Evaluation of the Ion AmpliSeq™ PhenoTrivium Panel: MPS-Based Assay for Ancestry and Phenotype Predictions Challenged by Casework Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Panel Design
2.2. Reference Samples
2.3. Sensitivity Study
2.4. Casework Samples
2.5. Library Preparation and Sequencing
2.6. Data Analysis
Phenotype and Ancestry Predictions
3. Results
3.1. Coverage and Sensitivity
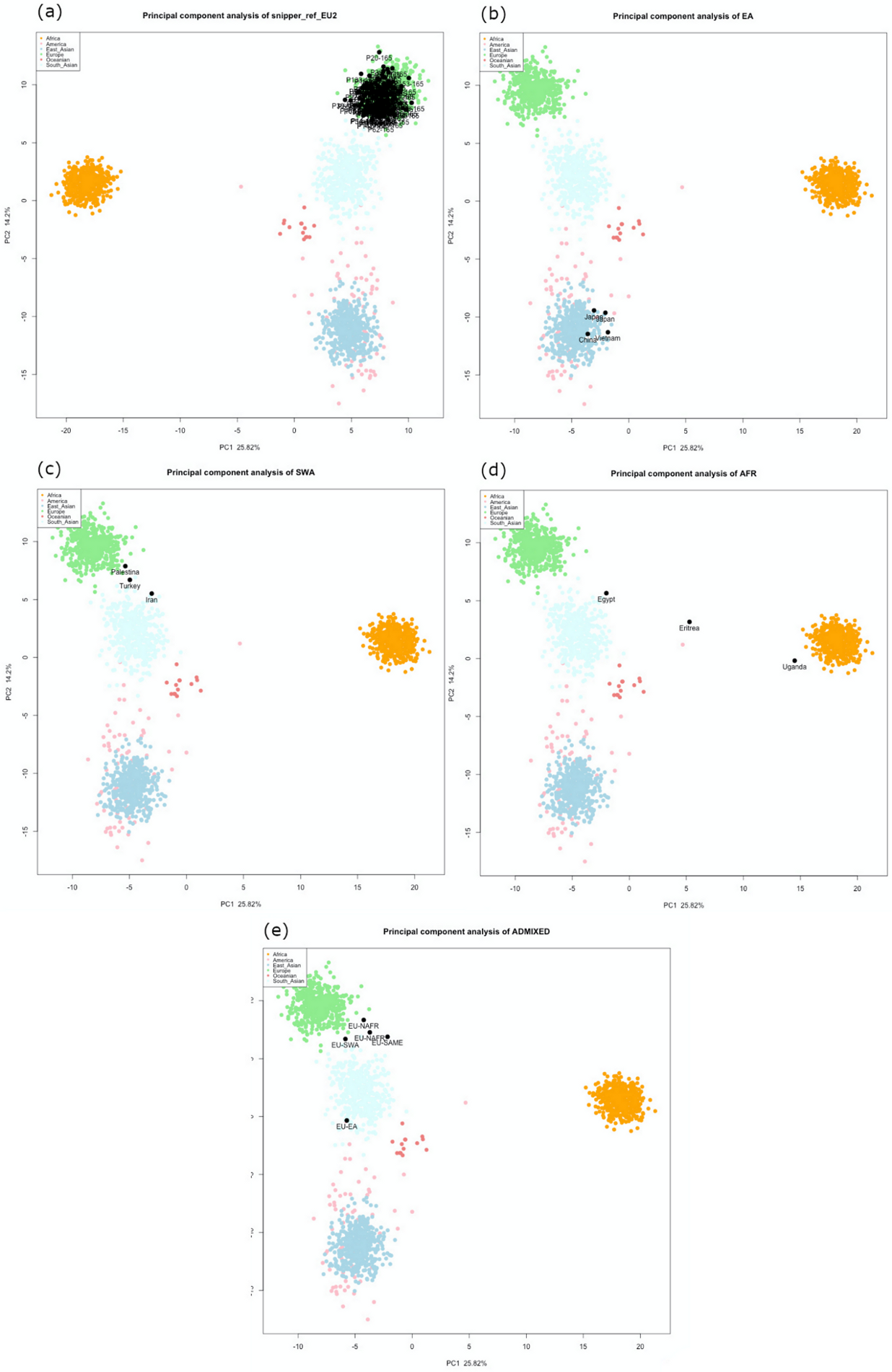
3.2. Reference Samples
3.2.1. Phenotype Predictions
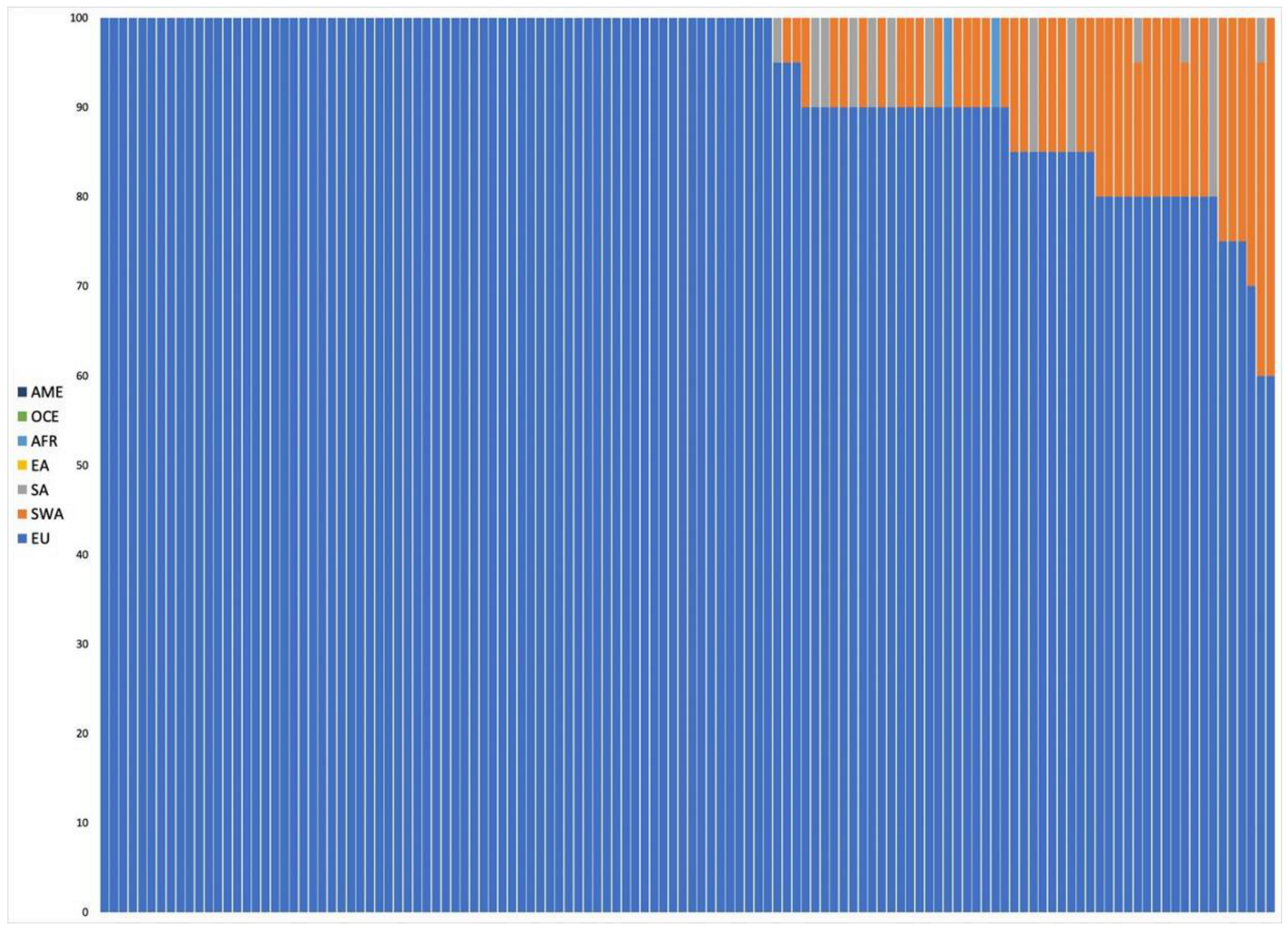
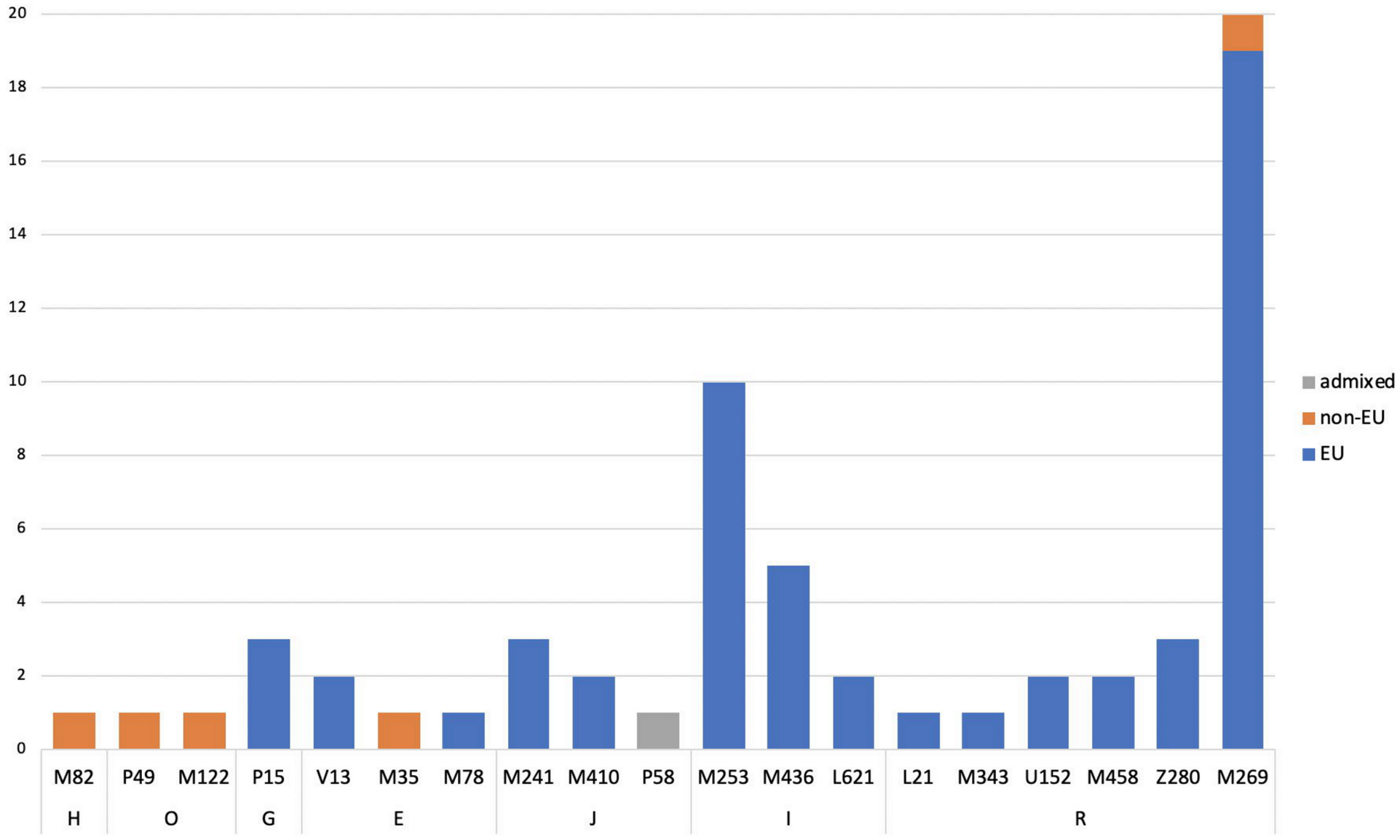
3.2.2. Ancestry Predictions
European Samples
Non-European Samples
Admixed Samples
3.3. Casework Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gettings, K.B.; Lai, R.; Johnson, J.L.; Peck, M.A.; Hart, J.A.; Gordish-Dressman, H.; Schanfield, M.S.; Podini, D.S. A 50-SNP assay for biogeographic ancestry and phenotype prediction in the U.S. population. Forensic Sci. Int. Genet. 2014, 8, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Nievergelt, C.M.; Maihofer, A.X.; Shekhtman, T.; Libiger, O.; Wang, X.; Kidd, K.K.; Kidd, J.R. Inference of human continental origin and admixture proportions using a highly discriminative ancestry informative 41-SNP panel. Investig. Genet. 2013, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.; Parson, W.; Lundsberg, B.; Santos, C.; Freire-Aradas, A.; Torres, M.; Eduardoff, M.; Børsting, C.; Johansen, P.; Fondevila, M.; et al. Building a forensic ancestry panel from the ground up: The EUROFORGEN Global AIM-SNP set. Forensic Sci. Int. Genet. 2014, 11, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Phillips, C.; Fondevila, M.; Daniel, R.; Van Oorschot, R.A.; Burchard, E.G.; Schanfield, M.S.; Souto, L.; Uacyisrael, J.; Via, M.; et al. Pacifiplex: An ancestry-informative SNP panel centred on Australia and the Pacific region. Forensic Sci. Int. Genet. 2016, 20, 71–80. [Google Scholar] [CrossRef]
- Pereira, V.; Mogensen, H.S.; Børsting, C.; Morling, N. Evaluation of the Precision ID Ancestry Panel for crime case work: A SNP typing assay developed for typing of 165 ancestral informative markers. Forensic Sci. Int. Genet. 2017, 28, 138–145. [Google Scholar] [CrossRef]
- Bulbul, O.; Filoglu, G. Development of a SNP panel for predicting biogeographical ancestry and phenotype using massively parallel sequencing. Electrophoresis 2018, 39, 2743–2751. [Google Scholar] [CrossRef]
