Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Measurement of Right-Ventricular Systolic Pressure (RVSP)
2.3. Assessment of Right-Ventricular Hypertrophy (RVH)
2.4. Histology
2.5. Microarray
2.6. Transcriptomic Analysis
2.7. Pathway Analysis
3. Results
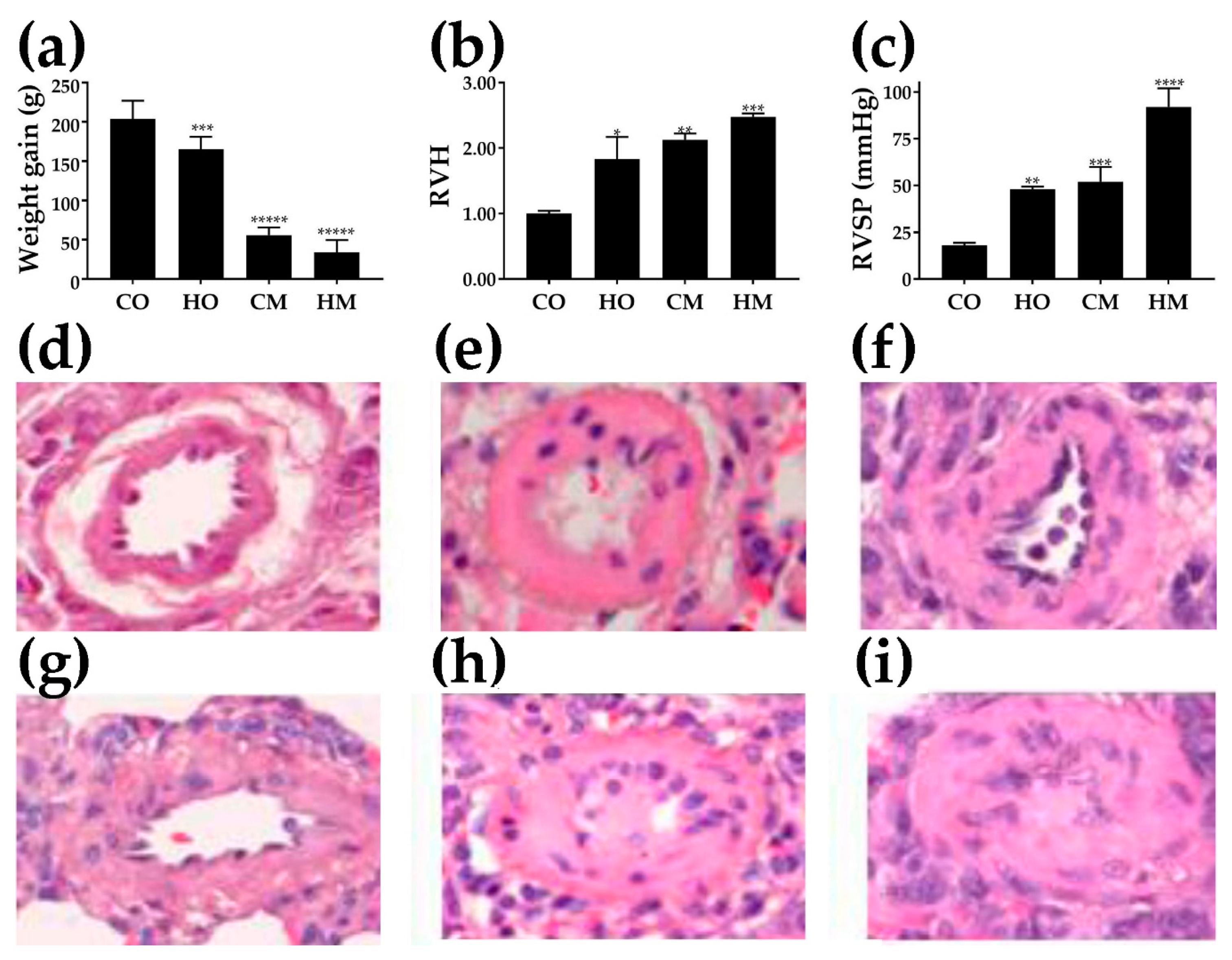
3.1. Decreased Weight Gain
3.2. Right-Ventricle Hypertrophy
3.3. Increased Right-Ventricle Systolic Pressure
3.4. Histological Alterations
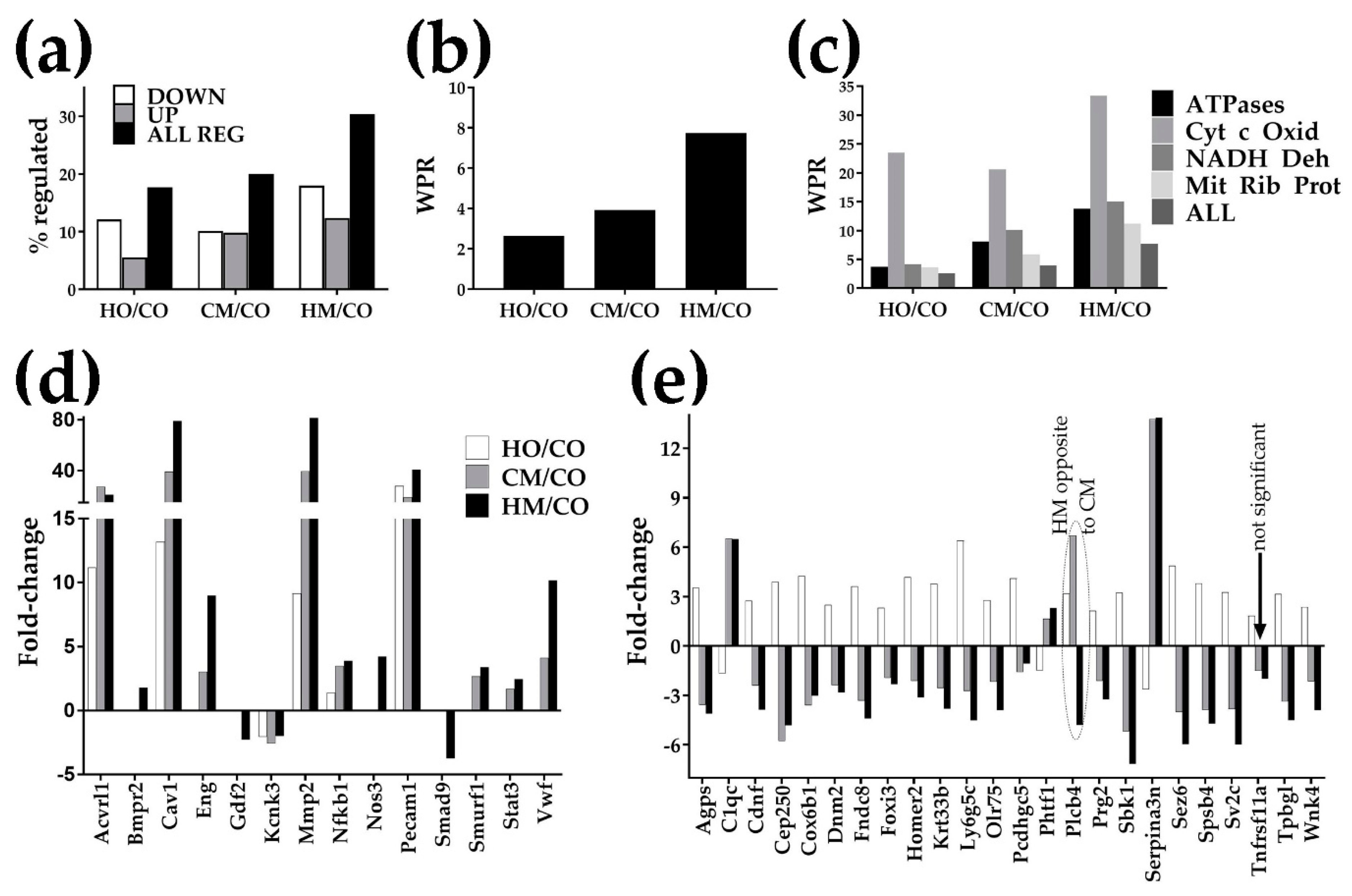
3.5. Overview of the Transcriptomic Alterations
3.6. Regulation of the Immune-Inflammatory Response
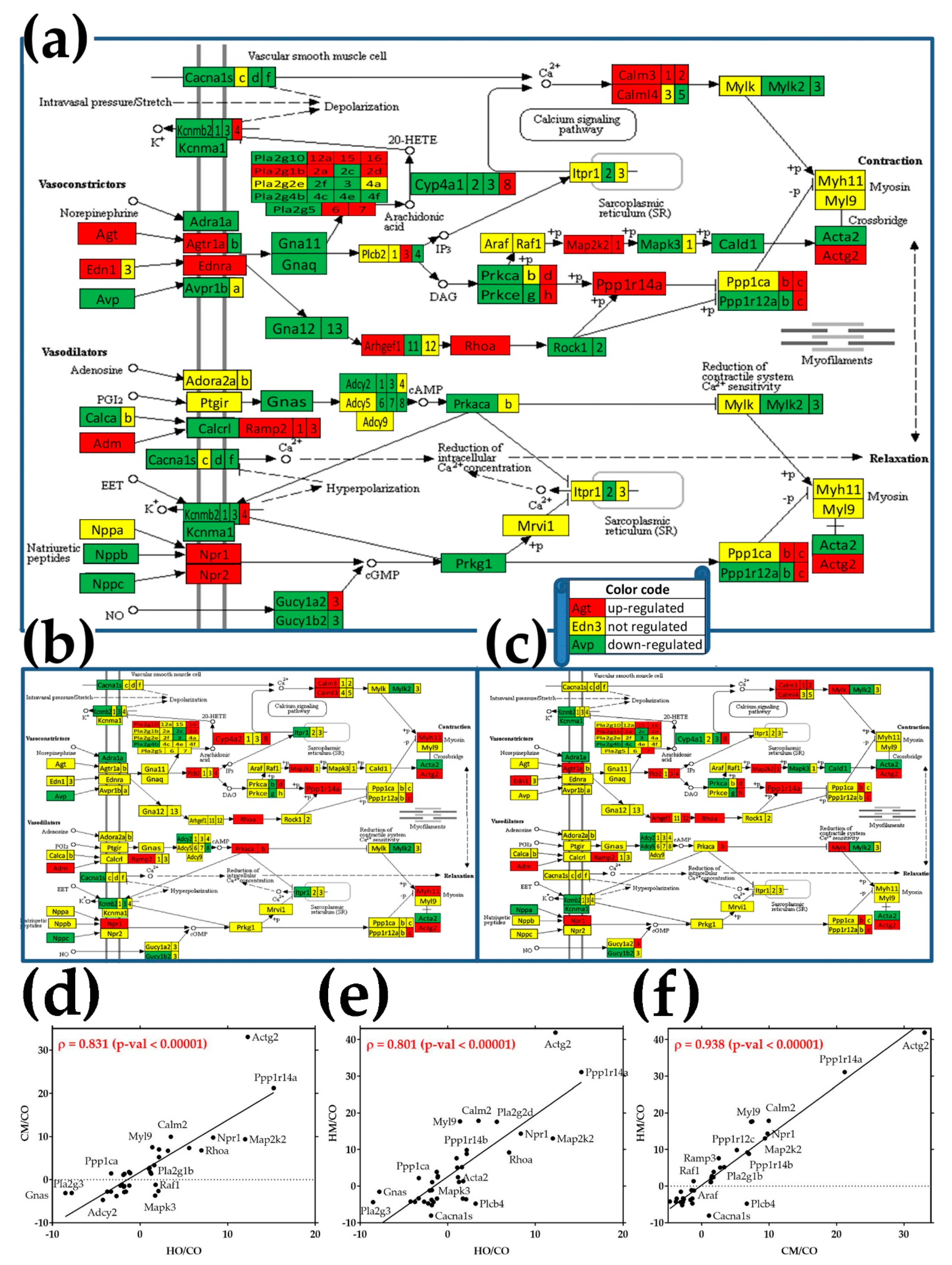
3.7. Regulation of the Vascular Smooth Muscle Contraction Pathway
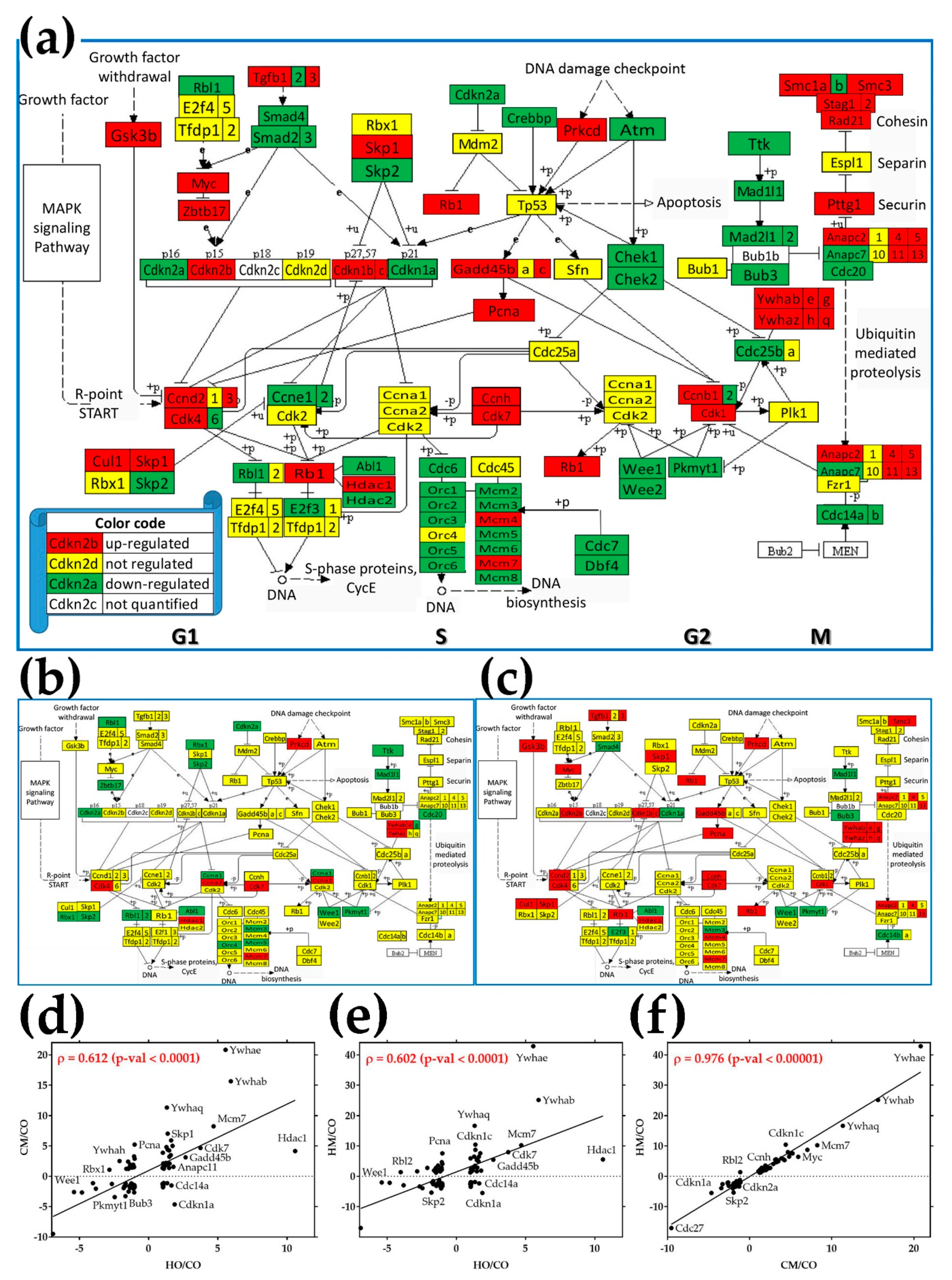
3.8. Regulation of the Cell-Cycle Pathway
3.9. Alteration of Cellular Respiration
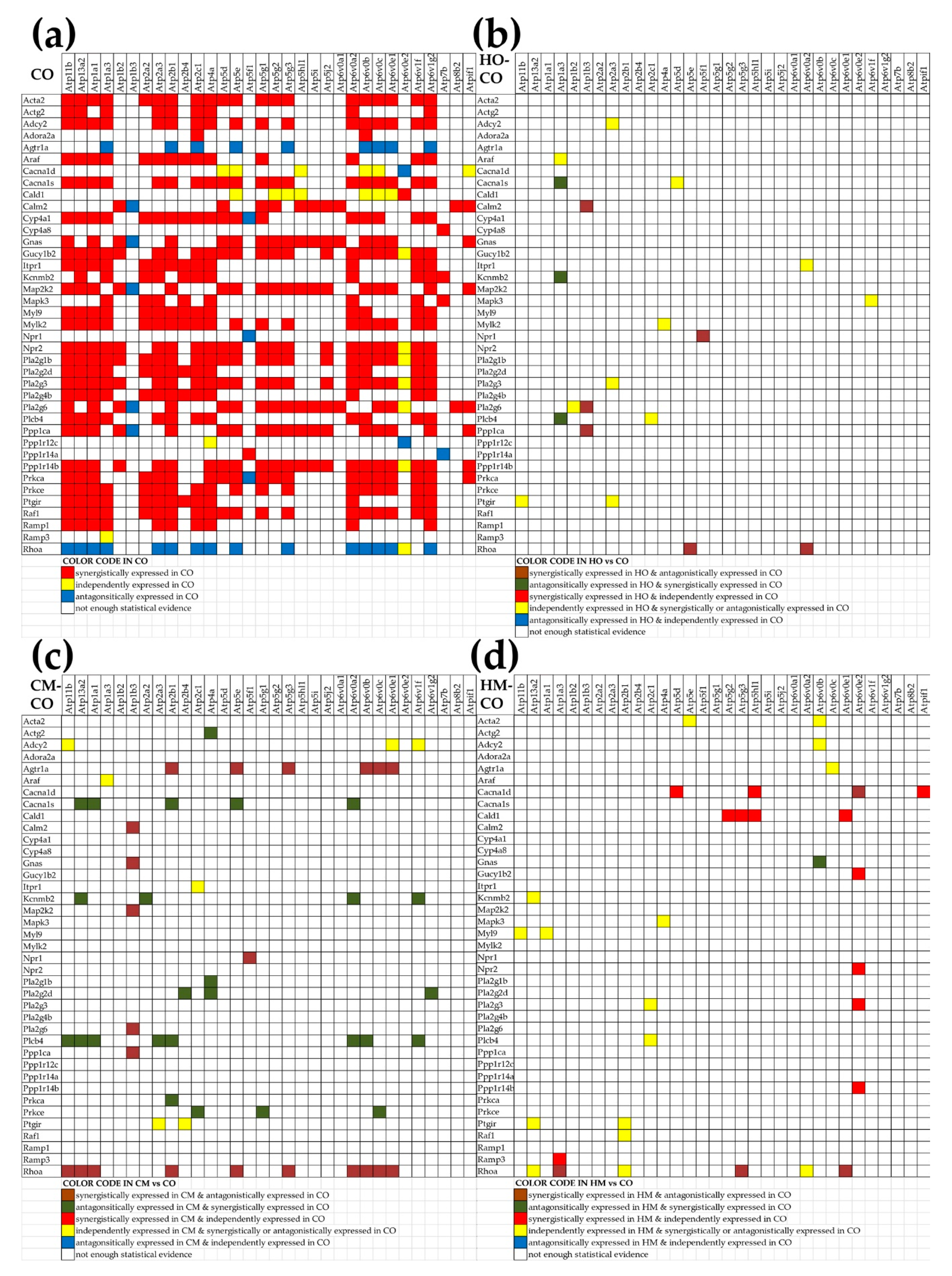
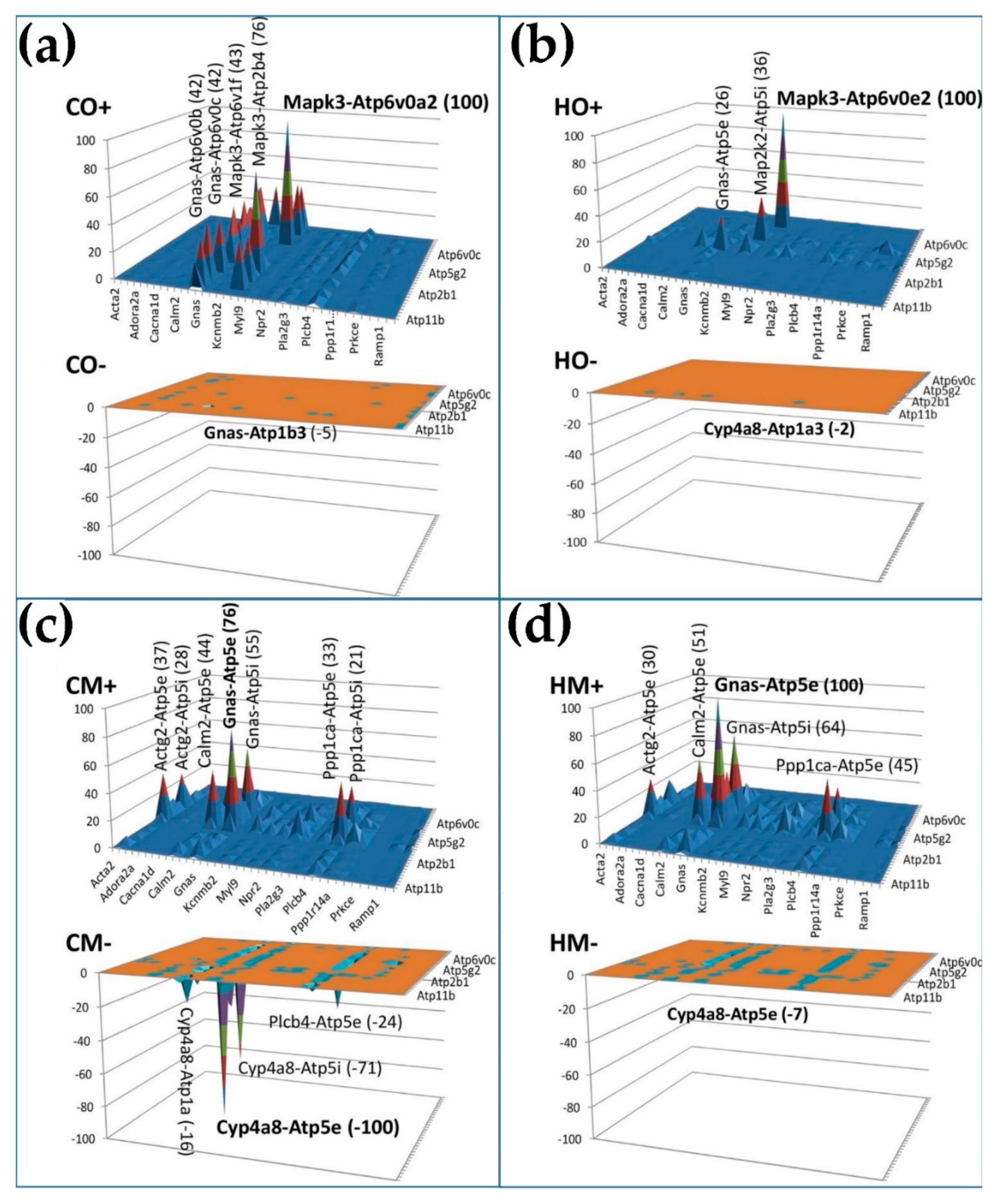
3.10. Remodeling of the Relationship between the Genes Involved in the Vascular Smooth Muscle Contraction and Oxidative Phosphorylation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Stearman, R.S.; Bui, Q.M.; Speyer, G.; Handen, A.; Cornelius, A.R.; Graham, B.B.; Geraci, M.W. Systems Analysis of the Human Pulmonary Arterial Hypertension Lung Transcriptome. Am. J. Respir. Cell Mol. Biol. 2019, 60, 637–649. [Google Scholar] [CrossRef]
- Galiè, N.; McLaughlin, V.V.; Rubin, L.J.; Simonneau, G. An overview of the 6th World Symposium on Pulmonary Hypertension. Eur. Respir. J. 2019, 53, 1802148. [Google Scholar] [CrossRef]
- D’Alonzo, G.C.; Barst, R.J.; Ayers, S.M.; Bergofsky, E.H.; Brundage, B.H.; Detre, K.M.; Levy, P.S. Survival in patients with primary pulmonary hypertension: Results from a national registry. Ann. Intern. Med. 1991, 115, 343–349. [Google Scholar] [CrossRef]
- Pogoriler, J.E.; Rich, S.; Archer, S.L.; Husain, A.N. Persistence of complex vascular lesions despite prolonged prostacyclin therapy of pulmonary arterial hypertension. Histopathology 2012, 61, 597–609. [Google Scholar] [CrossRef]
- Sakao, S.; Tatsumi, K.; Voelkel, N.F. Reversible or irreversible remodeling in pulmonary arterial hypertension. Am. J. Respir. Cell Mol. Biol. 2010, 43, 629–634. [Google Scholar] [CrossRef]
- Humbert, M.; Sitbon, O.; Yaïci, A.; Montani, D.; O’Callaghan, D.S.; Jaïs, X.; Chaouat, A. Survival in incident and prevalent cohorts of patients with pulmonary arterial hypertension. Eur. Respir. J. 2010, 36, 549–555. [Google Scholar] [CrossRef]
- Farber, H.W.; Miller, D.P.; Poms, A.D.; Badesch, D.B.; Frost, A.E.; Muros-Le Rouzic, E.; Benza, R.L. Five-Year outcomes of patients enrolled in the REVEAL Registry. Chest 2015, 148, 1043–1054. [Google Scholar] [CrossRef]
- Deng, Z.; Morse, J.H.; Slager, S.L.; Cuervo, N.; Moore, K.J.; Venetos, G.; Hodge, S.E. Familial primary pulmonary hypertension (gene PPH1) is caused by mutations in the bone morphogenetic protein receptor-II gene. Am. J. Hum. Genet. 2000, 67, 737–744. [Google Scholar] [CrossRef]
- Girerd, B.; Montani, M.; Coulet, F.; Sztrymf, B.; Yaici, A.; Jaïs, X.; Sitbon, O. Clinical Outcomes of Pulmonary Arterial Hypertension in Patients Carrying an ACVRL1 (ALK1) Mutation. Am. J. Respir. Crit. Care Med. 2010, 181, 851–861. [Google Scholar] [CrossRef]
- Chaouat, A.; Coulet, F.; Favre, C.; Simonneau, G.; Weitzenblum, E.; Soubrier, F.; Humbert, M. Endoglin germline mutation in a patient with hereditary haemorrhagic tlangiectasia and dexfenfluramine associated pulmonary arterial hypertension. Thorax 2004, 59, 446–448. [Google Scholar] [CrossRef]
- Drake, K.M.; Zygmunt, D.; Mavrakis, L.; Harbor, P.; Wang, L.; Comhair, S.A. Altered microRNA processing in heritable pulmonary arterial hypertension: An important role for Smad-8. Am. J. Respir. Crit. Care Med. 2011, 184, 1400–1408. [Google Scholar] [CrossRef]
- Austin, E.D.; Ma, L.; LeDuc, C.; Berman-Rozenzweig, E.; Borczuk, A.; Phillips, J.A.; West, J. Whole exon sequence to identify a novel gene (caveolin-1) associated with human pulmonary arterial hypertension. Circ. Cardiovasc. Genet. 2012, 5, 336–343. [Google Scholar] [CrossRef]
- Tejedor, P.N.; Castaño, J.T.; Doza, J.P.; Lajara, A.; Trujillo, G.G.; Meseguer, M.L.; Escribano Subía, P. A homozygous mutation in KCNK3 is associated with an aggressive form of hereditary pulmonary arterial hypertension. Clin. Genet. 2017, 91, 453–457. [Google Scholar] [CrossRef]
