The Personalisation of Glioblastoma Treatment Using Whole Exome Sequencing: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Biopsy Collection, Dispersion and Culturing of the Patient Derived GBM Cells
2.3. Whole Exome Sequencing of the GBM Tumour Tissue and Cells
2.4. Determination of the Cytotoxicity of Each Individual Drug and Drug Combination Against the Patient Derived GBM Cells
3. Results
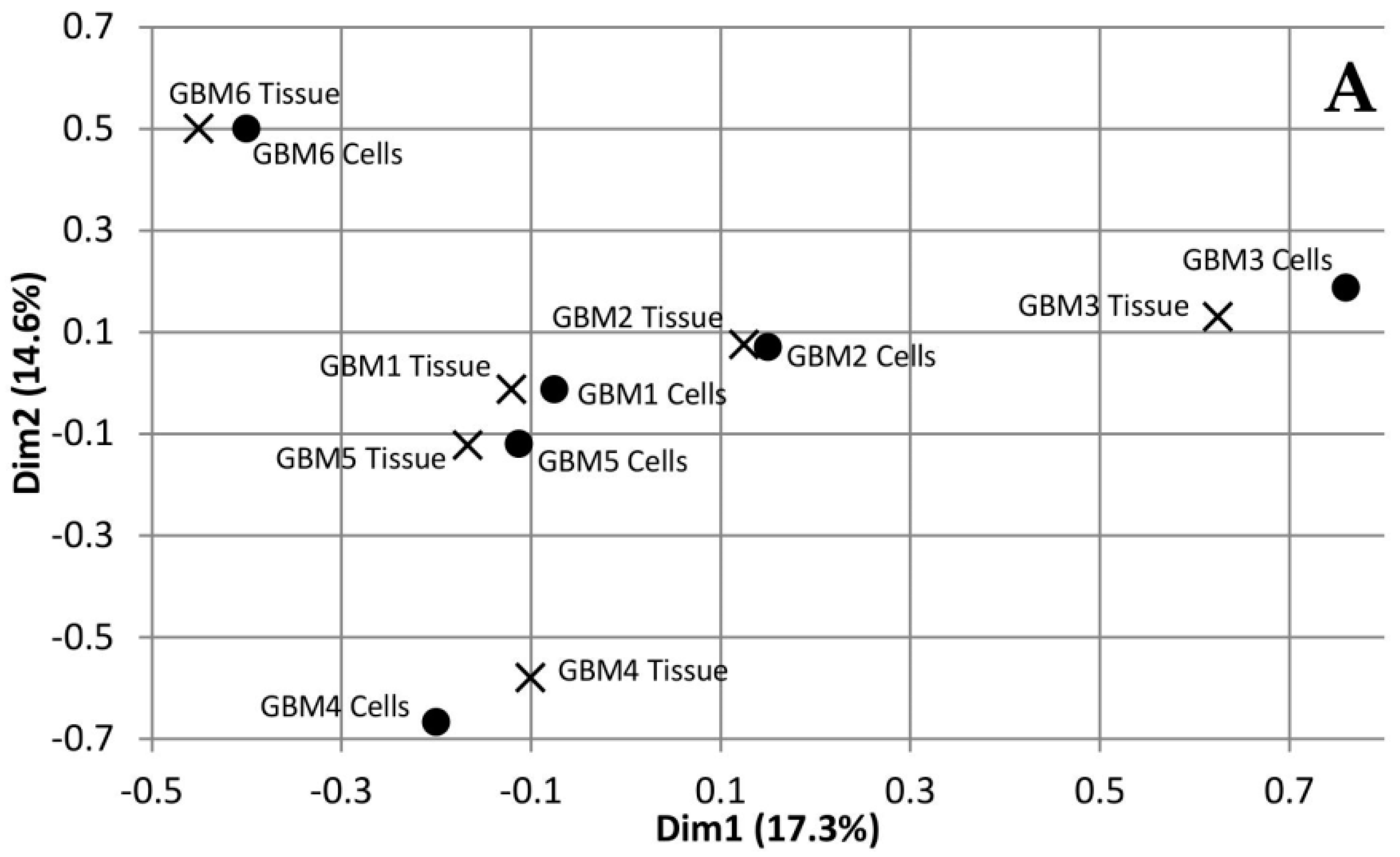
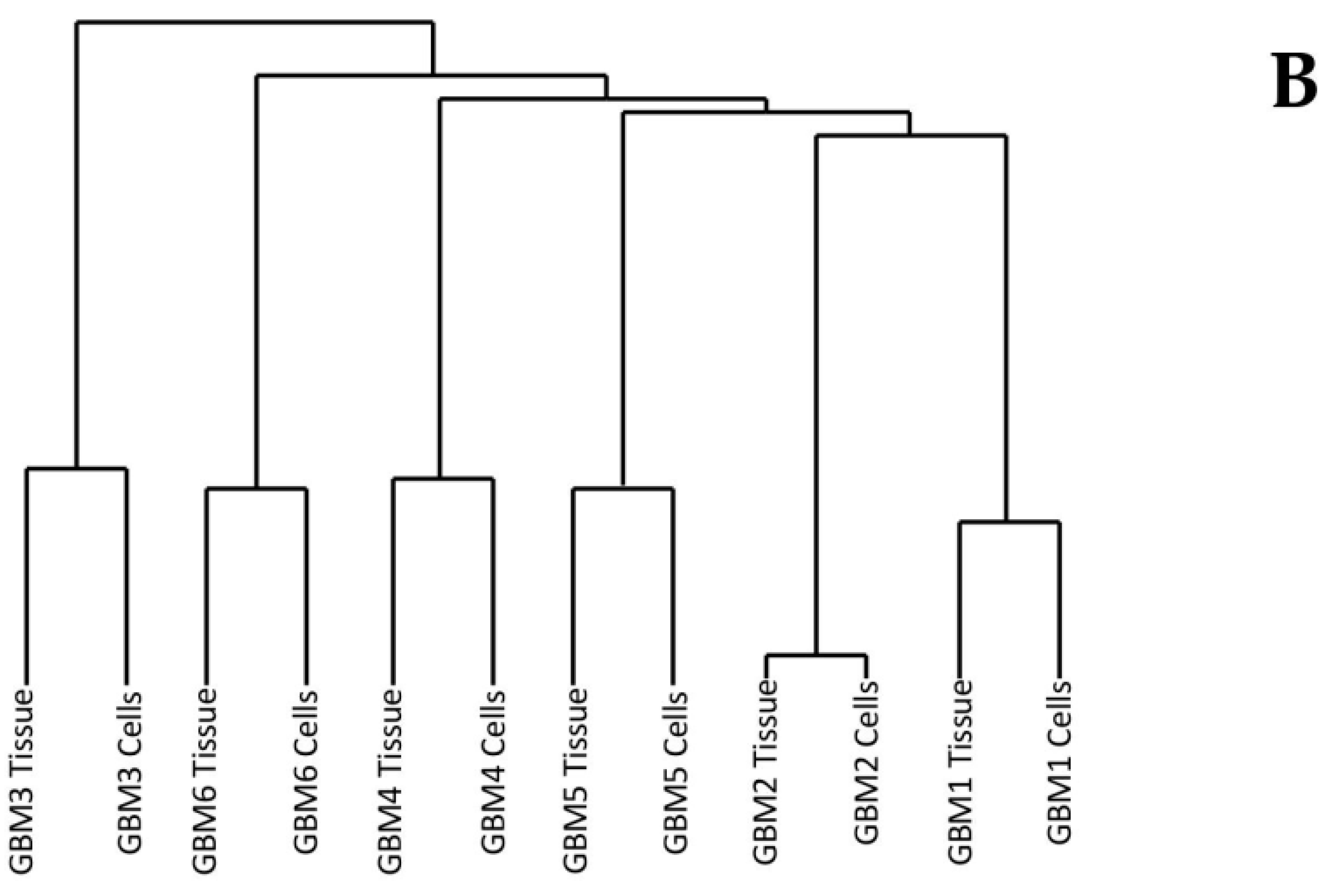
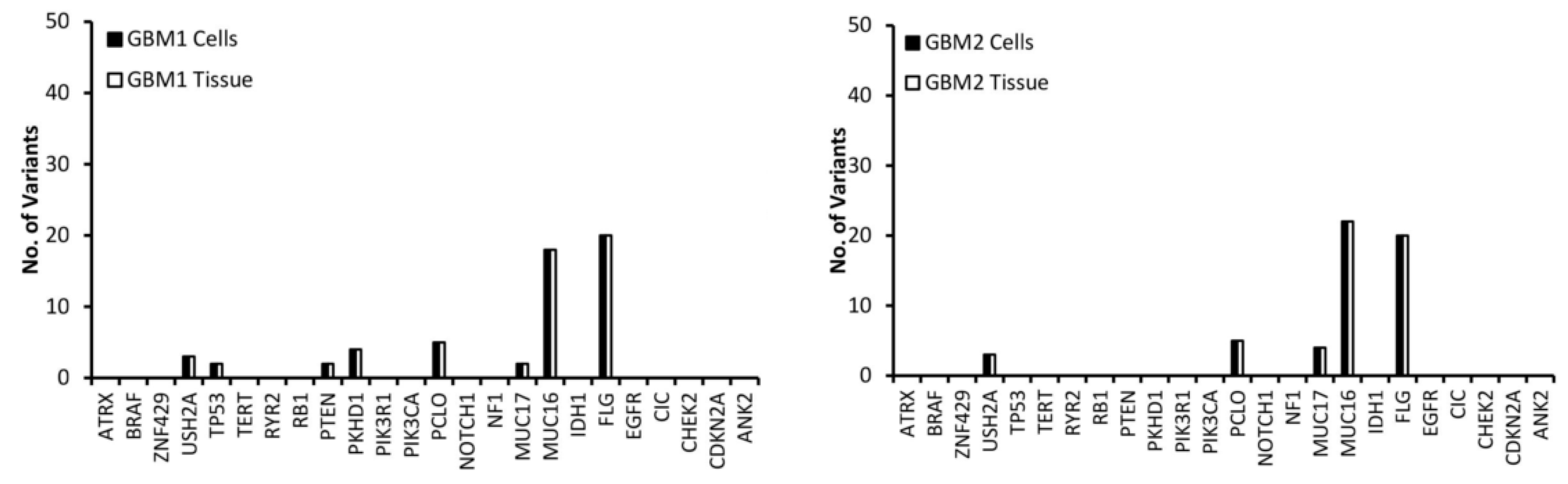
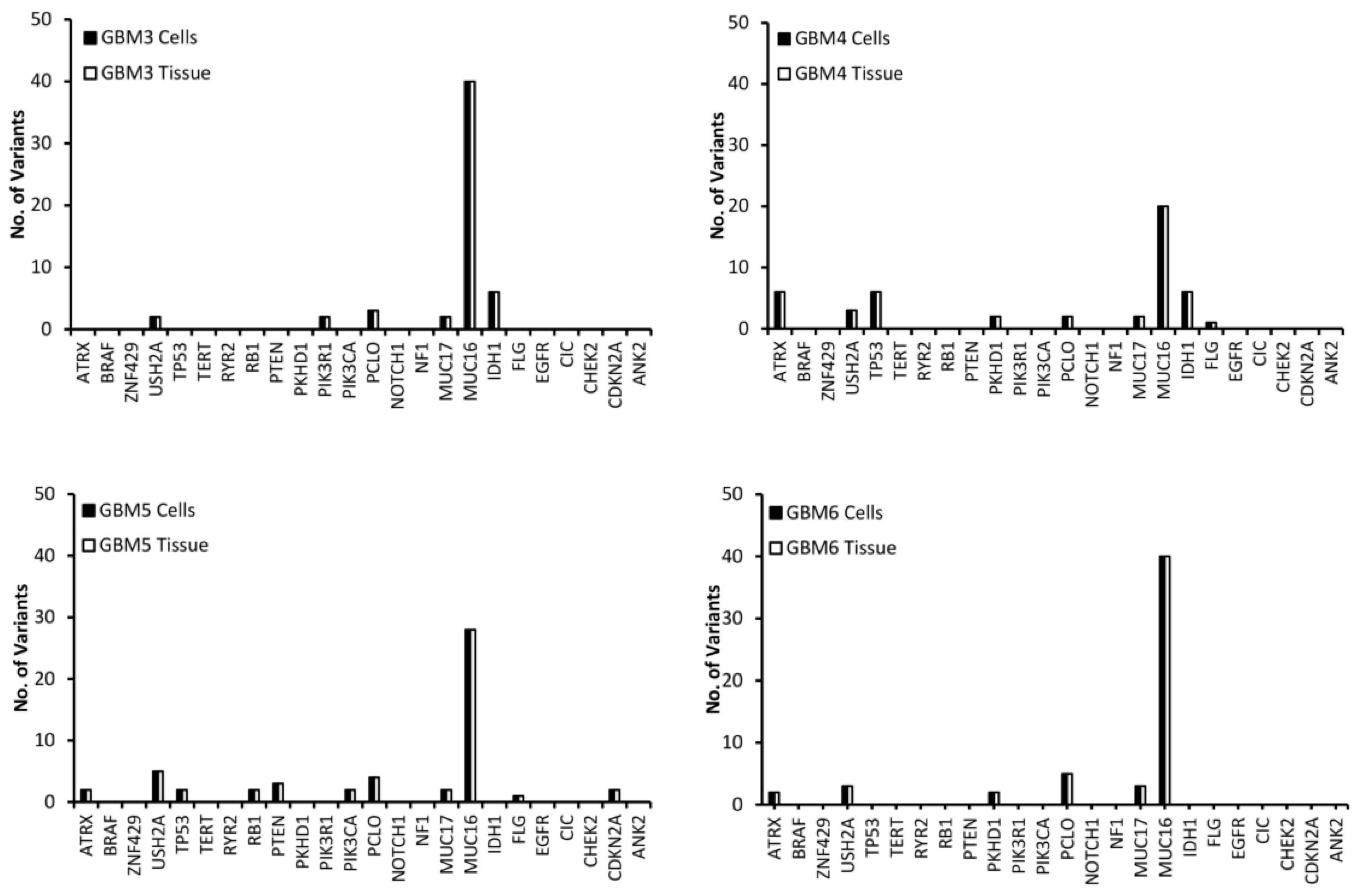
3.1. Comparison of Variants Between GBM Tumour Tissue and Associated Cells
3.2. Selection of Mutated Genes for Personalisation
3.3. Drug Response vs. Variant
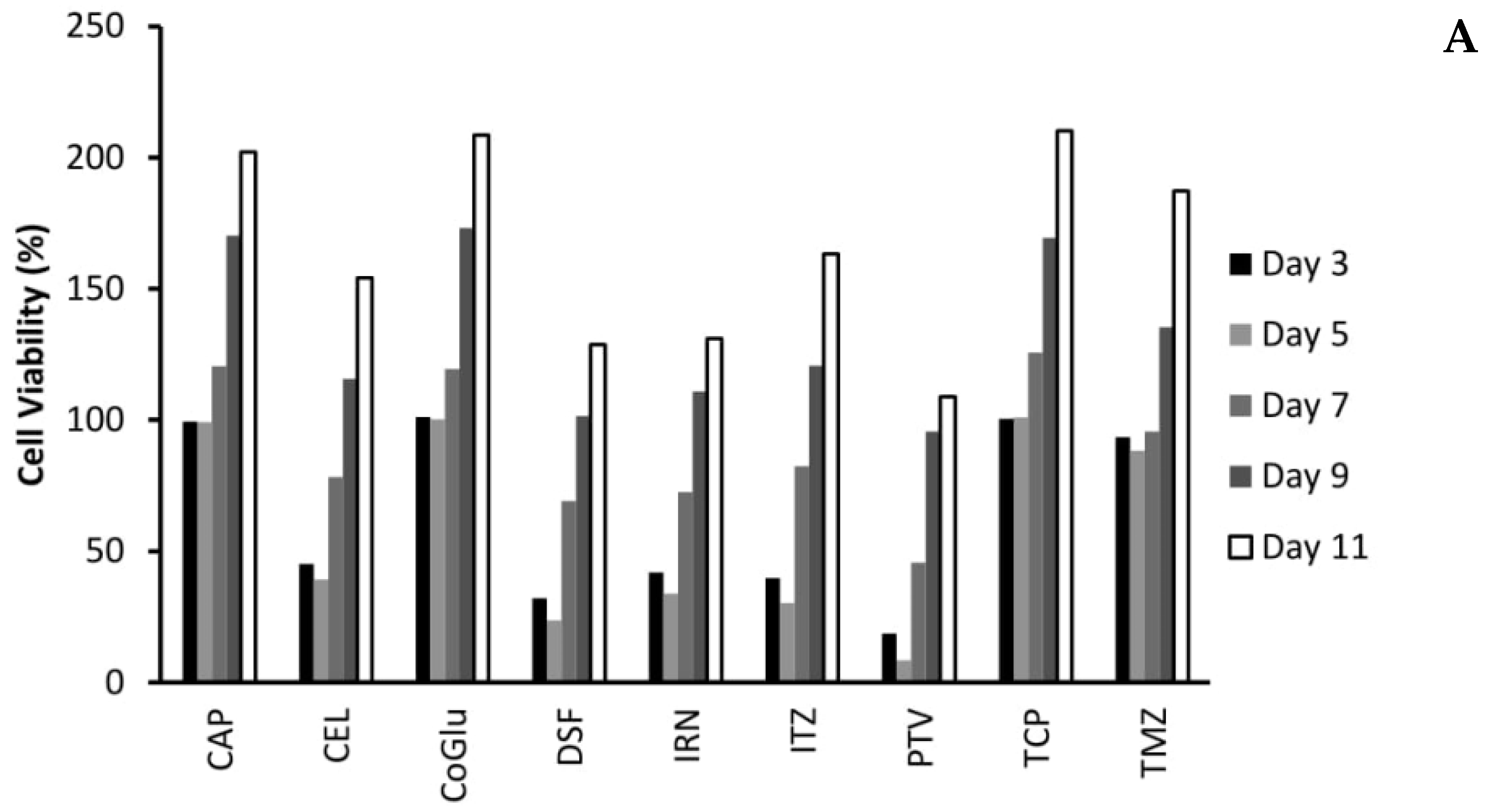
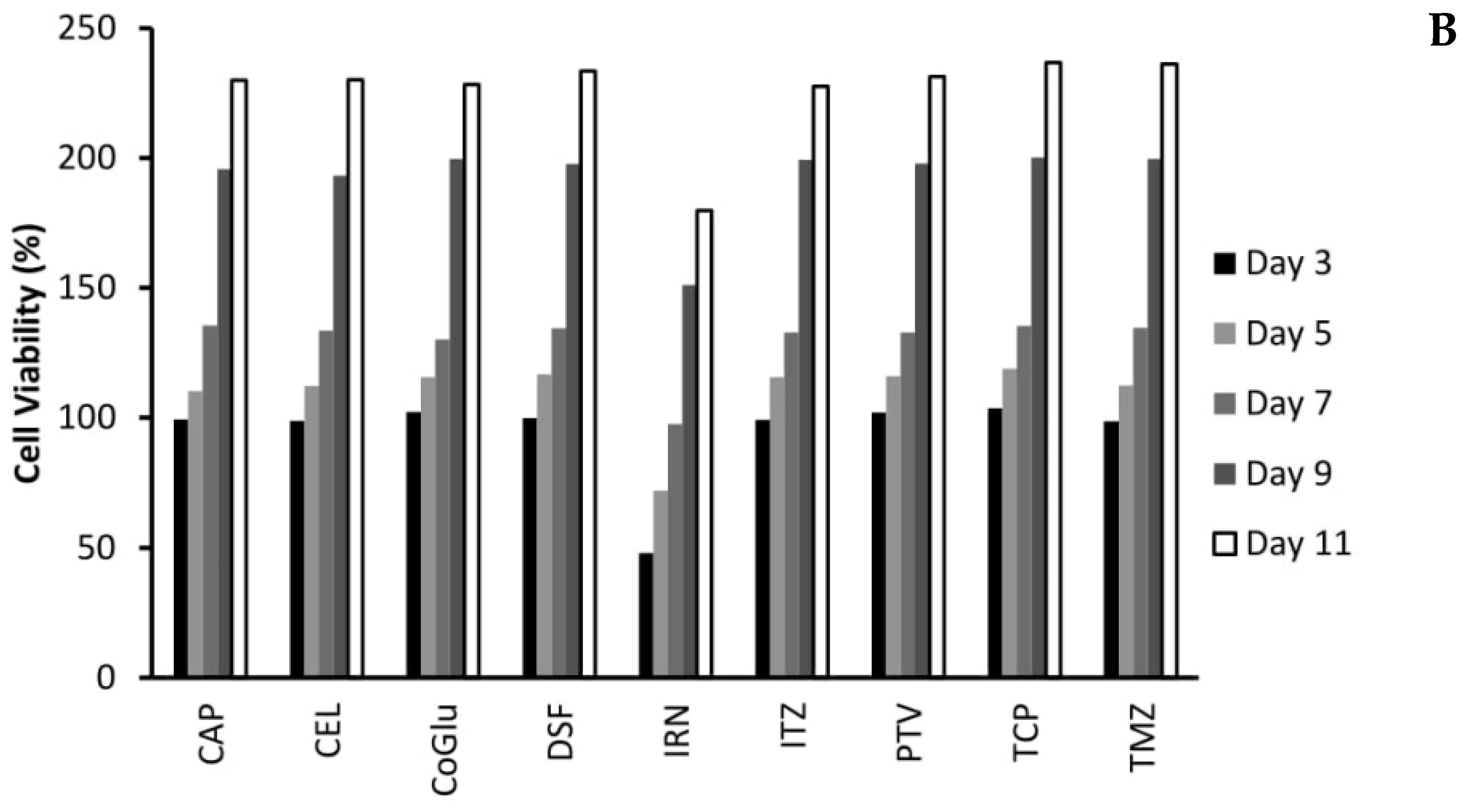
3.4. The Influence of Personalisation on Cell Viability
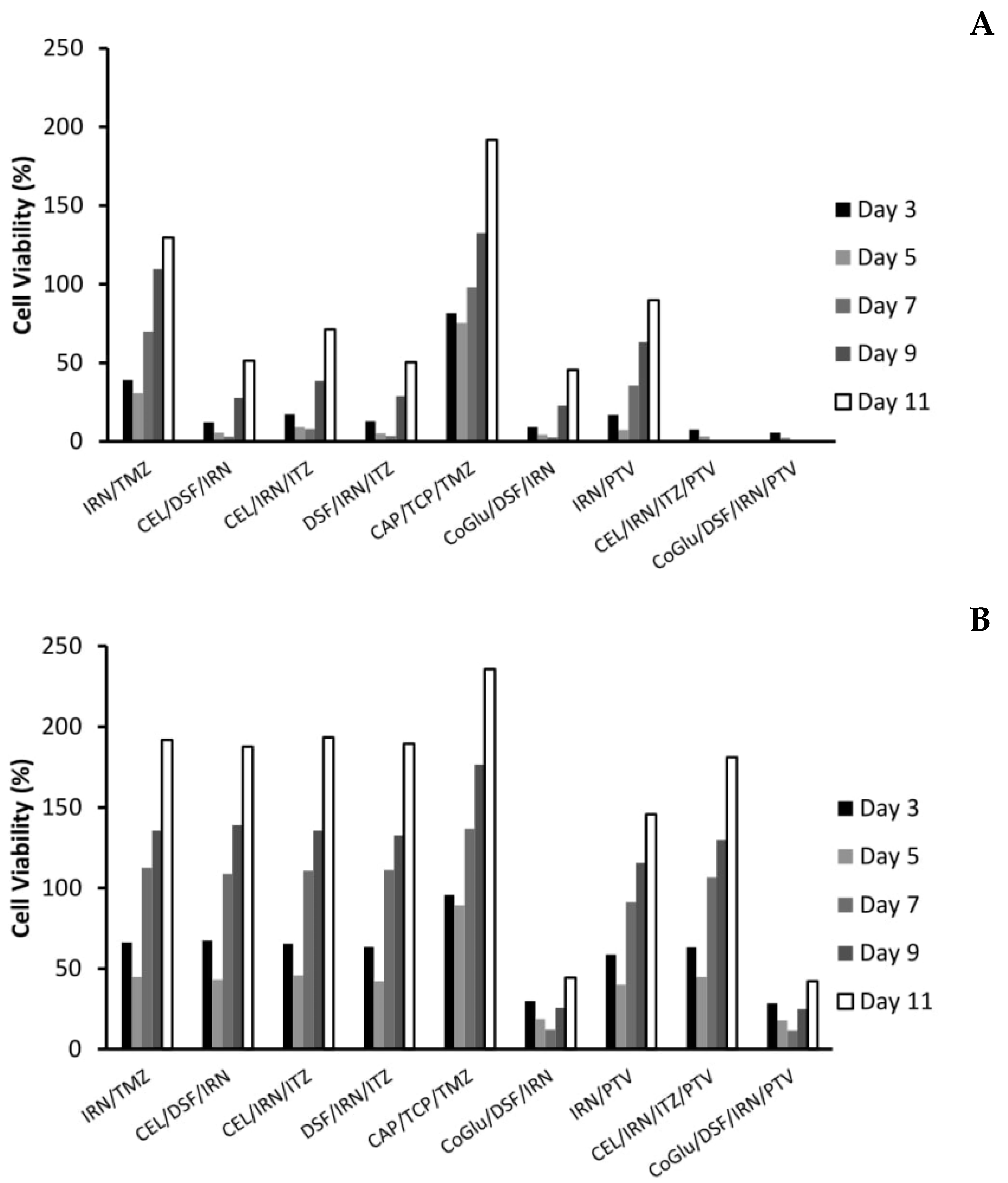
3.5. The Influence of Personalisation on GBM Recurrence
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Fulop, J.; Liu, M.; Blanda, R.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J. CBTRUS Statistical report: Primary brain and central Nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2015, 17, iv1–iv62. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 world health organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Snuderl, M.; Fazlollahi, L.; Le, L.P.; Nitta, M.; Zhelyazkova, B.H.; Davidson, C.J.; Akhavanfard, S.; Cahill, D.P.; Aldape, K.D.; Betensky, R.A.; et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell 2011, 20, 810–817. [Google Scholar] [CrossRef]
- Szerlip, N.J.; Pedraza, A.; Chakravarty, D.; Azim, M.; McGuire, J.; Fang, Y.; Ozawa, T.; Holland, E.C.; Huse, J.T.; Jhanwar, S.; et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc. Natl. Acad. Sci. USA 2012, 109, 3041–3046. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Friedmann-Morvinski, D. Glioblastoma heterogeneity and cancer cell plasticity. Crit. Rev. Oncog. 2014, 19, 327–336. [Google Scholar] [CrossRef]
- Nguyen, K.S.; Kobayashi, S.; Costa, D.B. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin. Lung Cancer 2009, 10, 281–289. [Google Scholar] [CrossRef]