- Phillips, C.; Freire-Aradas, A.; Kriegel, A.K.; Fondevila, M.; Bulbul, O.; Santos, C.; Serrulla Rech, F.; Perez Carceles, M.D.; Carracedo, A.; Schneider, P.M.; et al. Eurasiaplex: A forensic SNP assay for differentiating European and South Asian ancestries. Forensic Sci. Int. Genet. 2013, 7, 359–366. [Google Scholar] [CrossRef]
- Shi, C.-M.; Liu, Q.; Zhao, S.; Chen, H. Ancestry informative SNP panels for discriminating the major East Asian populations: Han Chinese, Japanese and Korean. Ann. Hum. Genet. 2019, 83, 348–354. [Google Scholar] [CrossRef]
- Phillips, C.; McNevin, D.; Kidd, K.; Lagacé, R.; Wootton, S.; De La Puente, M.; Freire-Aradas, A.; Mosquera-Miguel, A.; Eduardoff, M.; Gross, T.; et al. MAPlex—A massively parallel sequencing ancestry analysis multiplex for Asia-Pacific populations. Forensic Sci. Int. Genet. 2019, 42, 213–226. [Google Scholar] [CrossRef]
- Pakstis, A.J.; Speed, W.C.; Soundararajan, U.; Rajeevan, H.; Kidd, J.R.; Li, H.; Kidd, K.K. Population relationships based on 170 ancestry SNPs from the combined Kidd and Seldin panels. Sci. Rep. 2019, 9, 18874. [Google Scholar] [CrossRef]
- Chaitanya, L.; Breslin, K.; Zuñiga, S.; Wirken, L.; Pośpiech, E.; Kukla-Bartoszek, M.; Sijen, T.; De Knijff, P.; Liu, F.; Branicki, W.; et al. The HIrisPlex-S system for eye, hair and skin colour prediction from DNA: Introduction and forensic developmental validation. Forensic Sci. Int. Genet. 2018, 35, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Pośpiech, E.; Chen, Y.; Kukla-Bartoszek, M.; Breslin, K.; Aliferi, A.; Andersen, M.M.; Ballard, D.; Chaitanya, L.; Freire-Aradas, A.; Van Der Gaag, K.J.; et al. Towards broadening Forensic DNA Phenotyping beyond pigmentation: Improving the prediction of head hair shape from DNA. Forensic Sci. Int. Genet. 2018, 37, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Kukla-Bartoszek, M.; Pośpiech, E.; Spólnicka, M.; Karlowska-Pik, J.; Strapagiel, D.; Żądzińska, E.; Rosset, I.; Sobalska-Kwapis, M.; Słomka, M.; Walsh, S.; et al. Investigating the impact of age-depended hair colour darkening during childhood on DNA-based hair colour prediction with the HIrisPlex system. Forensic Sci. Int. Genet. 2018, 36, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Kukla-Bartoszek, M.; Pośpiech, E.; Woźniak, A.; Boroń, M.; Karłowska-Pik, J.; Teisseyre, P.; Zubańska, M.; Bronikowska, A.; Grzybowski, T.; Płoski, R.; et al. DNA-based predictive models for the presence of freckles. Forensic Sci. Int. Genet. 