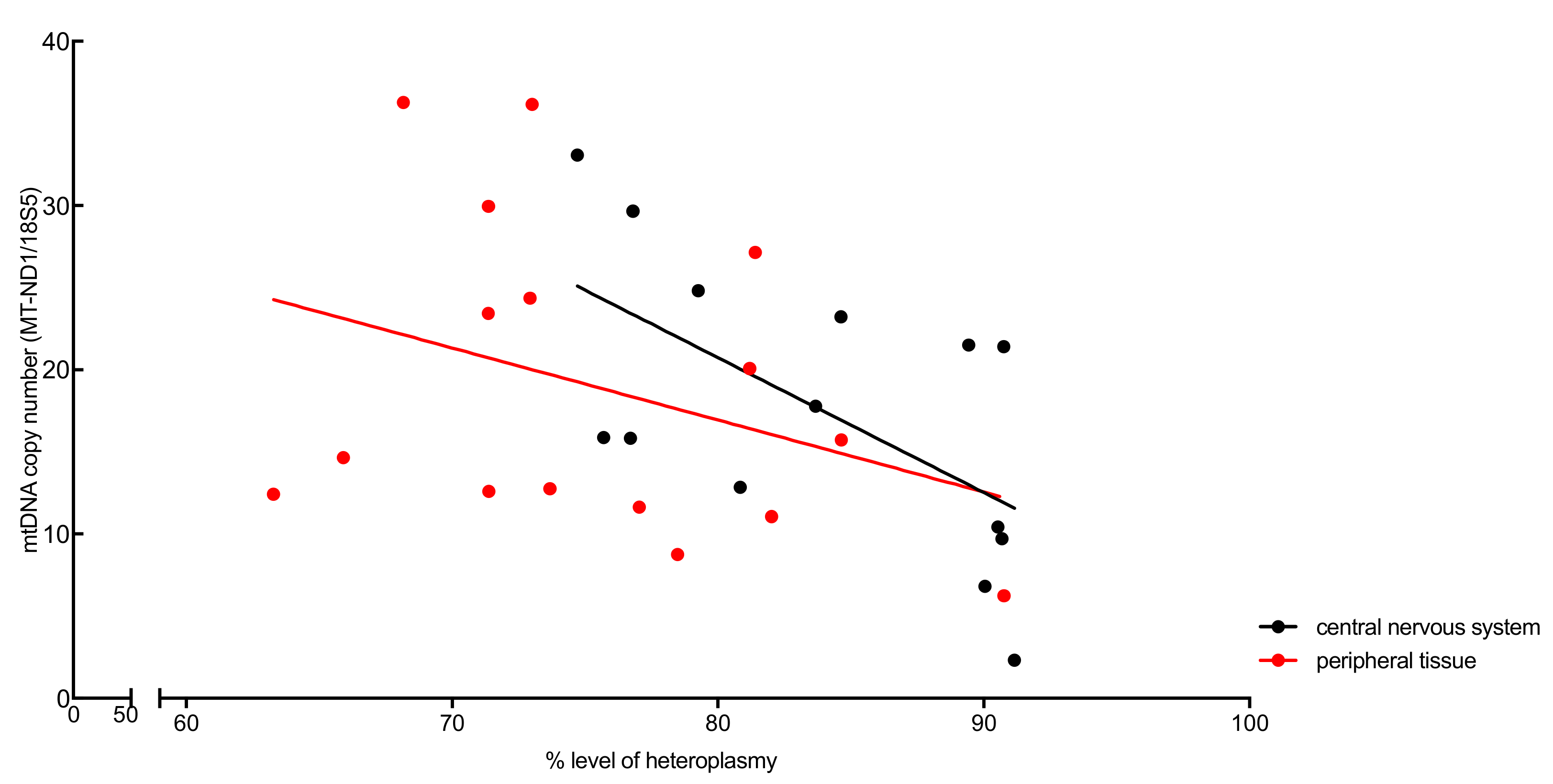
3.2. Analysis of Heteroplasmy Levels and Copy Number (Patient)
3.3. Analysis of Clinical Phenotype and Genotype in the Patient and Literature Cases
4. Discussion
5. Limitations
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- O’Hara, R.; Tedone, E.; Ludlow, A.; Huang, E.; Arosio, B.; Mari, D.; Shay, J.W. Quantitative mitochondrial DNA copy number determination using droplet digital PCR with single-cell resolution. Genome Res. 2019, 29, 1878–1888. [Google Scholar] [CrossRef] [PubMed]
- Schon, E.A.; DiMauro, S.; Hirano, M. Human mitochondrial DNA: Roles of inherited and somatic mutations. Nat. Rev. Genet. 2012, 13, 878–890. [Google Scholar] [CrossRef] [PubMed]
- Grady, J.P.; Murphy, J.L.; Blakely, E.L.; Haller, R.G.; Taylor, R.W.; Turnbull, D.M.; Tuppen, H.A. Accurate measurement of mitochondrial DNA deletion level and copy number differences in human skeletal muscle. PLoS ONE 2014, 9, e114462. [Google Scholar] [CrossRef] [PubMed]
- Majamaa, K.; Moilanen, J.S.; Uimonen, S.; Remes, A.M.; Salmela, P.I.; Karppa, M.; Majamaa-Voltti, K.A.; Rusanen, H.; Sorri, M.; Peuhkurinen, K.J.; et al. Epidemiology of A3243G, the mutation for mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes: Prevalence of the mutation in an adult population. Am. J. Hum. Genet. 1998, 63, 447–454. [Google Scholar] [CrossRef]
- Lehmann, D.; Schubert, K.; Joshi, P.R.; Baty, K.; Blakely, E.L.; Zierz, S.; Taylor, R.W.; Deschauer, M. A novel m.7539C>T point mutation in the mt-tRNAAsp gene associated with multisystemic mitochondrial disease. Neuromuscul. Disord. NMD 2015, 25, 81–84. [Google Scholar] [CrossRef]
- El-Hattab, A.W.; Scaglia, F. Mitochondrial DNA depletion syndromes: Review and updates of genetic basis, manifestations, and therapeutic options. Neurotherapeutics 2013, 10, 186–198. [Google Scholar] [CrossRef]
