Abstract
Carabidae are one of the most species-rich families of beetles, comprising more than 40,000 described species worldwide. Forty-three complete or partial mitochondrial genomes (mitogenomes) from this family have been published in GenBank to date. In this study, we sequenced a nearly complete mitogenome of Amara aulica (Carabidae), using a next-generation sequencing method. This mitogenome was 16,646 bp in length, which encoded the typical 13 mitochondrial protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a putative control region. Combining with the published mitogenomes of Carabidae and five outgroup species from Trachypachidae, Gyrinidae and Dytiscidae, we performed phylogenetic estimates under maximum likelihood and Bayesian inference criteria to investigate the phylogenetic relationships of carabid beetles. The results showed that the family Carabidae was a non-monophyletic assemblage. The subfamilies Cicindelinae, Elaphrinae, Carabinae, Trechinae and Harpalinae were recovered as monophyletic groups. Moreover, the clade (Trechinae + (Brachininae + Harpalinae)) was consistently recovered in all analyses.
1. Introduction
The Carabidae, also known as carabid beetles or ground beetles, are among the most species-rich families in Caraboidea. They currently comprise more than 40,000 described species worldwide, which can be classified into 16 subfamilies [1] and 86 tribes [2,3,4]. Carabid beetles are often considered as indicators of ecological changes, and are used as the biocontrol agents against insect pests in crops [5,6,7]. Furthermore, some researches indicated that carabids could contribute to weed management in agroecosystems (as reviewed in [8]).
The taxonomy of carabid beetles has been extensively studied. Traditionally, phylogenetic reconstructions of carabids are based on the morphological characters, for example, the male [9,10] and female genitalia [11] and the wing folding structures [12]. Liebherr and Will (1998) recovered Carabidae as a non-monophyletic assemblage, with the characters of the female reproductive tract [13]. By analyzing the larval morphology, Arndt (1998) retrieved Carabidae as a monophyletic group, with the members of Rhysodidae excluded [14]. Kavanaugh (1998) investigated the relationships among the basal carabids and recovered Trachypachidae as sister to all carabid taxa [15]. The Cicindelinae (tiger beetles) was found to be related to the tribes Carabini, Cychrini, Cicindelini and Omophronini [15]. Grebennikov and Maddison (2005) analyzed the phylogenetic relationships within the supertribe Trechitae based on larval morphology [16]. Beutel et al. (2006) applied morphological characters of adults and larvae to recover Carabidae as a sister to Omoglymmius (Rhysodidae), which together form a sister group of Trachypachidae [17]. Studies on morphology of defense glands [18,19,20] and those on karyotypes [21,22] of some carabid species also contributed to understanding of the phylogeny of Carabidae.
Molecular data can be used to address problems when morphological evidence have been conflicting or difficult to interpret. Based on 18S rDNA sequences, Maddison et al. (1999) supported Carabidae (including cicindelines, rhysodines and paussines) as monophyletic and that Brachinini probably was a sister group to Harpalinae [23]. Their results also assumed Harpalinae, Cicindelinae, Rhysodinae and Paussinae to be closely related to each other. However, their further analyses based on expanding molecular data (18S rDNA, 28S rDNA and wingless gene) recovered Carabidae as non-monophyletic, with respect to the trachypachid beetles [24].
Gough et al. (2019) recovered Cicindelinae as a sister group to the subfamily Rhysodinae, and placed the tribe Megacephalini nested within Platychilini in Cicidelinae [25]. Maddison et al. (2019) inferred the phylogeny of the supertribe Trechitae based on two nuclear ribosomal genes (18S rDNA and 28S rDNA) and four nuclear protein-coding genes (wingless gene, carbamoyl phosphate synthetase domain of the rudimentary gene, arginine kinase gene and muscle-specific protein 300 gene) [26]. In addition, some molecular studies had attempted the phylogenetic reconstructions at the genus or subgenus levels (Bembidion: Maddison, 2012; Carabus: Deuve et al., 2012; Ohomopterus: Sota and Vogler, 2003; Pamborus: Sota et al., 2005; Paraphaenops: Ortuño et al., 2017; Pterostichus: Sasakawa and Kubota, 2007) [27,28,29,30,31,32].
Recent studies on the suborders of Coleoptera or on the whole Coleoptera phylogeny also involved the exemplars of Carabidae. Hunt et al. (2007) [33] suggested that the monophyletic Geadephaga (comprising Trachypachidae, Rhysodidae and Carabidae including cicindelines) [34] formed a sister group to (Hydradephaga + Derodontoidea). At the subfamily level, the Harpalinae was strongly supported as a sister group to Paussinae, while the Cicindelinae was placed in a derived position and sister to a clade of (Rhysodinae + Migadopinae) [33]. Bocak et al. (2014) recovered the monophyletic Cicindelinae as a sister group to Haliplidae [35]. Timmermans et al. (2016) supported Carabidae as non-monophyletic and that tiger beetles were recognized as a separate family (namely Cicindelidae) [36]. Crampton-Platt et al. (2015) clustered the families Carabidae, Tenebrionidae, Coccinellidae and Ptilodactylidae in a clade to form the superfamily Caraboidea, which is sister to Archostemata [37]. In the study of Mckenna et al. (2015), the monophyletic Geadephaga was retrieved as sister to Hydradephaga, whereas the Carabidae was shown to be non-monophyletic with respect to Trachypachidae and Rhysodidae [38]. Baca et al. (2017) inferred Hydradephaga as a paraphyletic group, with Gyrinidae sister to Geadephaga (containing families Carabidae and Trachypachidae) [39]. López-López and Volger (2017) supported Geadephaga and Hydradephaga as two independent lineages based on the mitogenomic data, and recovered cicindelids and trachypachids as sister to all other Geadephaga [40]. Moreover, the authors suggested that the groups of cicindelids and trachypachids deserved the family status, namely, the Cicindelidae and Trachypachidae. Zhang et al. (2018) supported the monophyly of Carabidae and the most-basal position of Cicindela (Cicindelinae) within Carabidae [41]. In summary, resolving the phylogenetic relationships among these taxonomic groups is important and deserves further investigation.