2019, 42, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Pośpiech, E.; Kukla-Bartoszek, M.; Karłowska-Pik, J.; Zieliński, P.; Woźniak, A.; Boroń, M.; Dąbrowski, M.; Zubańska, M.; Jarosz, A.; Grzybowski, T.; et al. Exploring the possibility of predicting human head hair greying from DNA using whole-exome and targeted NGS data. BMC Genom. 2020, 21, 538. [Google Scholar] [CrossRef]
- Schneider, P.M.; Prainsack, B.; Kayser, M. The use of forensic DNA phenotyping in predicting appearance and biogeographic ancestry. Dtsch. Arztebl. Int. 2019, 116, 873–880. [Google Scholar] [CrossRef]
- Breslin, K.; Wills, B.; Ralf, A.; Garcia, M.V.; Kukla-Bartoszek, M.; Pospiech, E.; Freire-Aradas, A.; Xavier, C.; Ingold, S.; De La Puente, M.; et al. HIrisPlex-S system for eye, hair, and skin color prediction from DNA: Massively parallel sequencing solutions for two common forensically used platforms. Forensic Sci. Int. Genet. 2019, 43, 102152. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Walsh, S.; Liu, F.; Wollstein, A.; Kovatsi, L.; Ralf, A.; Kosiniak-Kamysz, A.; Branicki, W.; Kayser, M. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci. Int. Genet. 2013, 7, 98–115. [Google Scholar] [CrossRef]
- Walsh, S.; Liu, F.; Ballantyne, K.N.; van Oven, M.; Lao, O.; Kayser, M. IrisPlex: A sensitive DNA tool for accurate prediction of blue and brown eye colour in the absence of ancestry information. Forensic Sci. Int. Genet. 2011, 5, 170–180. [Google Scholar] [CrossRef]
- Jin, S.; Chase, M.; Henry, M.; Alderson, G.; Morrow, J.M.; Malik, S.; Ballard, D.; McGrory, J.; Fernandopulle, N.; Millman, J.; et al. Implementing a biogeographic ancestry inference service for forensic casework. Electrophoresis 2018, 39, 2757–2765. [Google Scholar] [CrossRef] [PubMed]
- Wootton, S.; Vijaychander, S.; Hasegawa, R.; Deng, J.; Lackey, A.; Gabriel, M.; Lagacé, R.; Lim, J. Analytical Improvements in Biogeographic Ancestry Inference. Presented at the 28th Congress of the International Society for Forensic Genetics. Available online: https://www.thermofisher.com/de/de/home/products-and-services/promotions/isfg.html (accessed on 1 February 2020).
- Converge™ Software 2.2 Software Release Notes. Available online: https://assets.thermofisher.com/TFS-Assets/GSD/Reference-Materials/converge-software-2-2-release-notes.pdf (accessed on 1 February 2020).