- Eichstaedt, C.A.; Song, J.; Benjamin, N.; Harutyunova, S.; Fischer, C.; Grünig, E.; Hinderhofer, K. EIF2AK4 Mutation as “Second Hit” in Hereditary Pulmonary Arterial Hypertension. Respir. Res. 2016, 17, 141. [Google Scholar] [CrossRef]
- Aldred, M.A.; Comhair, S.A.; Varella-Garcia, M.; Asosingh, K.; Xu, W.; Noon, G.P.; Coldren, C.D. Somatic chromosome abnormalities in the lungs of patients with pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2010, 182, 1153–1160. [Google Scholar] [CrossRef]
- Evans, J.D.; Girerd, B.; Montani, D.; Wang, X.-J.; Galiè, N.; Austin, E.D.; Jing, Z.C. BMPR2 mutations and survival in pulmonary arterial hypertension: An individual participant data meta-analysis. Lancet Respir. Med. 2016, 4, 129–137. [Google Scholar] [CrossRef]
- Yan, L.; Cogan, J.D.; Hedges, L.K.; Nunley, B.; Hamid, R.; Austin, E.D. The Y Chromosome regulates BMPR2 expression via SRY: A possible reason “why” fewer males develop pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2018, 198, 1581–1583. [Google Scholar] [CrossRef]
- Mathew, R. Possible mechanism/s of neointima formation in pulmonary arterial hypertension. Blood Heart Circ. 2018, 2, 1–5. [Google Scholar] [CrossRef]
- Murakami, K.; Mathew, R.; Huang, J.; Farahami, R.; Peng, H.; Olson, S.C.; Etlinger, J.D. Smurf1 ubiquitin ligase causes downregulation of BMP receptors and is induced in monocrotaline and hypoxia models of pulmonary arterial hypertension. Exp. Biol. Med. 2010, 235, 805–813. [Google Scholar] [CrossRef]
- McMurtry, M.S.; Moudgil, R.; Hashimoto, K.; Bonnet, S.; Michelakis, E.D.; Archer, S.L. Overexpression of human bone morphogenetic protein receptor 2 does not ameliorate monocrotaline pulmonary arterial hypertension. Am. J. Physiol. Lung Cell. Mol. Phsyiol. 2007, 292, L872–L878. [Google Scholar] [CrossRef]
- Long, L.; Ormiston, M.L.; Yang, X.; Southwood, M.; Gräf, S.; Machado, R.D.; Moore, S.D. Selective enhancement of endothelial BMPR-II with BMP9 reverses pulmonary arterial hypertension. Nat. Med. 2015, 21, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R. Pulmonary Hypertension: Endothelial cell Function. In Pulmonary Hypertension: From Bench Research to Clinical Challenge; Sulica, R., Preston, I., Eds.; IntechOpen: London, UK, 2011; pp. 1–24. [Google Scholar] [CrossRef][Green Version]
- Li, M.; Vattulainen, S.; Aho, J.; Orcholski, M.; Rojas, V.; Yuan, K.; Koskenvuo, J.W. Loss of bone morphogenetic protein receptor 2 is associated with abnormal DNA repair in pulmonary arterial hypertension. Am. J. Respir. Cell Mol. Biol. 2014, 50, 1118–1128. [Google Scholar] [CrossRef]
- Rothman, A.M.; Arnold, N.D.; Pickworth, J.A.; Iremonger, J.; Ciuclan, L.; Allen, R.M.; Francis, S.E. MicroRNA-140-5p and SMURF1 regulate pulmonary arterial hypertension. J. Clin. Investig. 2016, 126, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Mason, N.A.; Springall, D.R.; Burke, M.; Pollock, J.; Mikhail, G.; Yacoub, M.H.; Polak, J.M. High expression of endothelial nitric oxide synthase in plexiform lesions of pulmonary hypertension. J. Pathol. 1998, 185, 313–318. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Liu, Y.; Stan, R.V.; Fan, L.; Gu, Y.; Dalton, N.; Chien, K.R. Defects in caveolin-1 cause dilated cardiomyopathy and pulmonary hypertension in knockout mice. Proc. Natl. Acad. Sci. USA 2002, 99, 11375–11380. [Google Scholar] [CrossRef] [PubMed]
- Murata, T.; Lin, M.I.; Huang, Y.; Yu, J.; Bauer, P.M.; Giordano, F.G.; Sessa, W.C. Reexpression of caveolin-1 in endothelium rescues the vascular, cardiac, and pulmonary defects in global caveolin-1 knockout mice. J. Exp. Med. 2007, 204, 2373–2382. [Google Scholar] [CrossRef]
- Patel, H.H.; Zhang, S.; Murray, F.; Suda, R.Y.; Head, B.P.; Yokoyama, U.; Thistlethwaite, P.A. Increased smooth muscle cell expression of caveolin-1 and caveolae contribute to the pathophysiology of idiopathic pulmonary arterial hypertension. FASEB J. 2007, 21, 2970–2979. [Google Scholar] [CrossRef]
- Dereddy, N.; Hunag, J.; Erb, M.; Guzel, S.; Wolk, J.H.; Sett, S.S.; Mathew, R. Associated inflammation or increased flow-mediated shear stress, but not the pressure alone disrupts endothelial caveolin-1 in infants with pulmonary hypertension. Pulm. Circ. 2012, 2, 492–500. [Google Scholar] [CrossRef]