- Grady, J.P.; Pickett, S.J.; Ng, Y.S.; Alston, C.L.; Blakely, E.L.; Hardy, S.A.; Feeney, C.L.; Bright, A.A.; Schaefer, A.M.; Gorman, G.S.; et al. mtDNA heteroplasmy level and copy number indicate disease burden in m.3243A>G mitochondrial disease. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Pickett, S.J.; Grady, J.P.; Ng, Y.S.; Gorman, G.S.; Schaefer, A.M.; Wilson, I.J.; Cordell, H.J.; Turnbull, D.M.; Taylor, R.W.; McFarland, R. Phenotypic heterogeneity in m.3243A>G mitochondrial disease: The role of nuclear factors. Ann. Clin. Transl. Neurol. 2018, 5, 333–345. [Google Scholar] [CrossRef]
- Rahman, S.; Poulton, J.; Marchington, D.; Suomalainen, A. Decrease of 3243 A→G mtDNA mutation from blood in MELAS syndrome: A longitudinal study. Am. J. Hum. Genet. 2001, 68, 238–240. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ichihashi, K.; Ohta, S.; Nihei, K.; Kagawa, Y.; Yanagisawa, M.; Momoi, M.Y. The mutant mitochondrial genes in mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS) were selectively amplified through generations. J. Inherit. Metab. Dis. 1992, 15, 803–808. [Google Scholar] [CrossRef]
- Goto, Y.; Nonaka, I.; Horai, S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 1990, 348, 651–653. [Google Scholar] [CrossRef] [PubMed]
- Hamazaki, S.; Koshiba, M.; Sugiyama, T. Organ distribution of mutant mitochondrial tRNAleu(UUR) gene in a MELAS patient. Acta Pathol. Jpn. 1993, 43, 187–191. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Narayan, B.; Prasad, A.N.; Rupar, C.A.; Levin, S.; Kronick, J.; Ramsay, D.; Tay, K.Y.; Prasad, C. MELAS: A multigenerational impact of the MTTL1 A3243G MELAS mutation. Can. J. Neurol. Sci. 2014, 41, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Enter, C.; Muller-Hocker, J.; Zierz, S.; Kurlemann, G.; Pongratz, D.; Forster, C.; Obermaier-Kusser, B.; Gerbitz, K.D. A specific point mutation in the mitochondrial genome of Caucasians with MELAS. Hum. Genet. 1991, 88, 233–236. [Google Scholar] [CrossRef]
- Fornuskova, D.; Brantova, O.; Tesarova, M.; Stiburek, L.; Honzik, T.; Wenchich, L.; Tietzeova, E.; Hansikova, H.; Zeman, J. The impact of mitochondrial tRNA mutations on the amount of ATP synthase differs in the brain compared to other tissues. Biochim. Biophys. Acta 2008, 1782, 317–325. [Google Scholar] [CrossRef][Green Version]