The harpaline carabid beetles (Carabidae, Harpalinae) diversified rapidly during the Cretaceous period [42,43]. The Harpalinae includes more than 19,000 described species in the world [44], which is the largest subfamily of Carabidae. Harpalines are in appearance, anatomy, ecology and behavior a highly diverse group. The monophyly of Harpalinae seems uncontentious. Morphological characters uniting harpalines have been summarized in the study of Ober (2002) [45]. Some molecular studies recovered Harpalinae as a monophyletic group [4,23,45]. However, in the analysis of [24], Harpalinae was retrieved as non-monophyletic due to the embedded placement of Brachinini (Carabidae: Brachininae). In addition, the tribe Lebiini in Harpalinae was proposed as the rank of subfamily (Lebiinae) by some authors [46,47].
In recent years, sequences of mitochondrial genome (mitogenome) have been widely used to investigate insect phylogenetic relationships, molecular evolution and conservation genetics [36,37,48,49,50,51,52]. As a class of molecular marker, the mitogenome has the characteristics of maternal inheritance, rapid evolution rate, simple genetic structure and rare recombination [53]. The typical insect mitochondrial genome is a closed-circular and double-stranded DNA molecule of nearly 16 kb in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and one large AT-rich noncoding control region. The mitogenome provides an increasingly complete picture of phylogenetic relationships of insects through a large number of taxon sampling [51,54]. With the development of sequencing technology, next-generation sequencing (NGS) provides a much more cost-effective and time-saving method to generate a great number of mitogenome sequences simultaneously [37,51,55].
In this paper, we sequenced the nearly complete mitogenome of Amara aulica from the subfamily Harpalinae, by using a next-generation sequencing method. Combined with other 48 published mitogenome sequences, we reconstructed the phylogenetic relationships of the main lineages in Carabidae, under the maximum likelihood (ML) and Bayesian inference (BI) criteria.
2. Materials and Methods
2.1. Sampling and DNA Extraction
The species A. aulica is native to Europe and has been introduced to Asia and North America [56,57,58]. Adult specimens were collected from Zhengzhou, Henan Province (the geospatial coordinates: 34.723° N, 113.635° E). No specific permits were required for the insects sampled for this study.
After the samples were directly killed and preserved in absolute ethanol, they were stored in the dark at −20 °C in Entomological Museum of Henan Agricultural University (voucher number: EMHAU-2015-Zz122902) for further experiment. Total genomic DNA of the individual specimen was extracted from the thorax with the TIANamp Micro DNA kit (TIANGEN BIOTECH CO., LTD, Beijing, China), following the manufacturer’s instructions.
2.2. Mitochondrial Genome Sequencing and Assembly
Next-generation-sequencing (NGS) technology was applied to obtain the mitogenome sequences. Genomic DNA was pooled with other insect species, which had a distantly phylogenetic relationship to A. aulica. In the pool, the DNA concentrations were approximately equimolar. The library was constructed by using the Illumina TruSeqTM DNA Sample Prep Kit (Illumina, San Diego, CA, USA), with the insert size of 350 bp. Following sequencing was conducted on an Illumina HiSeq2500 platform at Shanghai OE Biotech CO., LTD, with the 150-base paired-end strategy.
NGS QC toolkit [59] was used to filter raw data for quality control. The high-quality reads were assembled using IDBA-UD v. 1.1.1 (Hong Kong, China) [60], with the following settings: the minimum size of contig of 200, an initial k-mer size of 40, an iteration size of 10 and a maximum k-mer size of 90. Three mitochondrial gene fragments (cox1, cob and rrnL) were pre-sequenced for bait sequences, by using traditional polymerase chain reaction and Sanger sequencing. The primers for the polymerase chain reactions were used as those in Song et al. (2016b) [61]. The local-blasting searches were implemented in BioEdit [62], in order to identify the mitochondrial contig.
2.3. Mitogenome Annotation and Analysis
The initial mitogenome annotation was conducted in MITOS web [63]. The start codon, stop codon and length of each protein-coding gene were further checked and adjusted by alignment to the published carabid beetle mitogenomes in GenBank. The mitogenome organization of A. aulica was presented in Table 1. The genome structure image was generated in CGView (http://stothard.afns.ualberta.ca/cgview_server/) (Figure 1). The composition skew was calculated based on the AT-skew = (A − T)/(A + T) and GC-skew = (G − C)/(G + C) formulas [64]. The newly determined mitogenome sequence of A. aulica was deposited in GenBank, accession number MN335930.

Table 1.
Organization of the Amara aulica mitochondrial genome.

Figure 1.
The structure of the mitochondrial genome of Amara aulica. Arrows indicate the direction of gene transcription. The inner circles show the GC content and the GC-skew values.
2.4. Sequence Alignment
Our taxon sample included 49 beetle mitogenome sequences representing 12 subfamilies of Carabidae (44 taxa) and three families of Trachypachidae, Gyrinidae and Dytiscidae as outgroups (five taxa) (Table 2). The protein-coding genes were aligned separately using TranslatorX [65] with the following parameters: Genetic code = “invertebrate mitochondrial”, Protein alignment = “MAFFT”, and the stop codons were excluded. Both the mitochondrial tRNA and rRNA genes were aligned using the program MAFFT under the iterative refinement method of “E-INS-i” [66]. The alignments were checked in MEGA 7 [67] and ambiguously aligned positions were manually excluded. Gaps were pruned using the online version of Gap Strip/Squeeze v2.1.0, with 40% Gap tolerance. Finally, the resulting alignments were concatenated together to make the dataset of PCGRNA (including 13 protein-coding genes, 22 tRNA genes two and rRNA genes), with the Perl script FASconCAT_v1.0 [68]. The mean ka (nonsynonymous substitution rate) and ks (synonymous substitution rate) values were calculated using DnaSP version 5 (Barcelona, Spain) [69].