- Huang, J.; Wolk, J.H.; Gewitz, M.H.; Loyd, J.E.; West, J.; Austin, E.D.; Mathew, R. Enhanced caveolin-1 expression in smooth muscle cells: Possible prelude to neointima formation. World J. Cardiol. 2015, 7, 671–684. [Google Scholar] [CrossRef]
- Codrici, E.; Albulescu, L.; Popescu, I.D.; Mihai, S.; Enciu, A.M.; Albulescu, R.; Hinescu, M.E. Caveolin-1-Knockout Mouse as a Model of Inflammatory Diseases. J. Immunol. Res. 2018, 2498576. [Google Scholar] [CrossRef]
- Jasmin, J.F.; Mercier, I.; Dupuis, J.; Tanowitz, H.B.; Lisanti, M.P. Short-term administration of a cell-permeable caveolin-1 peptide prevents the development of monocrotaline-induced pulmonary hypertension and right ventricular hypertrophy. Circulation 2006, 114, 912–920. [Google Scholar] [CrossRef]
- Huang, J.; Kaminski, P.M.; Edwards, J.G.; Yeh, A.; Wolin, M.S.; Frishman, W.H.; Mathew, R. Pyrrolidine dithiocarbamate restores endothelial cell membrane integrity and attenuates monocrotaline-induced pulmonary artery hypertension. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 294, L1250–L1259. [Google Scholar] [CrossRef]
- Bakhshi, F.R.; Mao, M.; Shajahan, A.N.; Piegeler, T.; Chen, Z.; Chernaya, O.; Comhair, S. Nitrosation-dependent caveolin 1 phosphorylation, ubiquitination, and degradation and its association with idiopathic pulmonary arterial hypertension. Pulm. Circ. 2013, 3, 816–830. [Google Scholar] [CrossRef]
- Shiroto, T.; Romero, N.; Sugiyama, T.; Sartoretto, J.L.; Kalwa, H.; Yan, Z.; Michel, T. Caveolin-1 is a critical determinant of autophagy, metabolic switching, and oxidative stress in vascular endothelium. PLoS ONE 2014, 9, e87871. [Google Scholar] [CrossRef]
- Huang, J.; Wolk, J.; Gewitz, M.H.; Mathew, R. Progressive Endothelial Cell Damage in an Inflammatory Model of Pulmonary Hypertension. Exp. Lung Res. 2010, 36, 57–66. [Google Scholar] [CrossRef]
- Kravchick, D.O.; Hrdinka, M.; Iacobas, S.; Iacobas, D.A.; Kreutz, M.R.; Jordan, B.A. Synaptonuclear messenger PRR7 inhibits c-Jun ubiquitination and regulates NMDA mediated excitotoxicity. EMBO J. 2016, 35, 1923–1934. [Google Scholar] [CrossRef]
- Iacobas, S.; Neal-Perry, G.; Iacobas, D.A. The Cytoskeleton: Imaging, Isolation, and Interaction; Springer International Publishing: New York, NY, USA, 2013; Volume 79, pp. 119–133. [Google Scholar]
- Fan, C.; Iacobas, D.A.; Zhou, D.; Chen, Q.; Lai, J.K.; Gavrialov, O.; Haddad, G.G. Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol. Genom. 2005, 22, 292–307. [Google Scholar] [CrossRef][Green Version]
- Iacobas, D.A.; Iacobas, S.; Nebieridze, N.; Velisek, L.; Veliskova, J. Estrogen protects neurotransmission transcriptome during status epilepticus. Front. Neurosci. 2018, 12, 332. [Google Scholar] [CrossRef]
- Iacobas, S.; Ede, N.; Iacobas, D.A. The Gene Master Regulators (GMR) Approach Provides Legitimate Targets for Personalized, Time-Sensitive Cancer Gene Therapy. Genes 2019, 10, 560. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Tanowitz, H.B.; de Carvalho, A.C.; Spray, D.C. Functional genomic fabrics are remodeled in a mouse model of Chagasic cardiomyopathy and restored following cell therapy. Microbes Infect. 2018, 20, 185–195. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Chachua, T.; Iacobas, S.; Benson, M.J.; Borges, K.; Veliskova, J.; Velisek, L. ACTH and PMX53 recover the normal synaptic transcriptome in a rat model of infantile spasms. Sci. Rep. 2018, 8, 5722. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Fan, C.; Iacobas, S.; Spray, D.C.; Haddad, G.G. Transcriptomic changes in developing kidney exposed to chronic hypoxia. Biochem. Biophys. Res. Commun. 2006, 349, 329–338. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Fan, C.; Iacobas, S.; Haddad, G.G. Integrated transcriptomic responses to cardiac chronic hypoxia: Translational regulators and response to stress in cell survival. Funct. Integr. Genom. 2008, 8, 265–275. [Google Scholar] [CrossRef][Green Version]
- Iacobas, D.A.; Iacobas, S.; Haddad, G.G. Heart rhythm genomic fabric in hypoxia. Biochem. Biophys. Res. Commun. 2010, 391, 1769–1774. [Google Scholar] [CrossRef]