- Phillips, C.; Salas, A.; Sánchez, J.J.; Fondevila, M.; Gómez-Tato, A.; Alvarez-Dios, J.; Calaza, M.; Casares de Cal, M.; Ballard, D.; Lareu, M.V.; et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci. Int. Genet. 2007, 1, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Pereira, R.; Phillips, C.; Pinto, N.; Santos, C.; Batista dos Santos, S.E.; Amorim, A.; Carracedo, A.; Gusmão, L. Straightforward Inference of Ancestry and Admixture Proportions through Ancestry-Informative Insertion Deletion Multiplexing. PLoS ONE 2012, 7, e29684. [Google Scholar] [CrossRef] [PubMed]
- Fondevila, M.; Phillips, C.; Santos, C.; Freire-Aradas, A.; Vallone, P.M.; Butler, J.M.; Lareu, M.V.; Carracedo, A. Revision of the SNPforID 34-plex forensic ancestry test: Assay enhancements, standard reference sample genotypes and extended population studies. Forensic Sci. Int. Genet. 2013, 7, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Rajeevan, H.; Soundararajan, U.; Pakstis, A.J.; Kidd, K.K. Introducing the Forensic Research/Reference on Genetics knowledge base, FROG-kb. Investig. Genet. 2012, 1, 18. [Google Scholar] [CrossRef]
- Kidd, K.K.; Soundararajan, U.; Rajeevana, H.; Pakstis, A.J.; Moore, K.N.; Ropero-Millerc, J.D. The redesigned Forensic Research/Reference on Genetics-knowledge base, FROG-kb. , Forensic Sci. Int. Genet. 2018, 33, 33–37. [Google Scholar] [CrossRef]
- Rajeevan, H.; Soundararajan, U.; Pakstis, A.J.; Kidd, K.K. FrogAncestryCalc: A standalone batch likelihood computation tool for ancestry inference panels catalogued in FROG-kb. Forensic Sci. Int. Genet. 2020, 46, 102237. [Google Scholar] [CrossRef]
- Liu, F.; van Duijn, K.; Vingerling, J.R.; Hofman, A.; Uitterlinden, A.G.; Cecile, A.; Janssens, J.W.; Kayser, M. Eye color and the prediction of complex phenotypes from genotypes. Curr. Biol. 2009, 19, 192–193. [Google Scholar] [CrossRef]
- Walsh, S.; Wollstein, A.; Liu, F.; Chakravarthy, U.; Rahu, M.; Seland, J.H.; Soubrane, G.; Tomazzoli, L.; Topouzis, F.; Vingerling, J.R.; et al. DNA-based eye colour prediction across Europe with the IrisPlex system. Forensic Sci. Int. Genet. 2012, 6, 330–340. [Google Scholar] [CrossRef]
- Walsh, S.; Chaitanya, L.; Breslin, K.; Muralidharan, C.; Bronikowska, A.; Pospiech, E.; Koller, J.; Kovatsi, L.; Wollstein, A.; Branicki, W.; et al. Global skin colour prediction from DNA. Hum. Genet. 2017, 136, 847–863. [Google Scholar] [CrossRef]
- Fóthi, E.; Gonzalez, A.; Fehér, T.; Gugora, A.; Fóthi, A.; Biró, O.; Keyser, C. Genetic analysis of male Hungarian Conquerors: European and Asian paternal lineages of the conquering Hungarian tribes. Archaeol. Anthropol. Sci. 2020, 12, 31. [Google Scholar] [CrossRef]
- Cassidy, L.M.; Martiniano, R.; Murphy, E.M.; Teasdale, M.D.; Mallory, J.; Hartwell, B.; Bradley, D.G. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc. Natl. Acad. Sci. USA 2016, 113, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Underhill, P.A.; Myres, N.M.; Rootsi, S.; Metspalu, M.; Zhivotovsky, L.A.; King, R.J.; Lin, A.A.; Chow, C.-E.T.; Semino, O.; Battaglia, V.; et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur. J. Hum. 2009, 18, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Grugni, V.; Battaglia, V.; Kashani, B.H.; Parolo, S.; Al-Zahery, N.; Achilli, A.; Olivieri, A.; Gandini, F.; Houshmand, M.; Sanati, M.H.; et al. Ancient Migratory Events in the Middle East: New Clues from the Y-Chromosome Variation of Modern Iranians. PLoS ONE 2012, 7, e41252. [Google Scholar] [CrossRef]
- Hammer, M.F.; Karafet, T.M.; Park, H.; Omoto, K.; Harihara, S.; Stoneking, M.; Horai, S. Dual origins of the Japanese: Common ground for hunter-gatherer and farmer Y chromosomes. J. Hum. Genet. 2006, 51, 47–58. [Google Scholar] [CrossRef]
- Behar, D.M.; Garrigan, D.; Kaplan, M.E.; Mobasher, Z.; Rosengarten, D.; Karafet, T.M.; Quintana-Murci, L.; Ostrer, H.; Skorecki, K.; Hammer, M.F. Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations. Hum. Genet. 2004, 114, 354–365. [Google Scholar] [CrossRef]
- Hammer, M.F.; Behar, D.M.; Karafet, T.M.; Mendez, F.L.; Hallmark, B.; Erez, T.; Zhivotovsky, L.A.; Rosset, S.; Skorecki, K. Extended Y chromosome haplotypes resolve multiple and unique lineages of the Jewish priesthood. Hum. Genet. 2009, 126, 707–717. [Google Scholar] [CrossRef]
- Kayser, M.; Schneider, P.M. DNA-based prediction of human externally visible characteristics in forensics: Motivations, scientific challenges, and ethical considerations. Forensic Sci. Int. Genet. 2009, 3, 154–161. [Google Scholar] [CrossRef]
- Kayser, M. Forensic DNA Phenotyping: Predicting human appearance from crime scene material for investigative purposes. Forensic Sci. Int. Genet. 2015, 18, 33–48. [Google Scholar] [CrossRef]
- Ruiz, Y.; Phillips, C.; Gomez-Tato, A.; Alvarez-Dios, J.; De Cal, M.C.; Cruz, R.; Maroñas, O.; Söchtig, J.; Fondevila, M.; Rodriguez-Cid, M.; et al. Further development of forensic eye color predictive tests. Forensic Sci. Int. Genet. 2013, 7, 28–40. [Google Scholar] [CrossRef]