- Cardaioli, E.; Dotti, M.T.; Hayek, G.; Zappella, M.; Federico, A. Studies on mitochondrial pathogenesis of Rett syndrome: Ultrastructural data from skin and muscle biopsies and mutational analysis at mtDNA nucleotides 10463 and 2835. J. Submicrosc. Cytol. Pathol. 1999, 31, 301–304. [Google Scholar]
- Ciafaloni, E.; Ricci, E.; Servidei, S.; Shanske, S.; Silvestri, G.; Manfredi, G.; Schon, E.A.; DiMauro, S. Widespread tissue distribution of a tRNALeu(UUR) mutation in the mitochondrial DNA of a patient with MELAS syndrome. Neurology 1991, 41, 1663–1664. [Google Scholar] [CrossRef]
- Macmillan, C.; Lach, B.; Shoubridge, E.A. Variable distribution of mutant mitochondrial DNAs (tRNA(Leu[3243])) in tissues of symptomatic relatives with MELAS: The role of mitotic segregation. Neurology 1993, 43, 1586–1590. [Google Scholar] [CrossRef]
- Obermaier-Kusser, B.; Paetzke-Brunner, I.; Enter, C.; Muller-Hocker, J.; Zierz, S.; Ruitenbeek, W.; Gerbitz, K.D. Respiratory chain activity in tissues from patients (MELAS) with a point mutation of the mitochondrial genome [tRNA(Leu(UUR))]. FEBS Lett. 1991, 286, 67–70. [Google Scholar] [CrossRef]
- Shiraiwa, N.; Ishii, A.; Iwamoto, H.; Mizusawa, H.; Kagawa, Y.; Ohta, S. Content of mutant mitochondrial DNA and organ dysfunction in a patient with a MELAS subgroup of mitochondrial encephalomyopathies. J. Neurol. Sci. 1993, 120, 174–179. [Google Scholar] [CrossRef]
- Shoji, Y.; Sato, W.; Hayasaka, K.; Takada, G. Tissue distribution of mutant mitochondrial DNA in mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS). J. Inherit. Metab. Dis. 1993, 16, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Koga, Y.; Akita, Y.; Takane, N.; Sato, Y.; Kato, H. Heterogeneous presentation in A3243G mutation in the mitochondrial tRNA(Leu(UUR)) gene. Arch. Dis. Child. 2000, 82, 407–411. [Google Scholar] [CrossRef] [PubMed]
- Matthews, P.M.; Hopkin, J.; Brown, R.M.; Stephenson, J.B.; Hilton-Jones, D.; Brown, G.K. Comparison of the relative levels of the 3243 (A→G) mtDNA mutation in heteroplasmic adult and fetal tissues. J. Med. Genet. 1994, 31, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Rossignol, R.; Faustin, B.; Rocher, C.; Malgat, M.; Mazat, J.P.; Letellier, T. Mitochondrial threshold effects. Biochem. J. 2003, 370, 751–762. [Google Scholar] [CrossRef] [PubMed]