- Iacobas, D.A.; Iacobas, S.; Spray, D.C. Connexin43 and the brain transcriptome of the newborn mice. Genomics 2007, 89, 113–123. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Iacobas, D.A.; Iacobas, S.; Lee, P.R.; Cohen, J.E.; Fields, R.D. Coordinated Activity of Transcriptional Networks Responding to the Pattern of Action Potential Firing in Neurons. Genes 2019, 10, 754. [Google Scholar] [CrossRef]
- Raw and Processed Gene Expression Results were Deposited at the National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72707 (accessed on 1 December 2020).
- Mapk3 Mitogen Activated Protein Kinase 3, National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/50689 (accessed on 1 December 2020).
- Huang, J.; Frid, M.; Gewitz, M.H.; Fallon, J.T.; Brown, D.; Krafuser, G.; Mathew, R. Hypoxia-induced Pulmonary Hypertension and Chronic Lung Disease: Caveolin-1 Dysfunction an Important Underlying Feature. Pulm. Circ. 2019, 9, 2045894019837876. [Google Scholar] [CrossRef]
- Huang, J.; Wolk, J.H.; Gewitz, M.H.; Mathew, R. Caveolin-1 expression during the progression of pulmonary hypertension. Exp. Biol. Med. 2012, 237, 956–965. [Google Scholar] [CrossRef]
- Mathew, R.; Huang, J.; Shah, M.; Patel, K.; Gewitz, M.; Sehgal, P.B. Disruption of endothelial-cell caveolin-1 α/raft scaffolding during development of monocrotaline-induced pulmonary hypertension. Circulation 2004, 110, 1499–1506. [Google Scholar] [CrossRef]
- Mathew, R. Cell-specific dual role of caveolin-1 in pulmonary hypertension. Pulm. Med. 2011, 2011, 573432. [Google Scholar] [CrossRef]
- Mathew, R. Pathogenesis of pulmonary hypertension: A case for caveolin-1 and cell membrane integrity. Am. J. Physiol. Heart Circ. Physiol. 2014, 306, H15–H25. [Google Scholar] [CrossRef]
- Lepetit, H.; Eddahibi, S.; Fadel, E.; Frisdal, E.; Munaut, C.; Noel, A.; Lafuma, C. Smooth muscle cell matrix metalloproteinases in idiopathic pulmonary arterial hypertension. Eur. Respir. J. 2005, 25, 834–842. [Google Scholar] [CrossRef]
- Tiede, S.L.; Wassenberg, M.; Christ, K.; Schermuly, R.T.; Seeger, W.; Grimminger, F.; Gall, H. Biomarkers of tissue remodeling predict survival in patients with pulmonary hypertension. Int. J. Cardiol. 2016, 223, 821–826. [Google Scholar] [CrossRef]
- Dorfmüller, P.; Zarka, V.; Durand-Gasselin, I.; Monti, G.; Balabanian, K.; Garcia, G.; Emilie, D. Chemokine RANTES in severe pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2002, 165, 534–539. [Google Scholar] [CrossRef]
- Nie, X.; Tan, J.; Dai, Y.; Liu, Y.; Zou, J.; Sun, J.; Bian, J.S. CCL5 deficiency rescues pulmonary vascular dysfunction, and reverses pulmonary hypertension via caveolin-1-dependent BMPR2 activation. J. Mol. Cell. Cardiol. 2018, 116, 41–56. [Google Scholar] [CrossRef]
- Toshner, M.; Voswinckel, R.; Southwood, M.; Al-Lamki, R.; Howard, L.S.; Marchesan, D.; Stewart, S. Evidence of dysfunction of endothelial progenitors in pulmonary arterial hypertension. Am. J. Respir. Crit. Care Med. 2009, 180, 780–787. [Google Scholar] [CrossRef]
- Zhang, T.; Kawaguchi, N.; Hayama, E.; Furutani, Y.; Nakanishi, T. High expression of CXCR4 and stem cell markers in a monocrotaline and chronic hypoxia-induced rat model of pulmonary arterial hypertension. Exp. Ther. Med. 2018, 15, 4615–4622. [Google Scholar] [CrossRef]
- Farkas, D.; Kraskauskas, D.; Drake, J.I.; Alhussaini, A.A.; Kraskauskiene, V.; Bogaard, H.J.; Farkas, L. CXCR4 inhibition ameliorates severe obliterative pulmonary hypertension and accumulation of C-kit⁺cells in rats. PLoS ONE 2014, 9, e89810. [Google Scholar] [CrossRef]
- Yang, J.X.; Zhang, N.; Wang, H.W.; Gao, P.; Yang, Q.P.; Wen, Q.P. CXCR4 receptor overexpression in mesenchymal stem cells facilitates treatment of acute lung injury in rats. J. Biol. Chem. 2015, 290, 1994–2006. [Google Scholar] [CrossRef]
- Huang, Z.; Su, G.F.; Hu, W.J.; Bi, X.X.; Zhang, L.; Wan, G. The study on expression of CIAPIN1 interfering hepatocellular carcinoma cell proliferation and its mechanisms. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3054–3060. [Google Scholar]
- Yang, Z.; Wang, W.E.; Zhang, Q. CIAPIN1 siRNA inhibits proliferation, migration and promotes apoptosis of VSMCs by regulating Bcl-2 and Bax. Curr. Neurovasc. Res. 2013, 10, 4–10. [Google Scholar] [CrossRef]