- Maroñas, O.; Phillips, C.; Söchtig, J.; Gomez-Tato, A.; Cruz, R.; Alvarez-Dios, J.; Casares de Cal, M.; Ruiz, Y.; Fondevila, M.; Carracedo, A.; et al. Development of a forensic skin colour predictive test. Forensic Sci. Int. Genet. 2014, 13, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Kidd, K.K.; Speed, W.C.; Pakstis, A.J.; Furtado, M.R.; Fang, R.; Madbouly, A.; Maiers, M.; Middha, M.; Friedlaender, F.R.; Kidd, J.R. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci. Int. Genet. 2014, 10, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Oldoni, F.; Hart, R.; Long, K.; Maddela, K.; Cisana, S.; Schanfield, M.; Wootton, S.; Chang, J.; Lagace, R.; Hasegawa, R.; et al. Microhaplotypes for ancestry prediction. Forensic Sci. Int. Genet. Suppl. Ser. 2017, 6, e513–e515. [Google Scholar] [CrossRef]
- Scudder, N.; McNevin, D.; Kelty, S.F.; Walsh, S.J.; Robertson, J. Forensic DNA phenotyping: Developing a model privacy impact assessment. Forensic Sci. Int. Genet. 2018, 34, 222–230. [Google Scholar] [CrossRef]
- Scudder, N.; Robertson, J.; Kelty, S.F.; Walsh, S.J.; McNevin, D. A law enforcement intelligence framework for use in predictive DNA phenotyping. Aust. J. Forensic. Sci. 2019, 51, 255–258. [Google Scholar] [CrossRef]
- Murphy, E.E. Legal and Ethical Issues in Forensic DNA Phenotyping. NYU Sch. Law Public Law 2013, 13–46. [Google Scholar] [CrossRef][Green Version]
- Slabbert, N.; Heathfield, L.J. Ethical, legal and social implications of forensic molecular phenotyping in South Africa. Dev. World. Bioeth. 2018, 18, 171–181. [Google Scholar] [CrossRef]
- Samuel, G.; Prainsack, B. Civil society stakeholder views on forensic DNA phenotyping: Balancing risks and benefits. Forensic Sci. Int. Genet. 2019, 43, 102157. [Google Scholar] [CrossRef]
- MacLean, C.E.; Lamparello, A. Forensic DNA phenotyping in criminal investigations and criminal courts: Assessing and mitigating the dilemmas inherent in the science. Recent Adv. DNA Gene Seq. 2014, 8, 104–112. [Google Scholar]
- Gepshtein, S.; Wang, Y.; He, F.; Diep, D.; Albright, T.D. A perceptual scaling approach to eyewitness identification. Nat. Commun. 2020, 11, 3380. [Google Scholar] [CrossRef]
- Clifford, C.W.G.; Watson, T.L.; White, D. Two sources of bias explain errors in facial age estimation. R. Soc. Open Sci. 2018, 5, 180841. [Google Scholar] [CrossRef] [PubMed]
- Palencia-Madrid, L.; Xavier, C.; de la Puente, M.; Hohoff, C.; Phillips, C.; Kayser, M.; Parson, W. Evaluation of the VISAGE Basic Tool for Appearance and Ancestry Prediction Using PowerSeq Chemistry on the MiSeq FGx System. Genes 2020, 11, 708. [Google Scholar] [CrossRef] [PubMed]
- Xavier, C.; De La Puente, M.; Mosquera-Miguel, A.; Freire-Aradas, A.; Kalamara, V.; Vidaki, A.; Gross, T.E.; Revoir, A.; Pośpiech, E.; Kartasińska, E.; et al. Development and validation of the VISAGE AmpliSeq basic tool to predict appearance and ancestry from DNA. Forensic Sci. Int. Genet. 2020, 48, 102336. [Google Scholar] [CrossRef] [PubMed]
- Kukla-Bartoszek, M.; Szargut, M.; Pośpiech, E.; Diepenbroek, M.; Zielińska, G.; Jarosz, A.; Piniewska-Róg, D.; Arciszewska, J.; Cytacka, S.; Spólnicka, M.; et al. The challenge of predicting human pigmentation traits in degraded bone samples with the MPS-based HIrisPlex-S system. Forensic Sci. Int. Genet. 2020, 47, 102301. [Google Scholar] [CrossRef] [PubMed]
- Royal, C.D.; Novembre, J.; Fullerton, S.M.; Goldstein, D.B.; Long, J.C.; Bamshad, M.J.; Clark, A.G. Inferring Genetic Ancestry: Opportunities, Challenges, and Implications. Am. J. Hum. Genet. 2010, 14, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Rotimi, C.N.; Shriner, D. Human ancestry correlates with language and reveals that race is not an objective genomic classifier. Sci. Rep. 2017, 7, 1572. [Google Scholar] [CrossRef] [PubMed]
- Kidd, J.R.; Friedlaender, F.R.; Speed, W.C.; Pakstis, A.J.; De La Vega, F.M.; Kidd, K.K. Analyses of a set of 128 ancestry informative single-nucleotide polymorphisms in a global set of 119 population samples. Investig. Genet. 2011, 5, 1. [Google Scholar] [CrossRef]
- Jia, J.; Wei, Y.-L.; Qin, C.-J.; Hu, L.; Wan, L.-H.; Li, C.-X. Developing a novel panel of genome-wide ancestry informative markers for bio-geographical ancestry estimates. Forensic Sci. Int. Genet. 2014, 8, 187–194. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Solovieff, N.; Hartley, S.W.; Baldwin, C.T.; Perls, T.T.; Steinberg, M.H.; Sebastiani, P. Clustering by genetic ancestry using genome-wide SNP data. BMC Genet. 2010, 11, 108. [Google Scholar] [CrossRef]
- Cheung, E.Y.Y.; Gahan, M.E.; McNevin, D. Prediction of biogeographical ancestry from genotype: A comparison of classifiers. Int. J. Legal Med. 2017, 131, 901–912. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, H.S.; Tvedebrink, T.; Børsting, C.; Pereira, V.; Morling, N. Ancestry prediction efficiency of the software GenoGeographer using a z-score method and the ancestry informative markers in the Precision ID Ancestry Panel. Forensic Sci. Int. Genet. 2020, 44, 102154. [Google Scholar] [CrossRef] [PubMed]
- García, O.; Ajuriagerra, J.A.; Alday, A.; Alonso, S.; Pérez, J.A.; Soto, A.; Uriarte, I.; Yurrebaso, I. Frequencies of the precision ID ancestry panel markers in Basques using the Ion Torrent PGM TM platform. Forensic Sci. Int. Genet. 2017, 31, e1–e4. [Google Scholar]