- Nickel, G.C.; Barnholtz-Sloan, J.; Gould, M.P.; McMahon, S.; Cohen, A.; Adams, M.D.; Guda, K.; Cohen, M.; Sloan, A.E.; LaFramboise, T. Characterizing mutational heterogeneity in a glioblastoma patient with double recurrence. PLoS ONE 2012, 7, e35262. [Google Scholar] [CrossRef]
- Prados, M.D.; Byron, S.A.; Tran, N.L.; Phillips, J.J.; Molinaro, A.M.; Ligon, K.L.; Wen, P.Y.; Kuhn, J.G.; Mellinghoff, I.K.; de Groot, J.F.; et al. Toward precision medicine in glioblastoma: The promise and the challenges. Neuro Oncol. 2015, 17, 1051–1063. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. An integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterised by abnormalities in PDGFRA, IDH1, EGFR and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Alifieris, C.; Trafalis, D.T. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol. Ther. 2015, 152, 63–82. [Google Scholar] [CrossRef] [PubMed]
- Chaurasia, A.; Park, S.H.; Seo, J.W.; Park, C.K. Immunohistochemical analysis of ATRX, IDH1 and p53 in glioblastoma and their correlations with patient survival. J. Korean Med. Sci. 2016, 31, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Hu, R.; Yang, H.; Liu, J.; Sui, J.; Xiang, X.; Wang, F.; Chu, L.; Song, S. PTEN gene mutations correlate to poor prognosis in glioma patients: A meta-analysis. Onco Targets Ther. 2016, 9, 3485–3492. [Google Scholar] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Weller, M.; Stupp, R.; Hegi, M.E.; van den Bent, M.; Tonn, J.C.; Sanson, M.; Wick, W.; Reifenberger, G. Personalized care in neuro-oncology coming of age: Why we need MGMT and 1p/19q testing for malignant glioma patients in clinical practice. Neuro Oncol. 2012, 14, 100–108. [Google Scholar] [CrossRef]
- Yuan, Y.; Qi, C.; Maling, G.; Xiang, W.; Yanhui, L.; Ruofei, L.; Yunhe, M.; Jiewen, L.; Qing, M. TERT mutation in glioma: Frequency, prognosis and risk. J. Clin. Neurosci. 2016, 26, 57–62. [Google Scholar] [CrossRef]
- Tanase, C.P.; Enciu, A.M.; Mihai, S.; Neagu, A.I.; Calenic, B.; Cruceru, M.L. Anti-cancer therapies in high grade gliomas. Curr. Proteom. 2013, 10, 246–260. [Google Scholar] [CrossRef]
- Domingo-Musibay, E.; Galanis, E. What next for newly diagnosed glioblastoma? Future Oncol. 2015, 11, 3273–3283. [Google Scholar] [CrossRef]