- Iacobas, S.; Iacobas, D.A. Astrocyte proximity modulates the myelination gene fabric of oligodendrocytes. Neuron Glia Biol. 2010, 6, 157–169. [Google Scholar] [CrossRef]
- Lee, P.R.; Cohen, J.E.; Iacobas, D.A.; Iacobas, S.; Fields, R.D. Gene networks activated by pattern-specific generation of action potentials in dorsal root ganglia neurons. Sci. Rep. 2017, 7, 43765. [Google Scholar] [CrossRef]
- Clavier, L.; Legchenko, E.; Grimm, L.; Sallmon, H.; Hatch, A.; Plouff, B.D.; Hansmann, G. Galectin 3 and aldosterone as potential tandem biomarkers in pulmonary arterial hypertension. Heart 2016, 102, 390–396. [Google Scholar] [CrossRef]
- Perretti, M.; Ingegnoli, F.; Wheller, S.K.; Blades, M.C.; Solito, E.; Pitzalis, C. Annexin 1 modulates monocyte-endothelial cell interaction in vitro and cell migration in vivo in the human SCID mouse transplantation model. J. Immunol. 2002, 169, 2085–2092. [Google Scholar] [CrossRef]
- Drechsler, M.; de Jong, R.; Rossaint, J.; Viola, J.R.; Leoni, G.; Wang, J.M.; Döring, Y. Annexin A1 counteracts chemokine-induced arterial myeloid cell recruitment. Circ. Res. 2005, 116, 827–835. [Google Scholar] [CrossRef]
- Sheikh, M.H.; Solito, E. Annexin A1: Uncovering the Many Talents of an Old Protein. Int. J. Mol. Sci. 2018, 19, 1045. [Google Scholar] [CrossRef]
- De Jong, R.J.; Paulin, N.; Lemnitzer, P.; Viola, J.R.; Winter, C.; Ferraro, B.; Soehnlein, O. Protective Aptitude of Annexin A1 in Arterial Neointima Formation in Atherosclerosis-Prone Mice-Brief Report. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 312–315. [Google Scholar] [CrossRef][Green Version]
- Gore, B.; Izikki, M.; Mercier, O.; Dewachter, L.; Fadel, E.; Humbert, M.; Eddahibi, S. Key role of the endothelial TGF-β/ALK1/endoglin signaling pathway in humans and rodents pulmonary hypertension. PLoS ONE 2014, 9, e100310. [Google Scholar] [CrossRef]
- Scherer, C.; Pfisterer, L.; Wagner, A.H.; Hödebeck, M.; Cattaruzza, M.; Hecker, M.; Korff, T. Arterial wall stress controls NFAT5 activity in vascular smooth muscle cells. J. Am. Heart Assoc. 2014, 10, e000626. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zhang, D.; Li, Q.; Liu, Y.; Jing, S.; Cui, J.; Yu, B. Biomechanical Stretch Induces Inflammation, Proliferation, and Migration by Activating NFAT5 in Arterial Smooth Muscle Cells. Inflammation 2017, 40, 2129–2136. [Google Scholar] [CrossRef]
- Weiss, A.; Neubauer, M.C.; Yerabolu, D.; Kojonazarov, B.; Schlueter, B.C.; Neubert, L.; Pullamsetti, S.S. Targeting cyclin-dependent kinases for the treatment of pulmonary arterial hypertension. Nat. Commun. 2019, 10, 2204. [Google Scholar] [CrossRef]
- Brunyanszki, A.; Szczesny, B.; Virág, L.; Szabo, C. Mitochondrial poly(ADP-ribose) polymerase: The Wizard of Oz at work. Free Radic. Biol. Med. 2016. [Google Scholar] [CrossRef]
- Elser, M.; Borsig, L.; Hassa, P.O.; Erener, S.; Messner, S.; Valovka, T.; Hottiger, M.O. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol. Cancer Res. 2008, 6, 282–290. [Google Scholar] [CrossRef]
- Meloche, J.; Pflieger, A.; Vaillancourt, M.; Paulin, R.; Potus, F.; Zervopoulos, S.; Couture, C. Role for DNA damage signaling in pulmonary arterial hypertension. Circulation 2014, 129, 786–797. [Google Scholar] [CrossRef]
- Park, J.B.; Nagar, H.; Choi, S.; Jung, S.B.; Kim, H.W.; Kang, S.K.; Jeon, B.H. IDH2 deficiency impairs mitochondrial function in endothelial cells and endothelium-dependent vasomotor function. Free Radic. Biol. Med. 2016, 94, 36–46. [Google Scholar] [CrossRef]
- Lugus, J.J.; Ngoh, G.A.; Bachschmid, M.M.; Kenneth, K. Mitofusins are required for angiogenic function and modulate different signaling pathways in cultured endothelial cells. J. Mol. Cell. Cardiol. 2011, 51, 885–893. [Google Scholar] [CrossRef]
- Chen, F.; Barman, S.; Yu, Y.; Haigh, S.; Wang, Y.; Dou, H.; Bagi, Z.; Han, W.; Su, Y.; Fu, D.J. Caveolin-1 is a negative regulator of NADPH oxidase-derived reactive oxygen species. Free Radic. Biol. Med. 2014, 73, 201–213. [Google Scholar] [CrossRef]
- Hart, P.C.; Ratti, B.A.; Mao, M.; Ansenberger-Fricano, K.; Shajahan-Haq, A.N.; Tyner, A.L.; Bonini, M.G. Caveolin-1 regulates cancer cell metabolism via scavenging Nrf2 and suppressing MnSOD-driven glycolysis. Oncotarget 2016, 7, 308–322. [Google Scholar] [CrossRef]