- Simayijiang, H.; Børsting, C.; Tvedebrink, T.; Morling, N. Analysis of Uyghur and Kazakh populations using the Precision ID Ancestry Panel. Forensic Sci. Int. Genet. 2019, 53, 102144. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Pereira, V.; Børsting, C.; Yamamoto, T.; Tvedebrink, T.; Hara, M.; Takada, A.; Saito, K.; Morling, N. Analysis of mainland Japanese and Okinawan Japanese populations using the precision ID Ancestry Panel. Forensic Sci. Int. Genet. 2018, 33, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, G.; Luo, T.; Zhao, X.; Liu, J.; Wang, M.; Zhou, D.; Chen, X.; Li, C.; Hou, Y. Massively parallel sequencing of 165 ancestry informative SNPs in two Chinese Tibetan-Burmese minority ethnicities. Forensic Sci. Int. Genet. 2018, 34, 141–147. [Google Scholar] [CrossRef]
- Sharma, V.; Jani, K.; Khosla, P.; Butler, E.; Siegel, D.; Wurmbach, E. Evaluation of ForenSeq™ Signature Prep Kit B on predicting eye and hair coloration as well as biogeographical ancestry by using Universal Analysis Software (UAS) and available web-tools. Electrophoresis 2019, 40, 1353–1364. [Google Scholar] [CrossRef]
- Ramani, A.; Wong, Y.; Zhen Tan, S.; Hong Shue, B.; Syn, C. Ancestry prediction in Singapore population samples using the Illumina ForenSeq kit. Forensic Sci. Int. Genet. 2017, 31, 171–179. [Google Scholar] [CrossRef]
- Kosoy, R.; Nassir, R.; Tian, C.; White, P.A.; Butler, L.M.; Silva, G.; Kittles, R.; Alarcon-Riquelme, M.E.; Gregersen, P.K.; Belmont, J.W.; et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum. Mutat. 2009, 30, 69–78. [Google Scholar] [CrossRef]
- Lawson, D.J.; van Dorp, L.; Falush, D. A tutorial on how not to over-interpret STRUCTURE and ADMIXTURE bar plots. Nat. Commun. 2018, 9, 3258. [Google Scholar] [CrossRef]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Olalde, I.; Broomandkhoshbacht, N.; Candilio, F.; Cheronet, O.; et al. The genomic history of southeastern Europe. Nature 2018, 555, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Pakstis, A.J.; Gurkan, C.; Dogan, M.; Balkaya, H.E.; Dogan, S.; Neophytou, P.I.; Cherni, L.; Boussetta, S.; Khodjet-El-Khil, H.; Elgaaied, A.B.A.; et al. Genetic relationships of European, Mediterranean, and SW Asian populations using a panel of 55 AISNPs. Eur. J. Hum. 2019, 27, 1885–1893. [Google Scholar] [CrossRef] [PubMed]
- Bulbul, O.; Cherni, L.; Khodjet-el-khil, H.; Rajeevan, H.; Kidd, K.K. Evaluating a subset of ancestry informative SNPs for discriminating among Southwest Asian and circum-Mediterranean populations. Forensic Sci. Int. Genet. 2016, 23, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Hinde, V.; Narasimhan, V.M.; Rohland, N.; Mallick, S.; Mah, M.; Lipson, M.; Nakatsuka, N.; Adamski, N.; Broomandkhoshbacht, N.; Ferry, M.; et al. An Ancient Harappan Genome Lacks Ancestry from Steppe Pastoralists or Iranian Farmers. Cell 2019, 179, 729–735. [Google Scholar]
- Terreros, M.C.; Rowold, D.J.; Mirabal, S.; Herrera, R.J. Mitochondrial DNA and Y-chromosomal stratification in Iran: Relationship between Iran and the Arabian Peninsula. J. Hum. Genet. 2011, 56, 235–246. [Google Scholar] [CrossRef][Green Version]
- Bánfai, Z.; Melegh, B.I.; Sümegi, K.; Hadzsiev, K.; Miseta, A.; Kásler, M.; Melegh, B. Revealing the Genetic Impact of the Ottoman Occupation on Ethnic Groups of East-Central Europe and on the Roma Population of the Area. Front. Genet. 2019, 10, 558. [Google Scholar]
- Hodoğlugil, U.; Mahley, R.W. Turkish Population Structure and Genetic Ancestry Reveal Relatedness among Eurasian Populations. Ann. Hum. Genet. 2012, 76, 128–141. [Google Scholar] [CrossRef]
- Alkan, C.; Kavak, P.; Somel, M.; Gokcumen, O.; Ugurlu, S.; Saygi, C.; Dal, E.; Bugra, K.; Güngör, T.; Sahinalp, S.C.; et al. Whole genome sequencing of Turkish genomes reveals functional private alleles and impact of genetic interactions with Europe, Asia and Africa. BMC Genom. 2014, 15, 963. [Google Scholar] [CrossRef]
- Dobon, B.; Hassan, H.Y.; Laayouni, H.; Luisi, P.; Ricaño-Ponce, I.; Zhernakova, A.; Wijmenga, C.; Tahir, H.; Comas, D.; Netea, M.G.; et al. The genetics of East African populations: A Nilo-Saharan component in the African genetic landscape. Sci. Rep. 2015, 5, 9996. [Google Scholar] [CrossRef]
- Ruiz-Linares, A.; Adhikari, K.; Acuña-Alonzo, V.; Quinto-Sanchez, M.; Jaramillo, C.; Arias, W.; Fuentes, M.; Pizarro, M.; Everardo, P.; De Avila, F.; et al. Admixture in Latin America: Geographic Structure, Phenotypic Diversity and Self-Perception of Ancestry Based on 7,342 Individuals. PLoS Genet. 2014, 10, e1004572. [Google Scholar] [CrossRef]
- Norris, E.T.; Wang, L.; Conley, A.B.; Rishishwar, L.; Mariño-Ramírez, L.; Valderrama-Aguirre, A.; King Jordan, I. Genetic ancestry, admixture and health determinants in Latin America. BMC Genom. 2018, 19, 861. [Google Scholar] [CrossRef] [PubMed]
| Mean p-Values for Each HIrisPlex-S Category among Tested Reference Samples | Example | Prediction | Number of Predictions per Category (Incorrect Ones in Red) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| EYE COLOR | Blue | Intermediate | Brown | ||||||
| 0.900 | 0.061 | 0.039 |  | Blue |  | ||||
| 0.097 | 0.123 | 0.780 |  | Brown | |||||
| 0.336 | 0.240 | 0.424 |  | Inconclusive | |||||
| HAIR COLOR | Color | Shade | |||||||
| Blond | Brown | Red | Black | Light | Dark | ||||
| 0.212 | 0.103 | 0.678 | 0.007 | 0.969 | 0.031 |  | Red |  | |
| 0.758 | 0.185 | 0.041 | 0.017 | 0.986 | 0.014 |  | Light blond to blond | ||
| 0.582 | 0.315 | 0.066 | 0.037 | 0.935 | 0.065 |  | Blond to dark blond | ||
| 0.302 | 0.553 | 0.058 | 0.088 | 0.821 | 0.179 |  | Light brown to brown | ||
| 0.206 | 0.619 | 0.012 | 0.163 | 0.525 | 0.475 |  | Brown to dark brown | ||