- Oh, Y.; Cho, H.J.; Kim, J.; Lee, J.H.; Rho, K.; Seo, Y.J.; Choi, Y.S.; Jung, H.J.; Song, H.S.; Kong, D.S.; et al. Translational validation of personalized treatment strategy based on genetic characteristics of glioblastoma. PLoS ONE 2014, 9, e103327. [Google Scholar] [CrossRef]
- Tan, H.; Bao, J.; Zhoua, X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci. Rep. 2015, 5, 12566. [Google Scholar] [CrossRef]
- Leeper, H.E.; Caron, A.A.; Decker, P.A.; Jenkins, R.B.; Lachance, D.H.; Giannini, C. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget 2015, 6, 30295–30305. [Google Scholar] [CrossRef] [PubMed]
- The Gene Ontology Consortium. Expansion of the Gene Ontology Knowledgebase and Resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; et al. Gene Ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2009, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- National Centre for Biotechnology Information. Gene2go [Data File]. The NCBI FTP Site. 2017. Available online: Ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/ (accessed on 19 July 2019).
- Chen, J.R.; Yao, Y.; Xu, H.Z.; Qin, Z.Y. Isocitrate dehydrogenase (IDH)1/2 mutations as prognostic markers in patients with glioblastomas. Medicine 2016, 95, e2583. [Google Scholar] [CrossRef] [PubMed]
- Juratli, T.A.; Kirsch, M.; Robel, K.; Soucek, S.; Geiger, K.; von Kummer, R.; Schackert, G.; Krex, D. IDH mutations as an early and consistent marker in lowgrade astrocytomas WHO grade II and their consecutive secondary high-grade gliomas. J. Neurooncol. 2012, 108, 403–410. [Google Scholar] [CrossRef] [PubMed]
- SongTao, Q.; Lei, Y.; Si, G.; YanQing, D.; HuiXia, H.; XueLin, Z.; LanXiao, W.; Fei, Y. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012, 103, 269–273. [Google Scholar] [CrossRef]
- Houillier, C.; Wang, X.; Kaloshi, G.; Mokhtari, K.; Guillevin, R.; Laffaire, J.; Paris, S.; Boisselier, B.; Idbaih, A.; Laigle-Donadey, F.; et al. IDH1 or IDH2 mutations predict longer survival and response to temozolomide in low-grade gliomas. Neurology 2010, 75, 1560–1566. [Google Scholar] [CrossRef]