- Lu, W.; Kang, J.; Hu, K.; Tang, S.; Zhou, X.; XU, L.; Yu, S. The role of the Nox4-derived ROS-mediated RhoA/Rho kinase pathway in rat hypertension induced by chronic intermittent hypoxia. Sleep Breath. 2017, 21, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Gräf, S.; Haimel, M.; Bleda, M.; Hadinnapola, C.; Southgate, L.; Li, W.; Machado, R.D. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat. Commun. 2018, 9, 1416. [Google Scholar] [CrossRef] [PubMed]
- Oka, M.; Fagan, K.A.; Jones, P.L.; McMurtry, I.F. Therapeutic potential of RhoA/Rho kinase inhibitors in pulmonary hypertension. Br. J. Pharmacol. 2008, 155, 444–454. [Google Scholar] [CrossRef] [PubMed]
| Gene | Description | HO/CO | CUT | CM/CO | CUT | HM/CO | CUT |
|---|---|---|---|---|---|---|---|
| Adcy2 | Adenylate cyclase 2 (brain) | −4.24 | 2.32 | −4.72 | 2.33 | −4.21 | 2.34 |
| Akt1 | V-akt murine thymoma viral oncogene homolog 1 | −1.42 | 1.61 | 1.09 | 1.94 | 1.80 | 1.78 |
| Arrb1 | Arrestin, β 1 | −9.55 | 2.86 | −41.04 | 2.39 | −52.82 | 2.53 |
| Arrb2 | Arrestin, β 2 | −1.38 | 2.11 | 3.37 | 2.54 | 2.95 | 2.24 |
| Bcar1 | Breast cancer anti-estrogen resistance 1 | 1.74 | 2.98 | 5.27 | 2.68 | 10.39 | 1.83 |
| Ccl21 | Chemokine (C-C motif) ligand 21 | 3.26 | 2.70 | 79.95 | 3.31 | 247.07 | 1.34 |
| Ccl24 | Chemokine (C-C motif) ligand 24 | 5.07 | 3.07 | 10.24 | 2.36 | 5.74 | 2.13 |
| Ccl27 | Chemokine (C-C motif) ligand 27 | 1.18 | 3.00 | −7.76 | 1.85 | −10.14 | 2.14 |
| Ccl5 | Chemokine (C-C motif) ligand 5 | −1.35 | 1.85 | 4.56 | 2.45 | 4.40 | 1.65 |
| Ccl6 | C-C motif chemokine 6 | 7.53 | 3.32 | 22.93 | 2.72 | 38.26 | 1.98 |
| Ccl9 | Chemokine (C-C motif) ligand 9 | −1.18 | 1.88 | 3.75 | 2.68 | 2.16 | 1.82 |
| Ccr1l1 | Chemokine (C-C motif) receptor 1-like 1 | −1.48 | 1.46 | −1.66 | 1.83 | −2.77 | 1.80 |
| Ccr9 | Chemokine (C-C motif) receptor 9 | 4.01 | 2.84 | 10.35 | 2.65 | 1.29 | 1.78 |
| Cx3cr1 | Chemokine (C-X3-C motif) receptor 1 | 2.48 | 2.96 | 2.65 | 2.99 | −2.53 | 2.35 |
| Cxcl12 | Chemokine (C-X-C motif) ligand 12 | 1.77 | 2.38 | 3.77 | 2.41 | 60.14 | 1.83 |
| Cxcl3 | Chemokine (C-X-C motif) ligand 3 | 1.61 | 2.33 | 3.93 | 3.13 | 3.33 | 1.93 |
| Gnai2 | Guanine nucleotide binding protein (G protein), α inhibiting 2 | 4.30 | 3.59 | 5.79 | 2.95 | 10.05 | 2.13 |
| Gnb1 | Guanine nucleotide binding protein (G protein), β polypeptide 1 | −4.22 | 2.37 | −1.47 | 2.80 | 1.23 | 2.19 |
| Gnb2 | Guanine nucleotide binding protein (G protein), β polypeptide 2 | 2.15 | 2.99 | 3.44 | 2.49 | 6.23 | 1.70 |
| Gnb3 | Guanine nucleotide binding protein (G protein), β polypeptide 3 | −3.33 | 2.17 | −6.18 | 2.40 | −11.78 | 2.42 |
| Gng13 | Guanine nucleotide binding protein (G protein), γ 13 | −2.64 | 2.43 | −3.56 | 2.36 | −2.33 | 2.10 |
| Gng3 | Guanine nucleotide binding protein (G protein), γ 3 | −22.18 | 2.46 | −21.14 | 2.51 | −32.06 | 2.44 |
| Gng4 | guanine nucleotide binding protein (G protein), γ 4 subunit | −1.24 | 1.46 | −1.29 | 1.77 | −2.19 | 1.71 |
| Gngt2 | Guanine nucleotide binding protein (G protein), γ transducing activity polypeptide 2 | 1.76 | 2.62 | 7.16 | 2.75 | 11.11 | 1.63 |
| Grb2 | Growth factor receptor bound protein 2 | 4.31 | 2.79 | 3.69 | 2.75 | 6.32 | 1.87 |
| Grk6 | G protein-coupled receptor kinase 6 | −3.00 | 2.59 | −1.04 | 2.69 | 1.69 | 2.07 |
| Gsk3a | Glycogen synthase kinase 3 α | −4.67 | 2.17 | −4.44 | 2.31 | −7.27 | 2.21 |
| Mapk3 | Mitogen activated protein kinase 3 | 1.72 | 2.92 | −3.71 | 2.05 | −3.42 | 1.89 |
| Nfkbia | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, α | 1.92 | 3.60 | 5.56 | 3.03 | 9.00 | 2.20 |
| Nfkbid | NF-κB inhibitor delta | −1.79 | 1.64 | −2.08 | 1.89 | −3.25 | 1.85 |
| Pak1 | P21 protein (Cdc42/Rac)-activated kinase 1 | −1.79 | 2.19 | −3.31 | 2.17 | −3.17 | 2.15 |
| Pik3r1 | Phosphoinositide-3-kinase, regulatory subunit 1 (α) | 1.20 | 2.03 | 2.77 | 2.28 | 4.25 | 1.28 |
| Pik3r5 | Phosphoinositide-3-kinase, regulatory subunit 5 | −11.53 | 2.66 | −5.87 | 2.40 | −7.66 | 2.38 |
| Plcb4 | Phospholipase C, β 4 | 3.18 | 2.86 | 6.70 | 2.99 | −4.79 | 2.12 |
| Ppbp | Pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) | −1.05 | 1.41 | −1.16 | 2.11 | −3.06 | 2.13 |
| Ptk2b | PTK2B protein tyrosine kinase 2 β | −12.47 | 2.45 | −6.89 | 2.36 | −8.51 | 2.34 |