- Wai, T.; Teoli, D.; Shoubridge, E.A. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat. Genet. 2008, 40, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Rajasimha, H.K.; Chinnery, P.F.; Samuels, D.C. Selection against pathogenic mtDNA mutations in a stem cell population leads to the loss of the 3243A→G mutation in blood. Am. J. Hum. Genet. 2008, 82, 333–343. [Google Scholar] [CrossRef]
- Sue, C.M.; Quigley, A.; Katsabanis, S.; Kapsa, R.; Crimmins, D.S.; Byrne, E.; Morris, J.G. Detection of MELAS A3243G point mutation in muscle, blood and hair follicles. J. Neurol. Sci. 1998, 161, 36–39. [Google Scholar] [CrossRef]
- Filograna, R.; Koolmeister, C.; Upadhyay, M.; Pajak, A.; Clemente, P.; Wibom, R.; Simard, M.L.; Wredenberg, A.; Freyer, C.; Stewart, J.B.; et al. Modulation of mtDNA copy number ameliorates the pathological consequences of a heteroplasmic mtDNA mutation in the mouse. Sci. Adv. 2019, 5, eaav9824. [Google Scholar] [CrossRef]
- Picard, M.; Zhang, J.; Hancock, S.; Derbeneva, O.; Golhar, R.; Golik, P.; O’Hearn, S.; Levy, S.; Potluri, P.; Lvova, M.; et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc. Natl. Acad. Sci. USA 2014, 111, E4033–E4042. [Google Scholar] [CrossRef]
- Wallace, D.C.; Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 2010, 10, 12–31. [Google Scholar] [CrossRef]
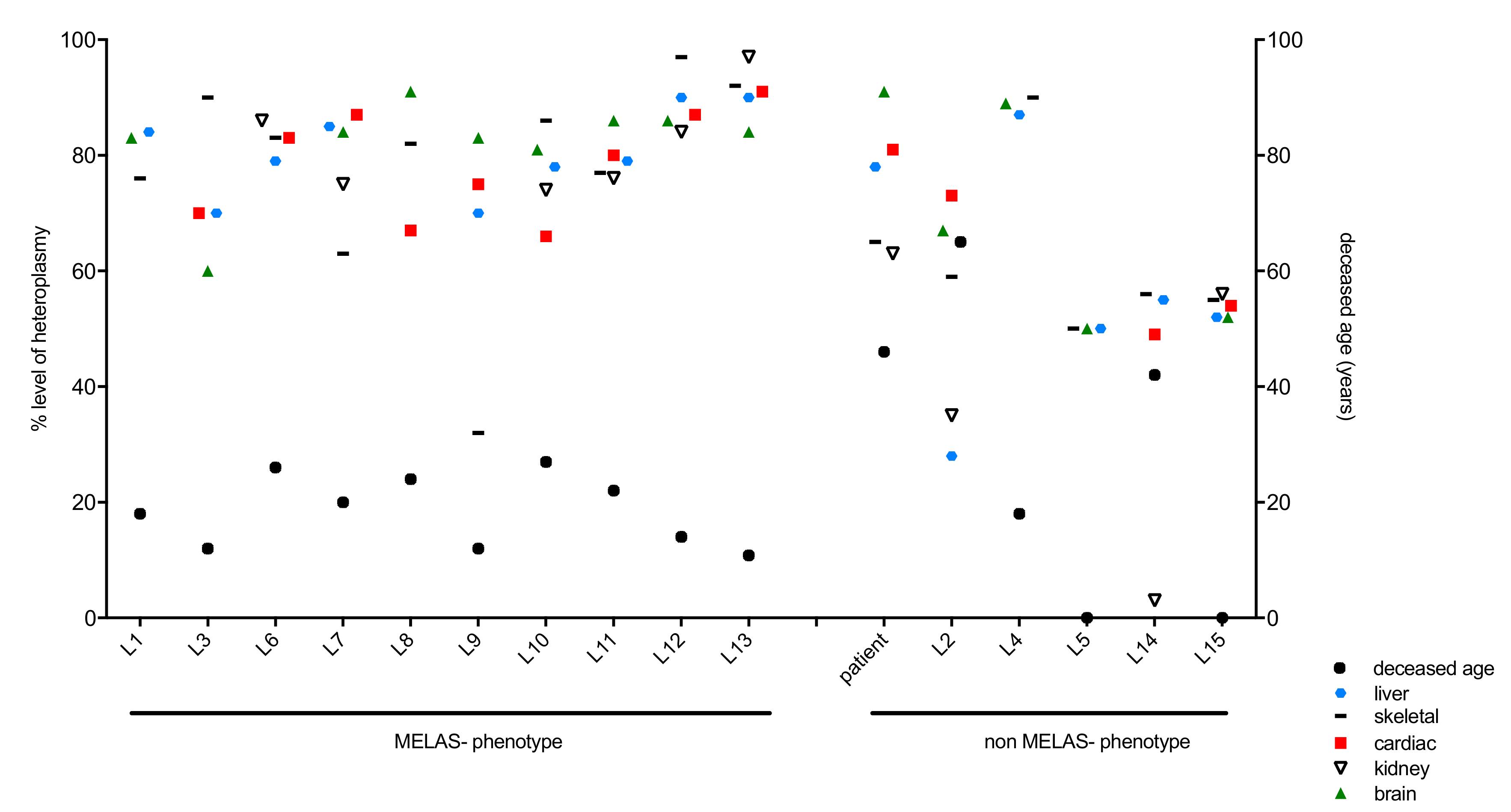


| Literature Case Number | Reference | Sex | MELAS-Phenotype | Deceased Age (Years) | Methods of mtDNA Heteroplasmy Determination |
|---|---|---|---|---|---|
| L1 | Prasad et al. 2014 [13] | Male | Yes | 18 | PCR, restriction enzyme ApaI; quantitative densitometry |
| L2 | Prasad et al. 2014 [13] | Female | No | 65 | PCR, restriction enzyme ApaI; quantitative densitometry |
| L3 | Enter et al. 1991 [14] | Female | Yes | 12 | PCR, restriction endonuclease ApaI; mutation detection by PCR |
| L4 | Fornuskova et al. 2008 [15] | Female | No | 18 | PCR, restriction enzyme ApaI; quantified with ImageQuant software |
