Deletion of Exon 1 in AMER1 in Osteopathia Striata with Cranial Sclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects and Ethical Consent
2.2. Sequence Analysis
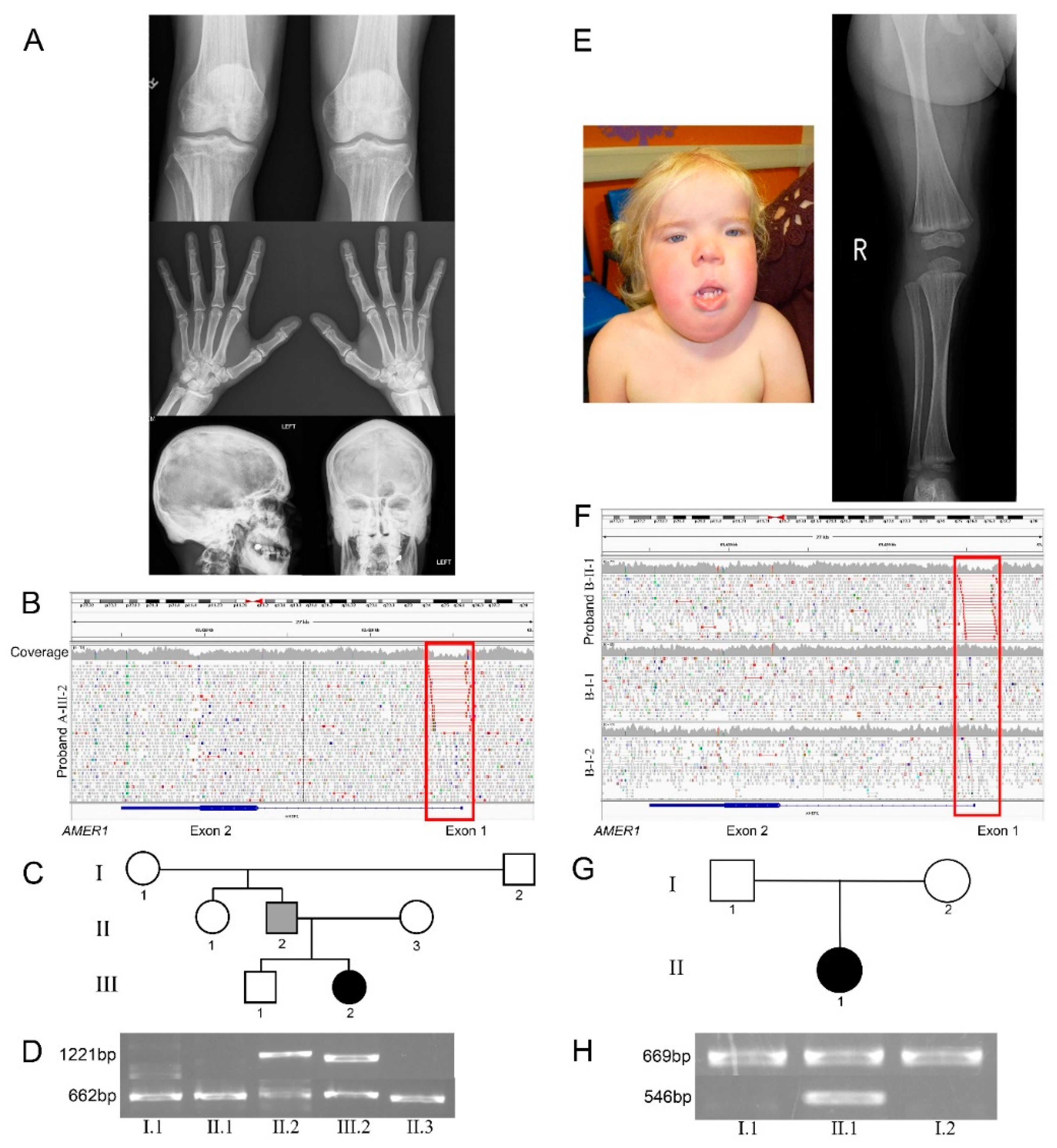
3. Results
3.1. Clinical Descriptions
3.1.1. Proband A
3.1.2. Proband B
3.2. Non-Coding Deletions in AMER1 Cause OSCS
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vuillaume, M.L.; Valard, A.G.; Houcinat, N.; Bouron, J.; Boucher, C.; Deves, S.; Toutain, J.; Schaub, B.; Adenet, C.; Lacombe, D.; et al. First female prenatal case of osteopathia striata with cranial sclerosis in a fetus carrying a de-novo 1.9 Mbp interstitial deletion at Xq11.1q11.2. Clin. Dysmorphol. 2017, 26, 231–234. [Google Scholar] [CrossRef]
- Enomoto, Y.; Tsurusaki, Y.; Harada, N.; Aida, N.; Kurosawa, K. Novel AMER1 frameshift mutation in a girl with osteopathia striata with cranial sclerosis. Congenit. Anom. (Kyoto) 2018, 58, 145–146. [Google Scholar] [CrossRef]
- Comai, G.; Boutet, A.; Tanneberger, K.; Massa, F.; Rocha, A.S.; Charlet, A.; Panzolini, C.; Motamedi, F.J.; Brommage, R.; Hans, W.; et al. Genetic and Molecular Insights Into Genotype-Phenotype Relationships in Osteopathia Striata With Cranial Sclerosis (OSCS) Through the Analysis of Novel Mouse Wtx Mutant Alleles. J. Bone Min. Res. 2018, 33, 875–887. [Google Scholar] [CrossRef] [PubMed]
- Holman, S.K.; Daniel, P.; Jenkins, Z.A.; Herron, R.L.; Morgan, T.; Savarirayan, R.; Chow, C.W.; Bohring, A.; Mosel, A.; Lacombe, D.; et al. The male phenotype in osteopathia striata congenita with cranial sclerosis. Am. J. Med. Genet. Part A 2011, 155, 2397–2408. [Google Scholar] [CrossRef] [PubMed]
- Major, M.B.; Camp, N.D.; Berndt, J.D.; Yi, X.; Goldenberg, S.J.; Hubbert, C.; Biechele, T.L.; Gingras, A.N.; Zheng, N.; Maccoss, M.J.; et al. Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science 2007, 316, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- Sperling, M. Pediatric Endocrinology E-Book: Expert Consult—Online and Print; Elsevier Health Sciences: Philadelphia, PA, USA, 2014. [Google Scholar]