| 0.013 | 0.330 | 0.000 | 0.657 | 0.026 | 0.974 |  | Dark brown to black | ||
| SKIN COLOR | Very Pale | Pale | Inter. | Dark | Dark/ Black | ||||
| 0.204 | 0.705 | 0.091 | 0.004 | 0.000 |  | Very pale to pale | 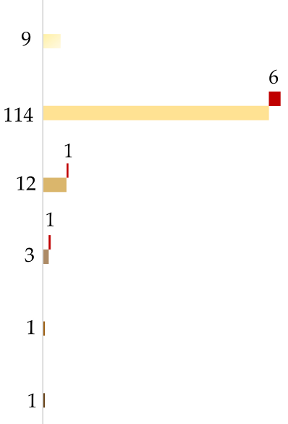 | ||
| 0.046 | 0.492 | 0.454 | 0.008 | 0.000 |  | Pale to intermediate | |||
| 0.005 | 0.068 | 0.878 | 0.050 | 0.004 |  | Intermediate | |||
| 0.003 | 0.021 | 0.497 | 0.458 | 0.024 |  | Intermediate to dark | |||
| 0.001 | 0.006 | 0.262 | 0.721 | 0.010 |  | Dark | |||
| 0.000 | 0.000 | 0.000 | 0.001 | 0.999 |  | Dark to black | |||
| Admixture (by Converge) | LR (by Snipper) | Population Likelihoods FROG (Highest) | Y-Lineage |
|---|---|---|---|
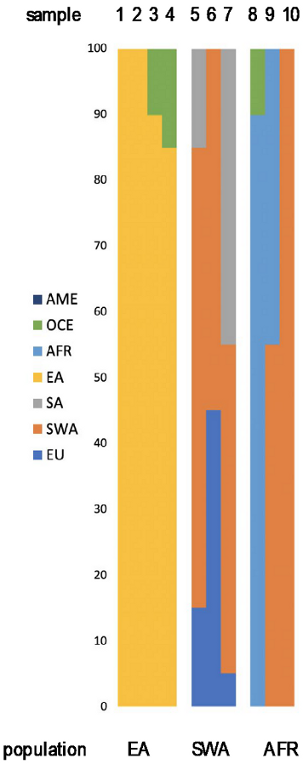 | Sample 1 (Japan) billion times more likely EA than SA and AME | Sample 1 (Japan) Japanese 1.3 × 10−51 Mainland Japanese 1.9 × 10−52 | O1b2 P49 |
| Sample 2 (China) billion times more likely EA than AME and SA | Sample 2 (China) Yi (Sichuan) 1.1 × 10−51 | O2 M122 | |
| Sample 3 (Vietnam) billion times more likely EA than AME and SA | Sample 3 (Vietnam) Hakka 1.1 × 10−46 Lao Long 8.3 × 10−47 Mainland Japanese 7.8 × 10−47 | N/A | |
| Sample 4 (Japan) billion times more likely EA than SA and AME | Sample 4 (Japan) Mainland Japanese 7.2 × 10−53 Okinawa Japanese 4.4 × 10−53 Japanese 2.2 × 10−53 | N/A | |
| Sample 5 (Turkey) 18.94 times more likely EU than SA and billion than OCE | Sample 5 (Turkey) Iranians 2.0 × 10−41 Pathan 6.9 × 10−42 Turks 3.1 × 10−42 | R1b1a1b M269 | |
| Sample 6 (Palestina) billion times more likely EU than SA and EA | Sample 6 (Palestina) Turkish Cypriots 6.1 × 10−49 | N/A | |
| Sample 7 (Iran) 458 times more likely SA than EU and billion than OCE | Sample 7 (Iran) Iranians 3.9 × 10−48 Turks 7.2 × 10−49 | H1a1a M82 | |
| Sample 8 (Uganda) billion times more likely AFR than SA and OCE | Sample 8 (Uganda) Lisongo 1.2 × 10−38 Hausa 1.1 × 10−39 | N/A | |
| Sample 9 (Eritrea) billion times more likely SA than EU and AFR | Sample 9 (Eritrea) Ethiopian Jews 9.1 × 10−51 Somalis 1.6 × 10−52 | E1b1b1 M35 | |
| Sample 10 (Egypt) 1.36 times more likely EU than SA and billion more than AME | Sample 10 (Egypt) Palestinian Arabs 1.7 × 10−51 | N/A |
| Expected Admixture (Based on Provided Data) | Predicted Admixture (Calculated by Converge) | LR (by SNIPPER) | Population Likelihoods FROG (Highest) | Y Lineage |
|---|---|---|---|---|
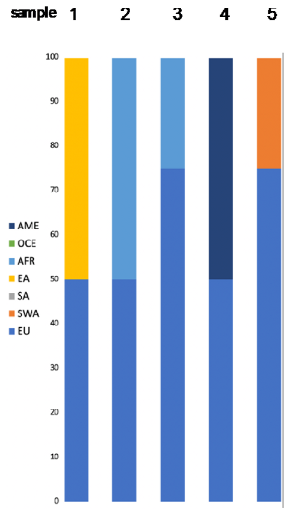 | 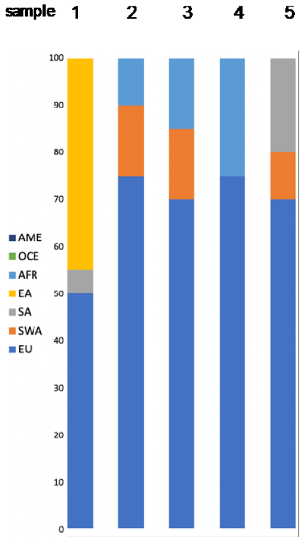 | billion times more likely SA than EU and EA | Sample 1 Chuvash 6.2 × 10–53 Qinghai Tibetans 1.1 × 10–53 Khazaks 7.8 × 10–54 | N/A |
| billion times more likely EU than SA and AME | Sample 2 Italians 7.0 × 10–48 Turks 5.7 × 10–48 Turkish Cypriots 1.2 × 10–48 | N/A | ||
| 128,027 times more likely EU than SA and billion more than AME | Sample 3 Kairoun,Tunisia 5.9 × 10–49 Smar,South Tunisia 5.9 × 10–50 | J1a P58 | ||
| 795 times more likely EU than SA and billion more than OCE | Sample 4 Sousse, Tunisia 1.1 × 10–53 Kairoun,Tunisia 1.8 × 10–54 Smar, South Tunisia 1.7 × 10–54 | N/A | ||
| 221,461 times more likely EU than SA and billion more than AME | Sample 5 Mixed EU 4.8 × 10–46 Russians 2.7 × 10–46 Finns 1.6 × 10–46 | N/A |
| Sample and Material | DNA Input (DI) | Used SNPs Maximum: | p-Values | Admixture Converge (% Mean) | Population Likelihoods FROG (Highest) | Y-Lineage | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Eye Color | Hair Color | Hair Shade | Skin Color | ||||||||
| 163 Ancestry | 41 Phenotype | 120 Y-SNPs | Blue Inter Brown | Blond Brown Red Black | Light Dark | V Pale Pale Inter Dark B-Dark | |||||
| C1 bone | 125 pg (1.4) | 163 | 30 | 110 | 0.001 0.017 0.982 | 0.097 0.645 0.001 0.257 | 0.052 0.948 | 0.000 0.000 0.001 0.192 0.807 | 51.50 SWA 48.50 AFR | Ethiopian Jews 5.7 × 10−52 | Major: E Subhap: E1b1b1 (M35) |
| C2 bone | 31 pg (1.2) | 66 | 12 | 29 | NA | NA | NA | NA | |||
| C3 trace | 62 pg (1.6) | 154 | 40 | 107 | The exact p-values cannot be published due to an ongoing investigation | ||||||
| C4 trace | 125 pg (1) | 163 | 40 | 113 | |||||||
| C5 blood | 1 ng (1.1) | 163 | 41 | 116 | 0.932 0.046 0.021 | 0.433 0.046 0.519 0.002 | 1.000 0.000 | 0.098 0.654 0.249 0.000 0.000 | 100 EU | Danes 4.5 × 10−45 Mixed EU 4.0 × 10−45 Irish 3.8 × 10−45 Hungarians 3.7 × 10−45 | Major: R Subhap: R1a1a1b1a2 (Z280) |
| C6 blood | 1 ng (0.9) | 162 | 41 | 116 | 0.000 0.002 0.998 | 0.002 0.301 0.000 0.697 | 0.002 0.998 | 0.000 0.000 0.000 0.003 0.997 | 100 AFR | Yoruba 3.1 × 10−34 Zaramo 4.7 × 10−35 Lisongo 3.5 × 10−35 | Major: E Subhap: E1b1a1 (M2) |
| C7 blood | 1ng 1 | 162 | 41 | 116 | 0.000 0.002 0.998 | 0.003 0.264 0.000 0.733 | 0.007 0.993 | 0.000 0.000 0.000 0.060 0.940 | 60.64 SWA 39.36 AFR | Ethiopian Jews 4.1 × 10−57 | Major: E Subhaplo: E1b1b1 (M35) |
| C8 blood | 1 ng 0.9 | 161 | 41 | 116 | 0.000 0.002 0.998 | 0.003 0.425 0.000 0.571 | 0.003 0.997 | 0.000 0.000 0.057 0.923 0.020 | 55.18 AFR 41.82 SWA | Somalis 6.7 × 10−57 Ethiopian Jews 6.6 × 10−57 | Major: T Subhaplo: T1a (M70) |
| C9 blood | 1 ng 1.6 | 163 | 41 | 115 | 0.000 0.007 0.993 | 0.007 0.246 0.000 0.747 | 0.014 0.986 | 0.000 0.000 0.998 0.002 0.000 | 92.40 EA 7.60 EU | Hakka 3.9 × 10−54 Taiwanese Han 1.0 × 10−54 SF Chinese 5.3 × 10−55 | Major: R Subhaplo: R1b1a1b (M269) |