Table 2.
List of the species included in this study.
2.5. Phylogenetic Analyses
In the phylogenetic analyses, our taxon sample included 46 beetle species representing 12 subfamilies of Carabidae, namely, Brachininae, Broscinae, Carabinae, Cicindelinae, Elaphrinae, Harpalinae, Nebriinae, Paussinae, Promecognathinae, Rhysodinae, Scaritinae and Trechinae. In addition, two mitogenome sequences from Dytiscidae and Gyrinidae respectively, and one from Trachypachidae were selected as outgroups. A total of 49 mitogenome sequences representing the taxa described above were compiled to make the data matrix of 49taxa_PCGRNA.
Phylogenetic trees were built based on the dataset of 49taxa_PCGRNA, under the maximum likelihood and Bayesian inferences. Maximum likelihood analysis was carried out using IQ-TREE [70] and applied the data partition schemes and best-fitting models pre-determined by PartitionFinder 2 [71] (Table S1). The data blocks were defined by genes and codon positions. Branch support was assessed using fast bootstrap analysis with 10,000 replicates. The Bayesian analysis was performed using PhyloBayes MPI v.1.5a [72]. Two parallel runs with four chains were performed, and started from a random topology. The site-heterogeneous CAT-GTR model was used for the analysis, which was originally developed to reduce long-branch attraction artifacts by modelling site-specific features of sequence evolution [73]. Convergence of runs was assessed using bpcomp program implemented in PhyloBayes to ensure that analyses had reached stationarity and that the maxdiff value was less than 0.1. Trees sampled after the burn-in from the two runs were combined and used to build a 50% majority rule consensus tree, with bpcomp program.
To investigate the potential effect of long-branch taxa on tree reconstruction, we compiled a reduced taxon dataset, namely the dataset of 48taxa_PCGRNA. In which, the long-branched Rhysodes sp. was removed. The same phylogenetic analyses were repeated with the dataset of 48taxa_PCGRNA. The sequence alignments supporting the phylogenetic results generated in this article are available in figshare (DOI: 10.6084/m9.figshare.11669280).
3. Results
3.1. Next-Generation Sequencing Output and Mitogenome Organization
In total, 4,110,380 mapped bases were generated by sequencing from the Illumina HiSeq2500. The mean base coverage of the mitochondrial contig was 248-fold. The nearly complete mitogenome of A. aulica was 16,646 bp in length. The only gap occurred in the putative control region.
The obtained mitogenome of A. aulica consisted of 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a partial control region (Figure 1). There are 23 genes encoded on the heavy strand, while the remaining 14 genes encoded on the light strand. The organization of A. aulica mitogenome was compact, because only 29 bp gene overlaps were identified in 11 gene junctions, with the length ranging from one to seven nucleotides. A total of 116 bp intergenic spacers were found in 16 positions, which had the lengths ranging from one to 32 bp. The largest intergenic regions (32 bp) lied between trnW and trnC. The average nucleotide composition of the full mitogenome sequence was: A = 41.2%, T = 39.2%, C = 11.5% and G = 8.0%, which shows a strong bias towards A and T nucleotides (80.4%). In the A. aulica mitogenome, the AT-skew is 0.025, whereas the GC-skew is −0.179 (Table 3).

Table 3.
Nucleotide composition of the Amara aulica mitochondrial genome.
3.2. Protein-Coding Gene
The protein-coding genes had a total length of 11,194 bp, which encoded 3719 amino acid residues and the 37 bp stop codons. Nine out of 13 protein-coding genes were encoded on the heavy strand, while the remaining four were encoded on the light strand. All the protein-coding genes started with the typical ATN codons, except for the cox1 gene. The start codon ATT was used for nad3, nad5, nad4l and atp8, ATG for cox2, cox3, atp6, nad4 and cob, ATA for nad1, nad2 and nad6. The cox1 gene was initiated with the unusual CGA. The cox2 gene used a single T as the stop codon, while the rest of protein-coding genes ended with the complete termination codon TAA or TAG. The relative synonymous codon usage (RSCU) of A. aulica mitogenome are presented in Figure S1 and Table S2. The results showed that UUA (Leu2), AUU (Ile), UUU (Phe), AUA (Met) and AAU (Asn) were the five most frequently used codons. It was obvious that all of them were AT-rich codons. The A+T content of protein-coding genes was 78.5%, and the third codon positions had the highest A + T content (Table 3).
3.3. Transfer RNAs
Twenty-two tRNA genes were identified in the mitogenome of A. aulica and ranged in length from 64 bp to 72 bp. The full length of tRNA genes was 1478 bp. Fourteen tRNA genes were located on the heavy strand, and the remaining eight were encoded on the light strand. The inferred secondary structures for tRNAs are provided in Figure S2. With the exception of trnS1, all tRNA genes can be folded into the typical cloverleaf secondary structure. In the structure of trnS1, the dihydrouridine arm was replaced by a simple loop, which is a common character in most of insect mitogenomes published.
3.4. Ribosomal RNAs
The large ribosomal gene (rrnL) was 1293 bp in length, which was located between the trnL (CUN) and trnV. The small ribosomal gene (rrnS) was 699 bp, which was located between trnV and the control region. The inferred secondary structures of both rrnL and rrnS are shown in Figures S3 and S4. The secondary structure of rrnL contained five domains (labeled I, II, IV, V and VI) and 50 helices. The rrnS gene was composed of three domains (labeled I, II, III) and 30 helices.