- Conte, D.; Huh, M.; Goodall, E.; Delorme, M.; Parks, R.J.; Picketts, D.J. Loss of Atrx sensitizes cells to DNA damaging agents through p53-mediated death pathways. PLoS ONE 2012, 7, e52167. [Google Scholar] [CrossRef]
- Zhang, Y.; Dube, C.; Gibert, M.; Cruickshanks, N.; Wang, B.; Coughlan, M.; Yang, Y.; Setiady, I.; Deveau, C.; Saoud, K.; et al. The p53 Pathway in glioblastoma. Cancers 2018, 10, 297. [Google Scholar] [CrossRef]
- Jiang, Z.; Pore, N.; Cerniglia, G.J.; Mick, R.; Georgescu, M.M.; Bernhard, E.J.; Hahn, S.M.; Gupta, A.K.; Maity, A. Phosphatase and tensin homologue deficiency in glioblastoma confers resistance to radiation and temozolomide that is reversed by the protease inhibitor nelfinavir. Cancer Res. 2007, 67, 4467–4473. [Google Scholar] [CrossRef]
- Benitez, J.A.; Ma, J.; D’Antonio, M.; Boyer, A.; Camargo, M.F.; Zanca, C.; Kelly, S.; Khodadadi-Jamayran, A.; Jameson, N.M.; Andersen, M.; et al. PTEN regulates glioblastoma oncogenesis through chromatin-associated complexes of DAXX and histone H3.3. Nat. Commun. 2017, 8, 15223. [Google Scholar] [CrossRef] [PubMed]
- Quayle, S.N.; Lee, J.Y.; Cheung, L.W.; Ding, L.; Wiedemeyer, R.; Dewan, R.W.; Huang-Hobbs, E.; Zhuang, L.; Wilson, R.K.; Ligon, K.L.; et al. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS ONE 2012, 7, e49466. [Google Scholar] [CrossRef] [PubMed]
- McNeill, R.S.; Stroobant, E.E.; Smithberger, E.; Canoutas, D.A.; Butler, M.K.; Shelton, A.K.; Patel, S.D.; Limas, J.C.; Skinner, K.R.; Bash, R.E.; et al. PIK3CA missense mutations promote glioblastoma pathogenesis, but do not enhance targeted PI3K inhibition. PLoS ONE 2018, 13, e0200014. [Google Scholar] [CrossRef] [PubMed]
- Draaisma, K.; Wijnenga, M.M.; Weenink, B.; Gao, Y.; Smid, M.; Robe, P.; van den Bent, M.J.; French, P.J. PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients. Acta Neuropathol. Commun. 2015, 3, 88. [Google Scholar] [CrossRef]
- Korshunov, A.; Sycheva, R.; Golanov, A. The prognostic relevance of molecular alterations in glioblastomas for patients age <50 years. Cancer 2005, 104, 825–832. [Google Scholar]
- Ohgaki, H.; Dessen, P.; Jourde, B.; Horstmann, S.; Nishikawa, T.; Di Patre, P.L.; Burkhard, C.; Schüler, D.; Probst-Hensch, N.M.; Maiorka, P.C.; et al. Genetic pathways to glioblastoma: A population-based study. Cancer Res. 2004, 64, 6892–6899. [Google Scholar] [CrossRef]