| L5 | Cardaioli et al. 1999 [16] | stillbirth | No | Miscarriage (25 weeks) | PCR, restriction enzyme ApaI, autoradiography–quantified with ultra-scan densitometer |
| L6 | Ciafaloni et al. 1991 [17] | Male | Yes | 26 | PCR, restriction enzyme HaeIII, quantified with Betascope 603 blot analyser. |
| L7 | Kobayashi et al. 1992 [10] | n.k. | Yes | 20 | PCR, restriction enzyme ApaI; autoradiography, quantified with Fujix bioimaging analyser (BAS2000) |
| L8 | Macmillan et al. 1993 [18] | Female | Yes | 24 | PCR, restriction enzyme ApaI; quantified on a molecular dyanmics series 4000 phosphorImager |
| L9 | Obermaier-Kusser et al. 1991 [19] | Female | Yes | 12 | PCR, restriction enzyme ApaI; densitometry using Elscript 400 UVR scanner |
| L10 | Shiraiwa et al. 1993 [20] | Female | Yes | 27 | PCR, restriction enzyme ApaI; autoradiography, quantified with Fujix bioimaging analyser (BAS2000) |
| L11 | Shoji et al. 1993 [21] | Female | Yes | 22 | PCR, restriction enzyme ApaI; autoradiography, quantified with a densitometer |
| L12 | Hamazaki et al. 1993 [12] | Female | Yes | 14 | PCR, restriction enzyme ApaI; quantified using laser densitometer |
| L13 | Koga et al. 2000 [22] | Male | Yes | 10.8 | PCR, restriction enzyme HaeIII, quantified using a Betascope 603 blot analyser. |
| L14 | Matthews et al. 1994 [23] | Female | No | 42 | PCR, restriction enzyme ApaI; analysed densitometrically from film negatives |
| L15 | Matthews et al. 1994 [23] | n.k. | No | stillborn (24 weeks) | PCR, restriction enzyme ApaI; analysed densitometrically from film negatives |
| Tissue Type (Origin) | Heteroplasmy Levels (%) by RFLP | mtDNA Copy Number MT-ND1/18S5 |
|---|---|---|
| Central nervous system | ||
| Putamen | 80.84 | 12.85 |
| Caudate nucleus | 79.26 | 24.81 |
| Mammillary body | 76.80 | 29.66 |
| Hypothalamus | 75.70 | 15.88 |
| Thalamus | 74.71 | 33.06 |
| Internal capsule | 76.71 | 15.83 |
| Globus pallidus | 84.63 | 23.22 |
| Visual cortex | 83.67 | 17.79 |
| Pons | 90.04 | 6.81 |
| Medulla oblongata | 90.75 | 21.41 |
| White matter | 90.69 | 9.72 |
| Cerebellar vermis | 89.43 | 21.51 |
