Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Microarray Data
2.2. Data Preprocessing and DEG Screening
2.3. Hierarchical Clustering Analysis
2.4. Functional and Pathway Enrichment Analysis
2.5. Protein–Protein Interaction Network Construction and Hub Gene Analysis
2.6. Validation and Survival Analysis of the Hub Genes in The Cancer Genome Atlas (TCGA) Dataset
3. Results
3.1. Identification of DEG
3.2. GO Functional and Pathway Enrichment Analysis
3.3. PPI Network Construction and Hub Genes Identification
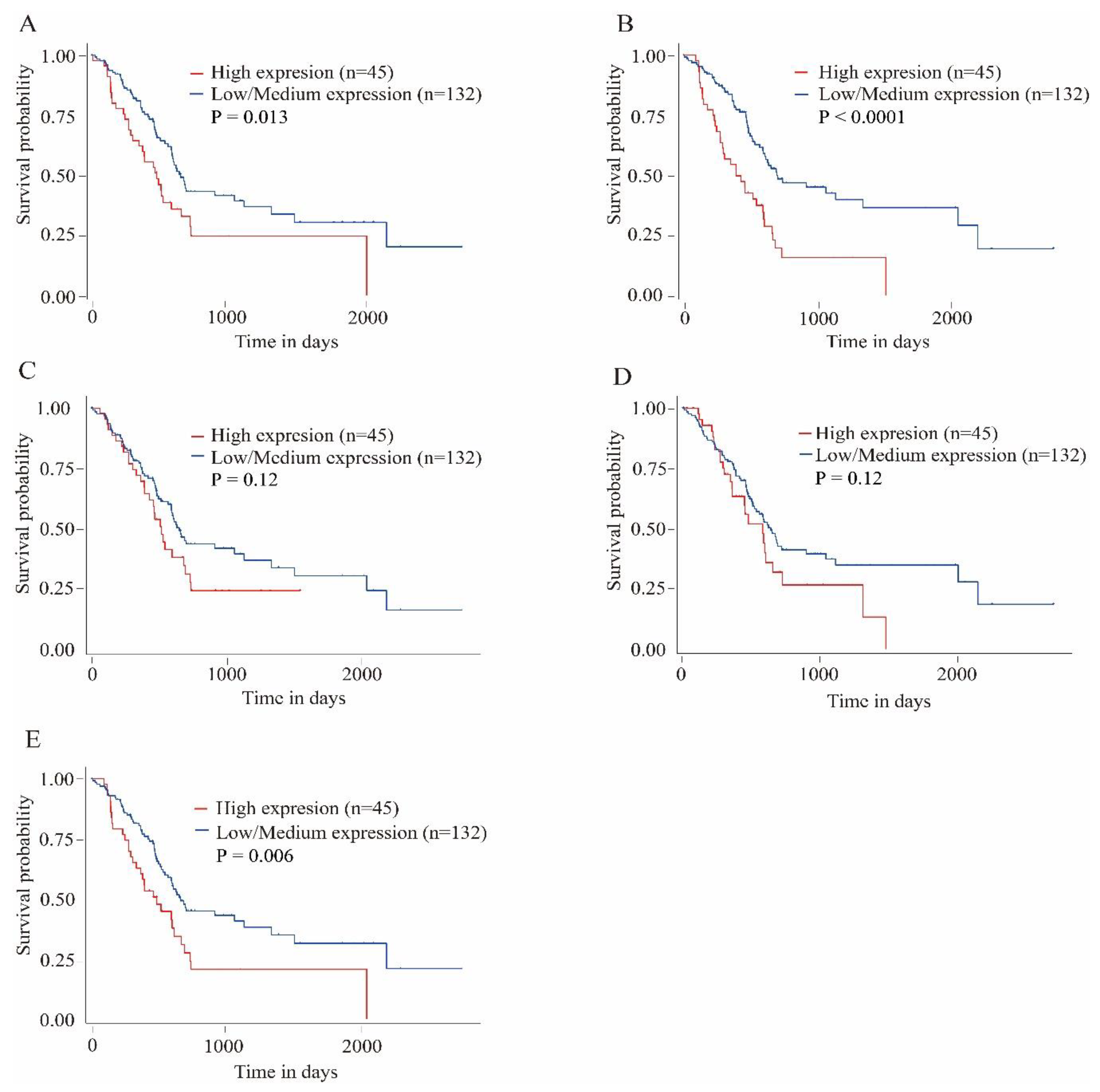
3.4. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA A Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Hoos, W.A.; James, P.M.; Rahib, L.; Talley, A.W.; Fleshman, J.M.; Matrisian, L.M. Pancreatic Cancer Clinical Trials and Accrual in the United States. J. Clin. Oncol. 2013, 31, 3432–3438. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, R.; Duraiyan, J.; Kaliyappan, K.; Palanisamy, M. Microarray and its applications. J. Pharm. Bioallied Sci. 2012, 4, 310–312. [Google Scholar]
- Han, D.Y.; Fu, D.; Xi, H.; Li, Q.Y.; Feng, L.J.; Zhang, W.; Ji, G.; Xiao, J.C.; Wei, Q. Genomic expression profiling and bioinformatics analysis of pancreatic cancer. Mol. Med. Rep. 2015, 12, 4133–4140. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Liu, Z.; Wu, X.; Xu, Y.; Ge, C. Gene expression profile analysis of pancreatic cancer based on microarray data. Mol. Med. Rep. 2016, 13, 3913–3919. [Google Scholar] [CrossRef]
- Malgerud, L.; Lindberg, J.; Wirta, V.; Gustafsson-Liljefors, M.; Karimi, M.; Moro, C.F.; Stecker, K.; Picker, A.; Huelsewig, C.; Stein, M.; et al. Bioinformatory-assisted analysis of next-generation sequencing data for precision medicine in pancreatic cancer. Mol. Oncol. 2017, 11, 1413–1429. [Google Scholar] [CrossRef] [PubMed]