- Kamiryo, T.; Tada, K.; Shiraishi, S.; Shinojima, N.; Nakamura, H.; Kochi, M.; Kuratsu, J.; Saya, H.; Ushio, Y. Analysis of homozygous deletion of the p16 gene and correlation with survival in patients with glioblastoma multiforme. J. Neurosurg. 2002, 96, 815–822. [Google Scholar] [CrossRef]
- Park, A.K.; Kim, P.; Ballester, L.Y.; Esquenazi, Y.; Zhao, Z. Subtype-specific signaling pathways and genomic aberrations associated with prognosis of glioblastoma. Neuro Oncol. 2019, 21, 59–70. [Google Scholar] [CrossRef]
- Jiang, P.; Mukthavaram, R.; Chao, Y.; Bharati, I.S.; Fogal, V.; Pastorino, S.; Cong, X.; Nomura, N.; Gallagher, M.; Abbasi, T.; et al. Novel anti-glioblastoma agents and therapeutic combinations identified from a collection of FDA approved drugs. J. Trans. Med. 2014, 12, 13. [Google Scholar] [CrossRef]
- Kast, R.E.; Boockvar, J.A.; Brüning, A.; Cappello, F.; Chang, W.W.; Cvek, B.; Dou, Q.P.; Duenas-Gonzalez, A.; Efferth, T.; Focosi, D.; et al. A conceptually new treatment approach for relapsed glioblastoma: Coordinated undermining of survival paths with nine repurposed drugs (CUSP9) by the International Initiative for Accelerated Improvement of Glioblastoma Care. Oncotarget 2013, 4, 502–530. [Google Scholar] [CrossRef]
- Kast, R.E.; Karpel-Massler, G.; Halatsch, M.E. CUSP9* treatment protocol for recurrent glioblastoma: Aprepitant, artesunate, auranofin, captopril, celecoxib, disulfiram, itraconazole, ritonavir, sertraline augmenting continuous low dose temozolomide. Oncotarget 2014, 5, 8052–8082. [Google Scholar] [CrossRef]
- Ramesh, M.; Ahlawat, P.; Srinivas, N. Irinotecan and its active metabolite, SN-38: Review of bioanalytical methods and recent update from clinical pharmacology perspectives. Biomed. Chromatogr. 2010, 24, 104–123. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y. Irinotecan: Mechanisms of tumor resistance and novel strategies for modulating its activity. Ann. Oncol. 