| Spinal cord | 90.53 | 10.42 |
| Dorsal root ganglion | 91.16 | 2.31 |
| Peripheral organs | ||
| Liver | 78.48 | 8.75 |
| Bladder | 82.02 | 11.06 |
| M. obliqus superior | 72.93 | 24.35 |
| M. rectus medialis | 71.37 | 29.95 |
| M. rectus superior | 73.02 | 36.17 |
| M. rectus inferior | 71.36 | 23.43 |
| M. iliopsoas | 68.16 | 36.29 |
| M. vastus lat. | 65.91 | 14.65 |
| M. pectoralis | 73.68 | 12.76 |
| M. biceps brachii | 81.20 | 20.08 |
| Tongue muscle base | 84.64 | 15.73 |
| Heart muscle (left) | 81.40 | 27.16 |
| Heart muscle (right) | 77.04 | 11.64 |
| AV-node | 71.38 | 12.60 |
| Kidney | 63.28 | 12.42 |
| Sural nerve | 90.76 | 6.25 |
| Significant | p-Value | MELAS (Mean) | Non-MELAS (Mean) | |
|---|---|---|---|---|
| Deceased Age | No | 0.266411820 | 18.58 | 28.53 |
| Skeletal | No | 0.108486887 | 77.8 | 62.5 |
| Cardiac | No | 0.059542563 | 78.44 | 64.25 |
| Liver | Yes | 0.011731624 | 80.56 | 58.33 |
| Kidney | Yes | 0.006090533 | 82 | 39.25 |
| Brain | No | 0.059855928 | 82.89 | 68.2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Motlagh Scholle, L.; Zierz, S.; Mawrin, C.; Wickenhauser, C.; Lehmann Urban, D. Heteroplasmy and Copy Number in the Common m.3243A>G Mutation—A Post-Mortem Genotype–Phenotype Analysis. Genes 2020, 11, 212. https://doi.org/10.3390/genes11020212
Motlagh Scholle L, Zierz S, Mawrin C, Wickenhauser C, Lehmann Urban D. Heteroplasmy and Copy Number in the Common m.3243A>G Mutation—A Post-Mortem Genotype–Phenotype Analysis. Genes. 2020; 11(2):212. https://doi.org/10.3390/genes11020212
Chicago/Turabian StyleMotlagh Scholle, Leila, Stephan Zierz, Christian Mawrin, Claudia Wickenhauser, and Diana Lehmann Urban. 2020. "Heteroplasmy and Copy Number in the Common m.3243A>G Mutation—A Post-Mortem Genotype–Phenotype Analysis" Genes 11, no. 2: 212. https://doi.org/10.3390/genes11020212
APA StyleMotlagh Scholle, L., Zierz, S., Mawrin, C., Wickenhauser, C., & Lehmann Urban, D. (2020). Heteroplasmy and Copy Number in the Common m.3243A>G Mutation—A Post-Mortem Genotype–Phenotype Analysis. Genes, 11(2), 212. https://doi.org/10.3390/genes11020212