| C10 blood | 1 ng 2.2 | 161 | 41 | 115 | 0.000 0.003 0.997 | 0.001 0.133 0.000 0.866 | 0.003 0.997 | 0.084 0.000 0.976 0.024 0.000 | 95.06 EA 4.94 SA | Lao Long 4.2 × 10−53 | Major: O Subhaplo: O1b1 (F2320) |
| C11 blood | 1 ng 7 | 162 | 41 | 108 | 0.911 0.057 0.032 | 0.576 0.379 0.003 0.042 | 0.917 0.083 | 0.021 0.489 0.475 0.011 0.004 | 92.84 EU 5.26 SWA | Irish 2.7 × 10−47 Danes 1.4 × 10−47 Russians 1.1 × 10−47 | Major: R Subhaplo: R1b1a1b (M269) |
| C12 blood | 1 ng 1 | 163 | 40 | 116 | 0.00 0.004 0.996 | 0.004 0.311 0.000 0.685 | 0.004 0.996 | 0.000 0.000 0.019 0.965 0.016 | 67.56 SWA 29.35 EU 3.09 SA | Iranians 2.4 × 10−42 Palestinian Arabs 2.1 × 10−42 | Major: I Subhaplo: I2 (M438) |
| C13 blood | 1 ng 1 | 161 | 40 | 116 | 0.000 0.002 0.998 | 0.002 0.301 0.000 0.697 | 0.002 0.998 | 0.000 0.054 0.000 0.005 0.995 | 100 AFR | Yoruba 1.1 × 10−29 Ibo 4.9 × 10−30 Lisongo 2.1 × 10−0 | Major: E Subhaplo: E1b1a1 (M2) |
| C14 blood | 1 ng 1.3 | 160 | 40 | 115 | 0.012 0.050 0.938 | 0.072 0.706 0.001 0.221 | 0.137 0.863 | 0.000 0.000 0.210 0.339 0.452 | 56.77 EU 27.40 SA 15.83 OCE | Iranians 9.1 × 10−53 | Major: R Subhaplo: R1a1a1b2 (Z93) |
| C15 blood | 1 ng 1 | 163 | 41 | 116 | 0.000 0.003 0.997 | 0.002 0.211 0.000 0.787 | 0.004 0.996 | 0.007 0.020 0.644 0.332 0.008 | 76.32 AME 15.06 SWA 8.62 AFR | Ecuadorian Mestizo 2.8 × 10−69 | Major: Q Subhaplo: Q1b1a1a (M3) |
| C16 blood | 1 ng 0.8 | 163 | 40 | 116 | 0.028 0.073 0.899 | 0.087 0.492 0.001 0.420 | 0.169 0.831 | 0.113 0.268 0.550 0.045 0.024 | 51.54 SWA 44.23 EU 4.23 SA | Druze 7.9 × 10−48 | Major: J Subhaplo: J2a (M410) |
| C17 blood | 1 ng 0.8 | 163 | 40 | 116 | 0.000 0.003 0.997 | 0.001 0.087 0.000 0.912 | 0.003 0.997 | 0.000 0.000 0.997 0.003 0.000 | 95.85 EA 4.15 OCE | Koreans 5.5 × 10−54 Japanese 3.0 × 10−54 | Major: D Subhaplo: D1b (M55) |
| Sample | Phenotype Prediction | Phenotype (Photo) | Ancestry Prediction | Place of Birth |
|---|---|---|---|---|
| C1 | Brown eyes Dark brown to black hair Black skin | No data (body skeletonized) | ADMIXED (AFR-SWA) Likely: East Africa | Eritrea |
| C2 | No prediction | No data (body skeletonized) | No prediction | Eritrea |
| C3 | Brown eyes Light brown to brown hair Pale to intermediate skin | No data (police investigation) | High: Europe | No data |
| C4 | Brown eyes Light brown to brown hair Pale to intermediate skin | No data (police investigation) | High: Europe | No data |
| C5 | Blue eyes Red hair Pale skin | No data (body decayed) | High: Europe | Russia |
| C6 | Brown eyes Black hair Black skin | No data Black hair Black skin | High: Africa Likely: Central/West | Burkina Faso |
| C7 | Brown eyes Black hair Black skin | Brown eyes Black hair Black skin | ADMIXED (SWA-AFR) Likely: East Africa | Eritrea |
| C8 | Brown eyes Black hair Dark skin | Brown eyes Black hair Dark skin | ADMIXED (SWA-AFR) Likely: East Africa | Ethiopia |
| C9 | Brown eyes Black hair Intermediate skin | Brown eyes Black hair Intermediate skin | High: Asia High: East Asia | China |
| C10 | Brown eyes Black hair Intermediate skin | Brown eyes Black hair Intermediate skin | High: Asia High: East Asia | Vietnam |
| C11 | Blue eyes Blond to light blond hair Pale to intermediate skin | No data (body decayed) | High: Europe | Brazil |
| C12 | Brown eyes Black hair Dark skin | No data (body decayed) | ADMIXED (SWA-EU-SA) Likely: Southwest Asia | Iraq |
| C13 | Brown eyes Black hair Black skin | No data Black hair Black skin | High; Africa Likely: Central/West | Nigeria |
| C14 | Brown eyes Brown to dark brown hair Dark skin to black skin | No data Dark greying hair No data | ADMIXED (EU-SA-OCE) | Afghanistan |
| C15 | Brown eyes Black hair Intermediate to dark skin | No data Black hair Intermediate skin | ADMIXED (AME-SWA-AFR) Likely: South America | Mexico |
| C16 | Brown eyes Dark brown to black hair Pale to intermediate skin | No data Dark greying hair Intermediate skin | ADMIXED (SWA-EU-SA) Likely: Southwest Asia | Iran |
| C17 | Brown eyes Black hair Intermediate skin | No data Dark greying hair Intermediate skin | High: Asia High: East Asia | Japan |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diepenbroek, M.; Bayer, B.; Schwender, K.; Schiller, R.; Lim, J.; Lagacé, R.; Anslinger, K. Evaluation of the Ion AmpliSeq™ PhenoTrivium Panel: MPS-Based Assay for Ancestry and Phenotype Predictions Challenged by Casework Samples. Genes 2020, 11, 1398. https://doi.org/10.3390/genes11121398
Diepenbroek M, Bayer B, Schwender K, Schiller R, Lim J, Lagacé R, Anslinger K. Evaluation of the Ion AmpliSeq™ PhenoTrivium Panel: MPS-Based Assay for Ancestry and Phenotype Predictions Challenged by Casework Samples. Genes. 2020; 11(12):1398. https://doi.org/10.3390/genes11121398
Chicago/Turabian StyleDiepenbroek, Marta, Birgit Bayer, Kristina Schwender, Roberta Schiller, Jessica Lim, Robert Lagacé, and Katja Anslinger. 2020. "Evaluation of the Ion AmpliSeq™ PhenoTrivium Panel: MPS-Based Assay for Ancestry and Phenotype Predictions Challenged by Casework Samples" Genes 11, no. 12: 1398. https://doi.org/10.3390/genes11121398
APA StyleDiepenbroek, M., Bayer, B., Schwender, K., Schiller, R., Lim, J., Lagacé, R., & Anslinger, K. (2020). Evaluation of the Ion AmpliSeq™ PhenoTrivium Panel: MPS-Based Assay for Ancestry and Phenotype Predictions Challenged by Casework Samples. Genes, 11(12), 1398. https://doi.org/10.3390/genes11121398