2002, 13, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Mukthavaram, R.; Chao, Y.; Nomura, N.; Bharati, I.S.; Fogal, V.; Pastorino, S.; Teng, D.; Cong, X.; Pingle, S.C.; et al. In vitro and in vivo anticancer effects of mevalonate pathway modulation on human cancer cells. Br. J. Cancer 2014, 111, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Pirmoradi, L.; Seyfizadeh, N.; Ghavami, S.; Zeki, A.A.; Shojaei, S. Targeting cholesterol metabolism in glioblastoma: A new therapeutic approach in cancer therapy. J. Investig. Med. 2019, 67, 715–719. [Google Scholar] [CrossRef]
- Srivastava, N.K.; Pradhan, S.; Gowda, G.A.; Kumar, R. In vitro, high-resolution 1H and 31P NMR based analysis of the lipid components in the tissue, serum, and CSF of the patients with primary brain tumors: One possible diagnostic view. NMR Biomed. 2010, 23, 113–122. [Google Scholar] [CrossRef]
- Yung, W.K.A.; Jiang, S.X.; Reardon, D.A.; Desjardins, A.; Vredenburgh, J.J.; Friedman, A.H.; Sampson, J.H.; McLendon, R.E.; Herndon, J.E.; Friedmancorresponding, H.S. Combination of temozolomide (TMZ) and irinotecan (CPT-11) showed enhanced activity for recurrent malignant gliomas: A North American Brain Tumor Consortium (NABTC) phase II study. J. Clin. Oncol. 2005, 23, 1521. [Google Scholar] [CrossRef]
- Gruber, M.L.; Buster, W.P. Temozolomide in combination with irinotecan for treatment of recurrent malignant glioma. Am. J. Clin. Oncol. 2004, 27, 33–38. [Google Scholar]
- Liu, M.; Li, C.M.; Chen, Z.F.; Ji, R.; Guo, Q.H.; Li, Q.; Zhang, H.L.; Zhou, Y.N. Celecoxib regulates apoptosis and autophagy via the PI3K/Akt signaling pathway in SGC-7901 gastric cancer cells. Int. J. Mol. Med. 2014, 33, 1451–1458. [Google Scholar] [CrossRef]
- Du, L.X.; Jia, Y.Q.; Meng, W.T.; Shi, F.F.; Zhong, X.S.; Ma, L.L.; Yuan, J.; Zeng, J.S. COX-2 inhibitor celecoxib can suppress the proliferation of FLT3-ITD positive acute myeloid leukemia cells with prominent down regulation of MEK/MCL-1 expression in vitro. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2013, 21, 1157–1161. [Google Scholar]
- Liu, P.; Brown, S.; Goktug, T.; Channathodiyil, P.; Kannappan, V.; Hugnot, J.P.; Guichet, P.O.; Bian, X.; Armesilla, A.L.; Darling, J.L.; et al. Cytotoxic effect of disulfiram/copper on human glioblastoma cell lines and ALDH-positive cancer-stem-like cells. Br. J. Cancer 2012, 107, 1488–1497. [Google Scholar] [CrossRef]