3.5. Phylogenetic Analysis
Based on the results from PartitonFinder, six partition schemes were selected for the dataset of 49taxa_PCGRNA, and the GTR+I+G or GTR+G model was the preferred model for the corresponding partition (Table S1). Both Bayesian trees and ML trees revealed an extremely long terminal branch corresponding to the Rhysodes (Figure 2 and Figure 3). Moreover, the placement of Rhysodes varied between analyses. In the ML tree under the site-homogeneous GTR model, the Rhysodes was retrieved as sister group to Cicindelinae, and both together were sister to the remaining carabid beetle lineages (including Trachypachidae). This branching pattern may be due to long-branch attraction effect. The substitution rate analyses indicated that the Rhysodes has been engaged in a process of accelerated rate of evolution, with the highest ka/ks values among the species analyzed (Table 4). In the long-branch extraction analyses, the removal of the Rhysodes did not change the tree topology greatly (Figures S5 and S6).
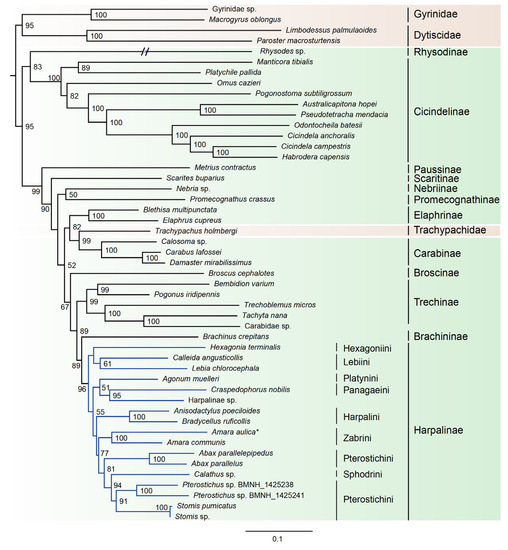
Figure 2.
Maximum likelihood tree inferred from the dataset of 49taxa_PCGRNA using IQ-TREE, under the partition schemes and best-fitting models selected by PartitionFinder. Bootstrap support values (≥50) are indicated near the nodes. The branch of Rhysodes is depicted as half of its original branch length. Green background indicates the ingroup taxa of Carabidae, and brown indicates the outgroups. Blue lines indicate the Harpalinae. The newly determined Amara aulica is emphasized by asterisk.
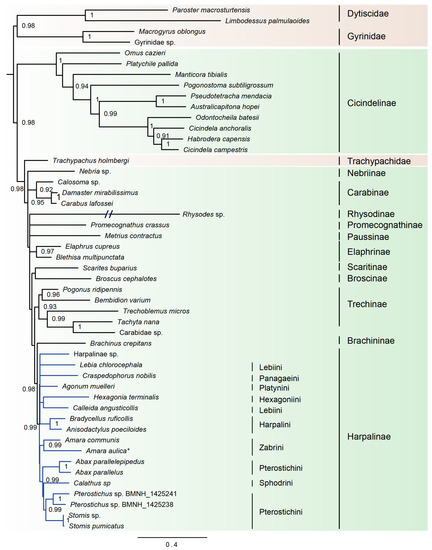
Figure 3.
Bayesian tree inferred from the dataset of 49taxa_PCGRNA using PhyloBayes under the site-heterogeneous CAT-GTR model. Poster probability values (≥50) are indicated near the nodes. The branch of Rhysodes is depicted as half of its original branch length. Green background indicates the ingroup taxa of Carabidae, and brown indicates the outgroups. Blue lines indicate the Harpalinae. The newly determined Amara aulica is emphasized by asterisk.

Table 4.
The substitution rate analyses conducted by DnaSP.
The family Trachypachidae always embedded within Carabidae, rendering the latter to be a non-monophyletic assemblage. In the ML analysis based on the dataset of 49taxa_PCGRNA, the Trachypachidae was the sister to the subfamily Carabinae, whereas in the Bayesian analysis based on the same dataset, the Trachypachidae was placed in an intermediate position between the subfamily Cicindelinae and the remaining carabid beetles.
Within the family Carabidae, four subfamilies with multiple taxon sampling (Cicindelinae, Carabinae, Elaphrinae and Harpalinae) were consistently recovered as monophyletic groups with high support (BP ≥ 96, PP ≥ 0.92). The Cicindelinae were placed as sister group to the remaining ingroup taxa. The monophyly of Trechinae remained elusive due to the ambiguous classification of the exemplar of Carabidae sp. (GenBank accession number: KT696200). A sister group relationship between Brachininae and Harpalinae was strongly supported (BP = 89, PP = 0.98). The phylogenetic positions of the remaining carabid subfamilies were unstable across phylogenetic analyses.
The subfamily Harpalinae had the largest taxon coverage in this study, which allowed us to address some lower taxonomic relationships within this group. The newly sequenced A. aulica was strongly supported as a sister to Amara communis (BP = 100, PP = 0.99). At the tribe level, the Pterostichini was found to be paraphyletic, with Sphodrini embedded therein. The Zabrini formed a sister group to the clade comprising Pterostichini and Sphodrini. The relationships between the rest of harpaline tribes remained largely unresolved in the Bayesian trees (Figure 3, Figure S6). In contrast, ML trees elucidated a clearer relationship: the intermediate position of Harpalini, and a sister-group relationship of Hexagoniini with Lebiini (Figure 2, Figure S5).
4. Discussion
Previous studies have shown that the site-heterogeneous CAT-GTR model implemented in Bayesian analysis can effectively suppress the long-branch attraction artefacts in the animal phylogeny [52,74,75,76,77]. The long-branched Rhysodinae was pulled toward a more derived position and away from the Cicindelinae in the PhyloBayes trees. However, analyses using the site-heterogeneous CAT-GTR model showed limited resolution on the subfamily relationships among Promecognathinae, Paussinae and Elaphrinae (Figure 3, Figure S6).