| Rap1b | RAP1B, member of RAS oncogene family | −3.04 | 2.84 | −1.84 | 3.17 | −1.17 | 2.37 |
| Rela | V-rel reticuloendotheliosis viral oncogene homolog A (avian) | −3.38 | 2.07 | −1.15 | 2.29 | 1.22 | 1.98 |
| Rhoa | Ras homolog gene family, member A | 7.00 | 2.80 | 6.78 | 2.52 | 9.21 | 1.24 |
| Shc1 | SHC (Src homology 2 domain containing) transforming protein 1 | −2.71 | 2.09 | −4.11 | 2.03 | −4.94 | 2.08 |
| Sos1 | Son of sevenless homolog 1 (Drosophila) | 1.02 | 1.99 | 2.07 | 2.49 | −2.86 | 1.77 |
| Tiam1 | T-cell lymphoma invasion and metastasis 1 | −2.79 | 1.75 | −2.67 | 2.13 | −4.85 | 1.95 |
| Vav2 | Vav 2 guanine nucleotide exchange factor | 2.06 | 2.74 | 3.44 | 2.43 | 6.02 | 1.45 |
| Xcl1 | Chemokine (C motif) ligand 1 | −1.51 | 1.96 | −1.53 | 2.20 | −3.49 | 2.03 |
| Gene | Description | HO/CO | CUT | CM/CO | CUT | HM/CO | CUT |
|---|---|---|---|---|---|---|---|
| Atp11b | ATPase phospholipid transporting 11B | −1.74 | 2.28 | 1.34 | 2.46 | 2.05 | 1.82 |
| Atp13a2 | ATPase type 13A2 | −1.67 | 2.18 | 1.55 | 2.40 | 2.11 | 2.00 |
| Atp1a1 | ATPase, Na+/K+ transporting, α 1 polypeptide | −1.78 | 3.34 | 4.01 | 3.08 | 6.65 | 2.25 |
| Atp1a3 | ATPase, Na+/K+ transporting, α 3 polypeptide | −1.52 | 1.71 | −1.86 | 1.95 | −3.48 | 2.07 |
| Atp1b2 | ATPase, Na+/K+ transporting, β 2 polypeptide | −146.48 | 3.21 | −257.37 | 2.51 | −111.20 | 2.47 |
| Atp1b3 | ATPase, Na+/K+ transporting, β 3 polypeptide | 2.14 | 2.32 | 12.00 | 2.35 | 15.88 | 1.26 |
| Atp2a2 | ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 | 1.56 | 2.48 | 4.83 | 2.46 | 10.66 | 1.49 |
| Atp2a3 | ATPase, Ca++ transporting, ubiquitous | 1.40 | 1.95 | 1.62 | 1.87 | 1.88 | 1.39 |
| Atp2b1 | ATPase, Ca++ transporting, plasma membrane 1 | −3.70 | 3.00 | −1.19 | 2.86 | 1.30 | 2.34 |
| Atp2b4 | ATPase, Ca++ transporting, plasma membrane 4 | −2.48 | 1.90 | −1.69 | 2.59 | −11.70 | 2.28 |
| Atp2c1 | ATPase, Ca++ transporting, type 2C, member 1 | −1.35 | 1.67 | 1.24 | 1.69 | 1.85 | 1.50 |
| Atp4a | ATPase, H+/K+ exchanging, α polypeptide | 1.13 | 1.59 | −1.12 | 1.94 | −2.35 | 1.61 |
| Atp5d | ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 2.27 | 3.21 | 6.84 | 2.82 | 9.15 | 1.92 |
| Atp5f1 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 | 4.36 | 3.01 | 27.45 | 2.92 | 51.30 | 1.67 |
| Atp5g1 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 | 4.71 | 3.18 | 10.84 | 2.78 | 16.08 | 2.08 |
| Atp5g2 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 | 3.21 | 2.91 | 14.44 | 2.84 | 26.97 | 1.86 |
| Atp5g3 | ATP synthase membrane subunit c locus 3 | −7.56 | 3.23 | −1.08 | 3.22 | 1.35 | 2.35 |
| Atp5i | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E | 17.18 | 2.97 | 11.45 | 2.89 | 15.65 | 2.19 |
| Atp5j2 | ATP synthase, H+ transporting, mitochondrial Fo complex, subunit F2 | −1.27 | 2.91 | 5.30 | 2.90 | 8.37 | 2.24 |
| Atp6v0a1 | ATPase, H+ transporting, lysosomal V0 subunit A1 | 4.14 | 2.57 | 3.11 | 2.35 | 4.83 | 1.65 |
| Atp6v0a2 | ATPase, H+ transporting, lysosomal V0 subunit A2 | −2.76 | 2.21 | −2.35 | 2.37 | −1.52 | 2.16 |
| Atp6v0b | ATPase, H+ transporting, lysosomal V0 subunit B | −6.71 | 3.10 | −1.45 | 3.18 | −1.31 | 2.59 |
| Atp6v0c | ATPase, H+ transporting, lysosomal V0 subunit C | −31.61 | 2.78 | −5.37 | 3.13 | −3.65 | 2.39 |
| Atp6v0e1 | ATPase, H+ transporting, lysosomal, V0 subunit e1 | 1.78 | 2.25 | 16.44 | 2.87 | 26.14 | 1.32 |
| Atp6v0e2 | ATPase, H+ transporting V0 subunit e2 | 1.84 | 2.59 | −3.40 | 1.79 | −5.62 | 1.95 |
| Atp6v1f | ATPase, H transporting, lysosomal V1 subunit F | 1.10 | 2.22 | 5.42 | 2.78 | 9.44 | 1.53 |
| Atp6v1g2 | ATPase, H+ transporting, lysosomal V1 subunit G2 | −11.80 | 2.62 | −16.92 | 2.68 | −39.54 | 2.66 |
| Atp7b | ATPase, Cu++ transporting, β polypeptide | −1.10 | 1.95 | −1.61 | 2.13 | −4.23 | 1.93 |
| Atpif1 | ATPase inhibitory factor 1 | 1.06 | 3.10 | 5.02 | 3.13 | 8.16 | 2.21 |
| Cox14 | cytochrome c oxidase assembly factor COX14 | −3.02 | 2.38 | 1.25 | 2.48 | 1.45 | 2.12 |
| Cox17 | cytochrome c oxidase assembly homolog 17 | −1.43 | 3.42 | 2.09 | 3.07 | 3.01 | 2.28 |
| Cox18 | cytochrome c oxidase assembly homolog 18 | −2.41 | 2.11 | −1.51 | 2.20 | −1.23 | 2.03 |