- Safi, R.; Nelson, E.R.; Chitneni, S.K.; Franz, K.J.; George, D.J.; Zalutsky, M.R.; McDonnell, D.P. Copper signaling axis as a target for prostate cancer therapeutics. Cancer Res. 2014, 74, 5819–5831. [Google Scholar] [CrossRef]
- Buac, D.; Schmitt, S.; Ventro, G.; Kona, F.R.; Dou, Q.P. Dithiocarbamate-based coordination compounds as potent proteasome inhibitors in human cancer cells. Mini Rev. Med. Chem. 2012, 12, 1193–1201. [Google Scholar] [CrossRef] [PubMed]
- Tawari, P.E.; Wang, Z.; Najlah, M.; Tsang, C.W.; Kannappan, V.; Liu, P.; McConville, C.; He, B.; Armesilla, A.L.; Wang, W. The cytotoxic mechanisms of disulfiram and copper(ii) in cancer cells. Toxicol. Res. 2015, 4, 1439–1442. [Google Scholar] [CrossRef] [PubMed]
- Birgersdotter, A.; Sandberg, R.; Ernberg, I. Gene expression perturbation in vitro—A growing case for three-dimensional (3D) culture systems. Semin. Cancer Biol. 2005, 15, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Harma, V.; Virtanen, J.; Makela, R.; Happonen, A.; Mpindi, J.P.; Knuuttila, M.; Kohonen, P.; Lötjönen, J.; Kallioniemi, O.; Nees, M. A Comprehensive panel of three-dimensional models for studies of prostate cancer growth, Invasion and Drug Responses. PLoS ONE 2010, 5, e10431. [Google Scholar] [CrossRef]
- Elliott, N.T.; Yuan, F. A Review of Three-dimensional in vitro tissue models for drug discovery and transport studies. J. Pharm. Sci. 2010, 100, 59–74. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A Three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]

| Gene | ||||
|---|---|---|---|---|
| Sample | IDH | ATRX | P53 | MGMT |
| GBM1 | Wild Type | Wild Type | Wild Type | Methylated |
| GBM2 | Wild Type | Wild Type | Unspecified | Unmethylated |
| GBM3 | Mutated | Wild Type | Wild Type | Unspecified |
| GBM4 | Mutated | Mutated | Mutated | Unspecified |
| GBM5 | Wild Type | Wild Type | Wild Type | Unmethylated |
| GBM6 | Wild Type | Wild Type | Wild Type | Unspecified |
| Gene | Full Name | Function |
|---|---|---|
| ATRX | Alpha Thalassemia/Mental Retardation Syndrome X-Linked | Involved in transcriptional regulation and chromatin remodelling |
| USH2A | Usher Syndrome 2A | Involved in hearing and vision |
| TP53 | Tumor Protein P53 | Tumour suppressor gene. |