- Maeda, K.; Kobayashi, Y.; Koide, M.; Uehara, S.; Okamoto, M.; Ishihara, A.; Kayama, T.; Saito, M.; Marumo, K. The Regulation of Bone Metabolism and Disorders by Wnt Signaling. Int. J. Mol. Sci. 2019, 20, 5525. [Google Scholar] [CrossRef] [PubMed]
- Glorieux, F.H.; Pettifor, J.M.; Juppner, H. Pediatric Bone: Biology and Diseases; Elsevier Science: Cambridge, MA, USA, 2011. [Google Scholar]
- Jenkins, Z.A.; van Kogelenberg, M.; Morgan, T.; Jeffs, A.; Fukuzawa, R.; Pearl, E.; Thaller, C.; Hing, A.V.; Porteous, M.E.; Garcia-Miñaur, S.; et al. Germline mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat. Genet. 2009, 41, 95–100. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 91–498. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Cingolani, P.; Platts, A.; Wang, L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 2012, 6, 80–92. [Google Scholar] [CrossRef] [PubMed]
- Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2015, 536, 285–291. [Google Scholar]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 2015, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mi, J.; Parthasarathy, P.; Halliday, B.J.; Morgan, T.; Dean, J.; Nowaczyk, M.J.M.; Markie, D.; Robertson, S.P.; Wade, E.M. Deletion of Exon 1 in AMER1 in Osteopathia Striata with Cranial Sclerosis. Genes 2020, 11, 1439. https://doi.org/10.3390/genes11121439
Mi J, Parthasarathy P, Halliday BJ, Morgan T, Dean J, Nowaczyk MJM, Markie D, Robertson SP, Wade EM. Deletion of Exon 1 in AMER1 in Osteopathia Striata with Cranial Sclerosis. Genes. 2020; 11(12):1439. https://doi.org/10.3390/genes11121439
Chicago/Turabian StyleMi, Jingyi, Padmini Parthasarathy, Benjamin J. Halliday, Tim Morgan, John Dean, Malgorzata J. M. Nowaczyk, David Markie, Stephen P. Robertson, and Emma M. Wade. 2020. "Deletion of Exon 1 in AMER1 in Osteopathia Striata with Cranial Sclerosis" Genes 11, no. 12: 1439. https://doi.org/10.3390/genes11121439
APA StyleMi, J., Parthasarathy, P., Halliday, B. J., Morgan, T., Dean, J., Nowaczyk, M. J. M., Markie, D., Robertson, S. P., & Wade, E. M. (2020). Deletion of Exon 1 in AMER1 in Osteopathia Striata with Cranial Sclerosis. Genes, 11(12), 1439. https://doi.org/10.3390/genes11121439