The family Carabidae was recovered as non-monophyletic, with respect to Trachypachidae (Figure 2 and Figure 3, Figures S5 and S6). Maddison et al. (2009) [24] supported the nested placement of Trachypachidae within a monophyletic Geadephaga, based on the nuclear gene sequences. However, the sister group of Trachypachidae within Geadephaga is undetermined. Trachypachids were placed with Carabitae, migadopines, elaphrines or a large clade comprising the majority of carabids [24]. In the study of Mckenna et al. (2015) [42] with expanding nuclear gene markers, the placement of Trachypachidae was still unclear. It clustered with Calosoma (Carabidae) or other Carabini [42]. These branching patterns resulted in a paraphyletic Carabidae. The similar situation was revealed in the current analyses based on the mitogenome sequence data.
In the Bayesian tree from 49taxa_PCGRNA, the Cicindelinae was placed as sister to all other carabids (including Trachypachidae). This reconstruction was consistent with some previous studies [23,35,36,41], but contradicted the more derived position recognized by the studies of Beutel et al. (2006) [17] and Hunt et al. (2007) [33]. The “CRPS quartet” (Cicindelidae + Rhysodinae + Paussinae + Scaritinae) inferred in the previous studies [23,24,38,40] was never recovered in the present study.
Within Carabidae, the subfamily relationships changed depending on analyses. Compared with ML trees, the deep divergences among several carabid subfamilies were unresolved in the Bayesian trees (Figure 3, Figure S6). Tree topology comprising very short internodes of early divergences occurred frequently in phylogenetic analysis [78,79]. Lack of resolution may be owing to non-optimal substitution rates, insufficient and conflicting phylogenetic signal. The short internal branches associated with the deep-level relationships of carabids (the large polytomy) also emerged in the prior studies [23]. The authors attributed this to inappropriate methods of inference. Rogue taxa may be another factor leading to weak nodal support and very short internal branches [38]. In addition, rapid radiation of beetle insects may explain the generally short diverging nodes between major groupings at the base of the carabid tree. A large clade comprising Trechinae, Brachininae and Harpalinae was consistently recovered in all analyses. The Brachininae formed a sister group to Harpalinae, both of which were sister to Trechinae. These two sister group relationships were strongly supported (BP ≥ 89, PP ≥ 0.98). This result was concordant with previous studies [23,45].
5. Conclusions
The Harpalinae is a megadiverse group within the family Carabidae. However, mitogenome sequences available for Harpaline are very limited. Here, we presented the detailed description of the nearly complete mitogenome of A. aulica (Carabidae, Harpalinae). In this mitogenome, gene order and content are consistent with the hypothesized ancestral insect [49]. The new mitogenome sequence was added to investigate the phylogenetic relationships among carabid beetles. The results supported the Carabidae to be a non-monophyletic group with respect to the Trachypachidae. Four subfamilies within Carabidae were strongly supported, namely Cicindelinae, Carabinae, Elaphrinae and Harpalinae. The Cicindelinae was retrieved as sister to all other carabid lineages. The Trechinae (including Carabidae sp.-KT696200) formed a sister group to the clade of (Brachininae + Harpalinae). These results demonstrated that mitogenome sequences can be useful for resolving the subfamily relationships of Carabidae.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/2/181/s1, Figure S1: Relative synonymous codon usage (RSCU) in the Amara aulica mitochondrial genome, Figure S2: Putative secondary structures of the 22 tRNA genes from Amara aulica, Figure S3: Putative rrnL secondary structure in the Amara aulica mitochondrial genome, Figure S4: Putative rrnS secondary structure in the Amara aulica mitochondrial genome, Figure S5: Maximum likelihood tree inferred from the dataset of 48taxa_PCGRNA using IQ-TREE under the best-fitting models, Figure S6: Bayesian tree inferred from the dataset of 48taxa_PCGRNA using PhyloBayes under the site-heterogeneous CAT-GTR model, Table S1: The best partitioning schemes selected by PartitionFinader for the dataset of (A) 49taxa_PCGRNA and (B) 48taxa_PCGRNA, Table S2: Codon usage of protein-coding genes in the Amara aulica mitochondrial genome.