| PTEN | Phosphatase And Tensin Homolog | Tumour suppressor gene |
| PKHD1 | Polycystic Kidney And Hepatic Disease 1 | Involved in cell adhesion, repulsion and proliferation. |
| PIK3R1 | Phosphoinositide-3-Kinase Regulatory Subunit 1 | Involved in cell proliferation and survival |
| PIK3CA | Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha | Involved in cell proliferation and survival |
| PCLO | Piccolo Presynaptic Cytomatrix Protein | Cell scaffolding protein |
| MUC17 | Mucin 17 | Maintains homeostasis on mucosal surfaces |
| MUC16 | Mucin 16 | Provides a protective and lubricating barrier at mucosal surfaces |
| IDH1 | Isocitrate Dehydrogenase 1 | Is necessary for many cellular processes |
| FLG | Filaggrin | Involved in the structure of the epidermis and plays an important role in the barrier function of the epidermis |
| CDKN2A | Cyclin Dependent Kinase Inhibitor 2A | Tumour suppressor gene. |
| Gene | Tumour Cells | Drug |
|---|---|---|
| ATRX | GBM 4, 5 and 6 | CEL, IRN, ITZ and PTV |
| TP53 | GBM 4 and 5 | CEL, IRN, ITZ and PTV |
| PTEN | GBM 5 | CEL, IRN, ITZ and PTV |
| PKHD1 | GBM 1, 4 and 6 | IRN |
| PIK3R1 | GBM 3 | CEL and PTV |
| PIK3CA | GBM 5 | CEL, IRN, ITZ and PTV |
| IDH1 | GBM 3 and 4 | CEL and PTV |
| CDKN2A | GBM 5 | CEL, IRN, ITZ and PTV |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garrett, A.-M.; Lastakchi, S.; McConville, C. The Personalisation of Glioblastoma Treatment Using Whole Exome Sequencing: A Pilot Study. Genes 2020, 11, 173. https://doi.org/10.3390/genes11020173
Garrett A-M, Lastakchi S, McConville C. The Personalisation of Glioblastoma Treatment Using Whole Exome Sequencing: A Pilot Study. Genes. 2020; 11(2):173. https://doi.org/10.3390/genes11020173
Chicago/Turabian StyleGarrett, Anne-Marie, Sarah Lastakchi, and Christopher McConville. 2020. "The Personalisation of Glioblastoma Treatment Using Whole Exome Sequencing: A Pilot Study" Genes 11, no. 2: 173. https://doi.org/10.3390/genes11020173
APA StyleGarrett, A.-M., Lastakchi, S., & McConville, C. (2020). The Personalisation of Glioblastoma Treatment Using Whole Exome Sequencing: A Pilot Study. Genes, 11(2), 173. https://doi.org/10.3390/genes11020173