| Cox4i1 | cytochrome c oxidase subunit IV isoform 1 | 11.99 | 2.91 | 30.01 | 2.81 | 52.39 | 1.89 |
| Cox4i2 | cytochrome c oxidase subunit IV isoform 2 (lung) | −6.30 | 3.02 | −4.07 | 2.77 | −2.89 | 2.32 |
| Cox5b | cytochrome c oxidase subunit Vb | −9.14 | 2.93 | −1.85 | 3.15 | −1.02 | 2.34 |
| Cox6a1 | cytochrome c oxidase, subunit VIa, polypeptide 1 | −1.85 | 3.68 | 1.66 | 3.10 | 2.87 | 2.24 |
| Cox6a2 | cytochrome c oxidase subunit VIa polypeptide 2 | 1.33 | 2.05 | 2.99 | 2.60 | 16.89 | 2.29 |
| Cox6b1 | cytochrome c oxidase subunit 6B1 | 4.27 | 2.82 | −3.59 | 2.54 | −3.02 | 2.29 |
| Cox6b2 | cytochrome c oxidase subunit VIb polypeptide 2 | −3.95 | 2.64 | −7.82 | 2.34 | −7.12 | 2.32 |
| Cox6c | cytochrome c oxidase, subunit VIc | 4.56 | 3.49 | 11.35 | 2.92 | 18.57 | 2.02 |
| Cox7a2 | cytochrome c oxidase subunit VIIa polypeptide 2 | 1.85 | 2.97 | 7.68 | 2.74 | 12.31 | 1.95 |
| Cox7b | cytochrome c oxidase subunit VIIb | 1.50 | 2.03 | 6.58 | 2.77 | 11.58 | 1.79 |
| Cox8a | cytochrome c oxidase subunit VIIIa | −4.07 | 3.07 | 1.27 | 3.18 | 2.50 | 2.33 |
| Cox8b | cytochrome c oxidase, subunit VIIIb | 2.96 | 2.01 | 2.35 | 1.93 | 6.01 | 2.37 |
| Coa5 | cytochrome C oxidase assembly factor 5 | −20.24 | 2.92 | −8.03 | 3.05 | −11.72 | 2.93 |
| Ndufa10 | NADH dehydrogenase (ubiquinone) 1 α subcomplex 10 | 1.24 | 2.99 | 1.81 | 2.56 | 3.27 | 1.92 |
| Ndufa11 | NADH dehydrogenase (ubiquinone) 1 α subcomplex 11 | −1.02 | 2.24 | 3.51 | 2.08 | 3.92 | 1.72 |
| Ndufa13 | NADH:ubiquinone oxidoreductase subunit A13 | −2.20 | 3.07 | 3.05 | 3.10 | 5.35 | 2.18 |
| Ndufa3 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 3 | −4.15 | 2.81 | −2.79 | 2.51 | −2.08 | 2.25 |
| Ndufa6 | NADH dehydrogenase (ubiquinone) 1 α subcomplex, 6 (B14) | 4.42 | 3.33 | 15.50 | 2.84 | 28.28 | 1.71 |
| Ndufaf5 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 | −1.16 | 2.21 | 1.50 | 2.45 | 2.21 | 1.94 |
| Ndufb10 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 10 | 2.67 | 3.13 | 4.78 | 2.67 | 7.57 | 1.63 |
| Ndufb2 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 2 | 7.38 | 3.22 | 20.76 | 2.78 | 35.55 | 1.73 |
| Ndufb3 | NADH dehydrogenase (ubiquinone) 1 β subcomplex 3 | 3.15 | 2.92 | 7.29 | 2.42 | 12.14 | 1.60 |
| Ndufb4 | NADH dehydrogenase (ubiquinone) 1 β subcomplex 4 | 3.52 | 3.35 | 11.33 | 2.85 | 21.34 | 1.89 |
| Ndufb5 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 5 | 1.31 | 2.83 | 4.31 | 2.77 | 6.50 | 1.94 |
| Ndufb6 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 6 | −6.75 | 2.75 | −1.20 | 3.14 | 1.33 | 2.28 |
| Ndufb7 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 7 | −4.28 | 3.63 | −1.59 | 3.12 | −1.12 | 2.32 |
| Ndufb8 | NADH dehydrogenase (ubiquinone) 1 β subcomplex 8 | 2.64 | 3.37 | 7.96 | 3.09 | 13.61 | 2.35 |
| Ndufb9 | NADH dehydrogenase (ubiquinone) 1 β subcomplex, 9 | 1.67 | 2.89 | 11.81 | 2.95 | 19.58 | 2.16 |
| Ndufs3 | NADH dehydrogenase (ubiquinone) Fe-S protein 3 | −5.54 | 3.14 | −1.39 | 3.14 | 1.08 | 2.42 |
| Ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | 3.54 | 2.87 | 12.91 | 2.84 | 23.34 | 1.94 |
| Ndufs7 | NADH dehydrogenase (ubiquinone) Fe-S protein 7 | 5.04 | 3.35 | 8.90 | 2.68 | 10.68 | 1.74 |
| Ndufv1 | NADH dehydrogenase (ubiquinone) flavoprotein 1 | 5.55 | 3.38 | 8.51 | 2.82 | 14.54 | 1.94 |
| Ndufv3 | NADH dehydrogenase (ubiquinone) flavoprotein 3 | 1.32 | 1.95 | 2.12 | 2.22 | 3.71 | 1.24 |
| Sdhaf2 | succinate dehydrogenase complex assembly factor 2 | 2.15 | 2.69 | 4.59 | 2.76 | 7.66 | 1.73 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathew, R.; Huang, J.; Iacobas, S.; Iacobas, D.A. Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways. Genes 2020, 11, 126. https://doi.org/10.3390/genes11020126
Mathew R, Huang J, Iacobas S, Iacobas DA. Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways. Genes. 2020; 11(2):126. https://doi.org/10.3390/genes11020126
Chicago/Turabian StyleMathew, Rajamma, Jing Huang, Sanda Iacobas, and Dumitru A. Iacobas. 2020. "Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways" Genes 11, no. 2: 126. https://doi.org/10.3390/genes11020126
APA StyleMathew, R., Huang, J., Iacobas, S., & Iacobas, D. A. (2020). Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways. Genes, 11(2), 126. https://doi.org/10.3390/genes11020126