Author Contributions
Conceptualization, N.S. and X.Y.; methodology, N.S.; software, N.S., X.L., Z.L. and H.T.; validation, Z.L., X.L., N.S., H.T. and X.Y.; formal analysis, N.S., X.L. and Z.L.; investigation, Z.L., X.L., H.T. and N.S.; resources, N.S., H.T. and X.Y.; data curation, Z.L., X.L. and N.S.; writing—original draft preparation, X.L. and N.S.; writing—review and editing, Z.L., X.L., N.S. and X.Y.; visualization, N.S., X.L. and Z.L.; supervision, N.S. and X.Y.; project, N.S. and X.Y.; funding acquisition, N.S. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science and Technology Innovation Fund of Henan Agricultural University (KJCX2019A10), Earmarked Fund for China Agriculture Research System (NO. CARS-27), Key Scientific Research Projects of Henan Province (18B210006) and the Science and Technology Innovation Fund of Henan Agricultural University (KJCX2019A10).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Erwin, T.L. The taxon pulse: A general pattern of lineage radiation and extinction among carabid beetles. In Taxonomy Phylogeny and Zoogeography of Beetles and Ants; A volume dedicated to the Memory of Philip Jackson Darlington Jr. (1904–1983); Dr W Junk: Dordrect, The Netherlands, 1985. [Google Scholar]
- Arnett, R.H.; Thomas, M.C. American Beetles, Volume 1: Archostemata, Myxophaga, Adephaga, Polyphaga: Staphyliniformia; CRC Press LLC: Boca Raton, FL, USA, 2000. [Google Scholar]
- Arndt, E.; Beutel, R.G.; Will, K. Carabidae Latreille, In Handbuch der Zoologie. In Handbook of Zoology Vol. IV Arthropoda: Insecta. Part 38; Walter de Gruyter: Berlin, Germany, 2005. [Google Scholar]
- Erwin, T.L. The taxon pulse: A general pattern of lineage radiation and extinction among carabid beetles. Taxon. Phylogeny Zoogeogr. Beetles Ants 1985, 30, 437–488. [Google Scholar]
- Frank, S.D.; Shrewsbury, P.M.; Denno, R.F. Effects of alternative food on cannibalism and herbivore suppression by carabid larvae. Ecol. Entomol. 2010, 35, 61–68. [Google Scholar] [CrossRef]
- Monzó, C.; Sabater-Muñoz, B.; Urbaneja, A.; Castañera, P. The ground beetle Pseudophonus rufipes revealed as predator of Ceratitis capitata in citrus orchards. Biol. Control 2011, 56, 17–21. [Google Scholar] [CrossRef]
- Suenaga, H.; Hamamura, T. Occurrence of carabid beetles (Coleoptera: Carabidae) in cabbage fields and their possible impact on lepidopteran pests. Appl. Entomol. Zool. 2001, 36, 151–160. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Dosdall, L.M.; Willenborg, C.J. The role of ground beetles (Coleoptera: Carabidae) in weed seed consumption: A review. Weed Sci. 2015, 63, 355–376. [Google Scholar] [CrossRef]
- Jeannel, R. Faune de France No. 40: Coléoptères Carabiques (II); Paris: Lib. Fac. Sci; Lechevalier: Paris, France, 1942. [Google Scholar]
- Lindroth, C.H. A revision of the genus Synuchus Gyllenhal (Coleoptera, Carabidae) in the widest sense, with notes on Pristosia and Calathus. Trans. R. Entomol. Soc. Lond. 1956, 108, 485–585. [Google Scholar] [CrossRef]
- Habu, A. Fauna Japonica. In Carabidae: Platynini (Insecta: Coleoptera); Keigaku: Tokyo, Japan, 1978. [Google Scholar]
- Hammond, P.M. Wing-folding mechanisms of beetles, with special reference to investigations of Adephagan phylogeny (Coleoptera). In Carabid Beetles; Springer: Dordrecht, The Netherlands, 1979; pp. 113–180. [Google Scholar]
- Liebherr, J.K.; Will, K.W. Inferring phylogenetic relationships within the Carabidae (Insecta, Coleoptera) from characters of the female reproductive tract. In Phylogeny and classification of Caraboidea (Coleoptera: Adephaga); Ball, G.E., Casale, A., Vigna Taglianti, A., Eds.; Museo Regionale di Scienze Naturali: Torino, Italy, 1998. [Google Scholar]
- Arndt, E. Phylogenetic investigation of Carabidae (Coleoptera) using larval chaetotaxy. In Phylogeny and Classifcation of Caraboidea (Coleoptera: Adephaga); Ball, G.E., Casale, A., Vigna Taglianti, A., Eds.; Museo Regionale di Scienze Naturali: Torino, Italy, 1998. [Google Scholar]
- Kavanaugh, D.H. Investigations of phylogenetic relationships among some basal grade Carabidae (Coleoptera): A report on work in progress. In Phylogeny and Classification of Caraboidea (Coleoptera: Adephaga); Ball, G.E., Casale, A., Vigna Taglianti, A., Eds.; Museo Regionale di Scienze Naturali: Torino, Italy, 1998. [Google Scholar]
- Grebennikov, V.V.; Maddison, D.R. Phylogenetic analysis of Trechitae (Coleoptera: Carabidae) based on larval morphology, with a description of first-instar Phrypeus and a key to genera. Syst. Entomol. 2005, 30, 38–59. [Google Scholar] [CrossRef]
- Beutel, R.G.; Balke, M.; Steiner, W.E., Jr. The systematic position of Meruidae (Coleoptera, Adephaga) and the phylogeny of the smaller aquatic adephagan beetle families. Cladistics 2006, 22, 102–131. [Google Scholar] [CrossRef]
- Moore, B.P. Chemical defense in carabids and its bearing on phylogeny. In Carabid Beetles: Their Evolution, Natural History, and Classification; Erwin, T.L., Ball, G.E., Whitehead, D.R., Halpern, A.L., Eds.; Dr. W. Junk Publishers: The Hague, The Netherlands, 1979. [Google Scholar]
- Balestrazzi, E.; Dazzini, M.L.V.; Bernardi, M.D.; Vidari, G.; Vita-Finzi, P.; Mellerio, G. Morphological and chemical studies on the pygidial defence glands of some Carabidae (Coleoptera). Naturwissenschaften 1985, 72, 482–484. [Google Scholar] [CrossRef]
- Brandmayr, P.; Bonacci, T.; Giglio, A.; Talarico, F.F.; Brandmayr, T.Z. The evolution of defence mechanisms in carabid beetles: A review. In Life and Time: The Evolution of Life and Its History; Cleup: Padova, Italy, 2009. [Google Scholar]
- Serrano, J.; Galián, J.; Ortiz, A.S. Karyotypic data and current taxonomic ideas of the tribe Harpalini (Coleoptera, Carabidae). In Carabid Beetles: Ecology and Evolution; Desender, K., Dufrêne, M., Loreau, M., Luff, M.L., Maelfait, J.-P., Eds.; Kluwer Academic Press: Dordrecht, The Netherlands, 1994. [Google Scholar]
- Serrano, J.; Galián, J. A review of karyotypic evolution and phylogeny of carabid beetles (Coleoptera). In Phylogeny and Classification of Caraboidea (Coleoptera: Adephaga); Ball, G.E., Casale, A., Vigna Taglianti, A., Eds.; Museo Regionale di Scienze Naturali: Torino, Italy, 1998. [Google Scholar]
- Maddison, D.R.; Baker, M.D.; Ober, K.A. Phylogeny of carabid beetles as inferred from 18S ribosomal DNA (Coleoptera: Carabidae). Syst. Entomol. 1999, 24, 103–138. [Google Scholar] [CrossRef]
- Maddison, D.R.; Moore, W.; Baker, M.D.; Ellis, T.M.; Ober, K.A.; Cannone, J.J.; Gutell, R.R. Monophyly of terrestrial adephagan beetles as indicated by three nuclear genes (Coleoptera: Carabidae and Trachypachidae). Zool. Scr. 2009, 38, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Gough, H.M.; Duran, D.P.; Kawahara, A.Y.; Toussaint, E.F. A comprehensive molecular phylogeny of tiger beetles (Coleoptera, Carabidae, Cicindelinae). Syst. Entomol. 2019, 44, 305–321. [Google Scholar] [CrossRef]
- Maddison, D.R.; Kanda, K.; Boyd, O.F.; Faille, A.; Porch, N.; Erwin, T.L.; Roig-Juñent, S. Phylogeny of the beetle supertribe Trechitae (Coleoptera: Carabidae): Unexpected clades, isolated lineages, and morphological convergence. Mol. Phylogenet. Evol. 2019, 132, 151–176. [Google Scholar] [CrossRef]
- Maddison, D.R. Phylogeny of Bembidion and related ground beetles (Coleoptera: Carabidae: Trechinae: Bembidiini: Bembidiina). Mol. Phylogenet. Evol. 2012, 63, 533–576. [Google Scholar] [CrossRef] [PubMed]
- Deuve, T.; Cruaud, A.; Genson, G.; Rasplus, J.Y. Molecular systematics and evolutionary history of the genus Carabus (Col. Carabidae). Mol. Phylogenet. Evol. 2012, 65, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Sota, T.; Vogler, A.P. Reconstructing species phylogeny of the carabid beetles Ohomopterus using multiple nuclear DNA sequences: Heterogeneous information content and the performance of simultaneous analyses. Mol. Phylogenet. Evol. 2003, 26, 139–154. [Google Scholar] [CrossRef]
- Sota, T.; Takami, Y.; Monteith, G.B.; Moore, B.P. Phylogeny and character evolution of endemic Australian carabid beetles of the genus Pamborus based on mitochondrial and nuclear gene sequences. Mol. Phylogenet. Evol. 2005, 36, 391–404. [Google Scholar] [CrossRef]
- Ortuño, V.M.; Sendra, A.; Reboleira, A.S.P.; Fadrique, F.; Faille, A. The Iberian genus Paraphaenops Jeannel, 1916 (Coleoptera: Carabidae: Trechini): Morphology, phylogeny and geographical distribution. Zool. Anz. 2017, 266, 71–88. [Google Scholar] [CrossRef]
- Sasakawa, K.; Kubota, K. Phylogeny and genital evolution of carabid beetles in the genus Pterostichus and its allied genera (Coleoptera: Carabidae) inferred from two nuclear gene sequences. Ann. Entomol. Soc. Am. 2007, 100, 100–109. [Google Scholar] [CrossRef]
- Hunt, T.; Bergsten, J.; Levkanicova, Z.; Papadopoulou, A.; John, O.S.; Wild, R.; Hammond, P.M.; Ahrens, D.; Balke, M.; Caterino, M.S.; et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science 2007, 318, 1913–1916. [Google Scholar] [CrossRef]
- Bousquet, Y. Catalogue of the Geadephaga (Coleptera: Trachypachidae, Rhysodidae, Caraidae including Cicindelini) of America north of Mexico. Mem. Entomol. Soc. Can. 1993, 167, 1–397. [Google Scholar] [CrossRef]
- Bocak, L.; Barton, C.; Crampton-Platt, A.L.E.X.; Chesters, D.; Ahrens, D.; Vogler, A.P. Building the Coleoptera tree-of-life for > 8000 species: Composition of public DNA data and fit with Linnaean classification. Syst. Entomol. 2014, 39, 97–110. [Google Scholar] [CrossRef]
- Timmermans, M.J.T.N.; Barton, C.; Haran, J.; Ahrens, D.; Culverwell, C.L.; Ollikainen, A.; Dodsworth, S.; Foster, P.G.; Bocak, L.; Vogler, A.P. Family-level sampling of mitochondrial genomes in Coleoptera: Compositional heterogeneity and phylogenetics. Genome Biol. Evol. 2016, 8, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Crampton-Platt, A.; Timmermans, M.J.; Gimmel, M.L.; Kutty, S.N.; Cockerill, T.D.; Khen, C.V.; Vogler, A.P. Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, D.D.; Wild, A.L.; Kanda, K.; Bellamy, C.L.; Beutel, R.G.; Caterino, M.S.; Farnum, C.W.; Hawks, D.C.; Ivie, M.A.; Jameson, M.L.; et al. The beetle tree of life reveals that Coleoptera survived end-Permian mass extinction to diversify during the Cretaceous terrestrial revolution. Syst. Entomol. 2015, 40, 835–880. [Google Scholar] [CrossRef]
- Baca, S.M.; Alexander, A.; Gustafson, G.T.; Short, A.E. Ultraconserved elements show utility in phylogenetic inference of Adephaga (Coleoptera) and suggest paraphyly of ‘Hydradephaga’. Syst. Entomol. 2017, 42, 786–795. [Google Scholar] [CrossRef]
- López-López, A.; Vogler, A.P. The mitogenome phylogeny of Adephaga (Coleoptera). Mol. Phylogenet. Evol. 2017, 114, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Q.; Che, L.H.; Li, Y.; Liang, D.; Pang, H.; Ślipiński, A.; Zhang, P. Evolutionary history of Coleoptera revealed by extensive sampling of genes and species. Nat. Commu. 2018, 9, 205. [Google Scholar] [CrossRef]
- Lindroth, C.H. On elytral microsculpture of carabid beetles (Coleoptera: Carabidae). Insect. Syst. Evol. 1974, 5, 251–264. [Google Scholar] [CrossRef]
- Ponomarenko, A.G. Suborder Adephaga. In Mesozoic Coleoptera (Arnoldi, L.V., Zherikhin, V.V., Nikritin, L.M., Ponomarenko, A.G., EDs.). Proc. Inst. Acad. Sci. USSR 1977, 161, 17–104. [Google Scholar]
- Lorenz, W. Systematic List of Extant Ground Beetles of the World; W. Lorenz: Tutzing, Germany, 2005. [Google Scholar]
- Ober, K.A. Phylogenetic relationships of the carabid subfamily Harpalinae (Coleoptera) based on molecular sequence data. Mol. Phylogenet. Evol. 2002, 24, 228–248. [Google Scholar] [CrossRef]
- Jedlicka, A. Monographie der Truncatipennen aus Ostasien. Lebiinae, Odacanthinae, Brachyninae (Coleoptera, Carabidae). Entomol. Abh. Staatl. Mus. Tierkd. Dresd. 1963, 28, 269–579. [Google Scholar]
- Basilewsky, P. Essai d’une classification supragénérique naturelle des Carabides Lébiens d’Afrique et de Madagascar (Coleoptera Carabidae Lebiinae). Rev. Zool. Afr. 1984, 98, 525–559. [Google Scholar]
- Boore, J.L. Survey and summary: Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef]
- Cameron, S.L. How to sequence and annotate insect mitochondrial genomes for systematic and comparative genomics research. Syst. Entomol. 2014, 39, 400–411. [Google Scholar] [CrossRef]
- Gillett, C.P.; Crampton-Platt, A.; Timmermans, M.J.; Jordal, B.H.; Emerson, B.C.; Vogler, A.P. Bulk de novo mitogenome assembly from pooled total DNA elucidates the phylogeny of weevils (Coleoptera: Curculionoidea). Mol. Biol. Evol. 2014, 31, 2223–2237. [Google Scholar] [CrossRef]
- Song, F.; Li, H.; Jiang, P.; Zhou, X.; Liu, J.; Sun, C.; Cai, W. Capturing the phylogeny of Holometabola with mitochondrial genome data and Bayesian site-heterogeneous mixture models. Genome Biol. Evol. 2016, 8, 1411–1426. [Google Scholar] [CrossRef]
- Avise, J.C. Mitochondrial DNA and the evolutionary genetics of higher animals. Philos. Trans. R. Soc. Lond. 1986, 312, 325–342. [Google Scholar]
- Simon, S.; Hadrys, H. A comparative analysis of complete mitochondrial genomes among Hexapoda. Mol. Phylogenet. Evol. 2013, 69, 393–403. [Google Scholar] [CrossRef]
- Tang, M.; Tan, M.; Meng, G.; Yang, S.; Su, X. Multiplex sequencing of pooled mitochondrial genomes a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 2014, 42, e166. [Google Scholar] [CrossRef] [PubMed]
- Bousquet, Y. Bembidion femoratum Sturm and Amara communis (Panzer) (Coleoptera: Carabidae) new to North America. J. N. Y. Entomol. Soc. 1992, 1, 503–509. [Google Scholar]
- Majka, C.G. The Palearctic species Bembidion femoratum and Amara communis (Coleoptera: Carabidae): New records and notes on modes of introduction to North America. Can. Entomol. 2005, 137, 532–538. [Google Scholar] [CrossRef]
- Gokturk, T.; Celik, A. The Family of Carabidae (Coleoptera) in Artvin Hatila National Park of Turkey. Int. J. Environ. Agric. Biotechnol. 2017, 2, 983–990. [Google Scholar] [CrossRef]
- Patel, R.K.; Jain, M. NGS QC Toolkit: A toolkit for quality control of next generation sequencing data. PLoS ONE 2012, 7, e30619. [Google Scholar] [CrossRef]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Song, N.; Li, H.; Song, F.; Cai, W. Molecular phylogeny of Polyneoptera (Insecta) inferred from expanded mitogenomic data. Sci. Rep. 2016, 6, 36175. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Junqueira, A.C.M.; Lessinger, A.C.; Torres, T.T.; Silva, F.R.D.; Vettore, A.L.; Arruda, P.; Espin, A.M.L.A. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene 2004, 339, 7–15. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Telford, M.J. TranslatorX: Multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38, W7–W13. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kuck, P.; Meusemann, K. FASconCAT: Convenient handling of data matrices. Mol. Phylogenet. Evol. 2010, 56, 1115–1118. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef]
- Lartillot, N.; Philippe, H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004, 21, 1095–1109. [Google Scholar] [CrossRef]
- Lartillot, N.; Brinkmann, H.; Philippe, H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 2007, 7, S4. [Google Scholar] [CrossRef]
- Lartillot, N.; Lepage, T.; Blanquart, S. PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 2009, 25, 2286–2288. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Shao, R.; Song, N.; Song, F.; Jiang, P.; Li, Z.; Cai, W. Higher-level phylogeny of paraneopteran insects inferred from mitochondrial genome sequences. Sci. Rep. 2015, 5, 8527. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Leavengood, J.M.; Chapman, E.G.; Burkhardt, D.; Song, F.; Jiang, P.; Liu, J.; Zhou, X.; Cai, W. Mitochondrial phylogenomics of Hemiptera reveals adaptive innovations driving the diversification of true bugs. Proc. R. Soc. B Biol. Sci. 2017, 284. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.B.; Kjer, K.M. Ancient rapid radiations of insects: Challenges for phylogenetic analysis. Annu. Rev. Entomol. 2008, 53, 449–472. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.B.; Lockhart, P.J. Deciphering ancient rapid radiations. Trends Ecol. Evol. 2007, 22, 258–265. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).