- Jun, I.; Park, H.S.; Piao, H.; Han, J.W.; An, M.J.; Yun, B.G.; Zhang, X.; Cha, Y.H.; Shin, Y.K.; Yook, J.I.; et al. ANO9/TMEM16J promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br. J. Cancer 2017, 117, 1798–1809. [Google Scholar] [CrossRef] [PubMed]
- Hu, J. PYK2 as a Novel Therapeutic Target for Pancreatic Cancer. Pancreas 2017, 46, 1407. [Google Scholar]
- Kretz, A.L.; Schaum, M.; Richter, J.; Kitzig, E.F.; Engler, C.C.; Leithauser, F.; Henne-Bruns, D.; Knippschild, U.; Lemke, J. CDK9 is a prognostic marker and therapeutic target in pancreatic cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Hiraoka, N.; Yamazaki-Itoh, R.; Ino, Y.; Mizuguchi, Y.; Yamada, T.; Hirohashi, S.; Kanai, Y. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology 2011, 140, 310–321. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparmeter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, 3. [Google Scholar] [CrossRef]
- Von Mering, C.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003, 31, 258–261. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Crnogorac-Jurcevic, T.; Chelala, C.; Barry, S.; Harada, T.; Bhakta, V.; Lattimore, S.; Jurcevic, S.; Bronner, M.; Lemoine, N.R.; Brentnall, T.A. Molecular analysis of precursor lesions in familial pancreatic cancer. PLoS ONE 2013, 8, e54830. [Google Scholar] [CrossRef]
- Zhang, G.; Schetter, A.; He, P.; Funamizu, N.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Hassan, R.; Yfantis, H.G.; Lee, D.H.; et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS ONE 2012, 7, e31507. [Google Scholar] [CrossRef]
- Zhang, G.; He, P.; Tan, H.; Budhu, A.; Gaedcke, J.; Ghadimi, B.M.; Ried, T.; Yfantis, H.G.; Lee, D.H.; Maitra, A.; et al. Integration of metabolomics and transcriptomics revealed a fatty acid network exerting growth inhibitory effects in human pancreatic cancer. Clin. Cancer Res. 2013, 19, 4983–4993. [Google Scholar] [CrossRef]
- Hu, B.; Shi, C.; Jiang, H.X.; Qin, S.Y. Identification of novel therapeutic target genes and pathway in pancreatic cancer by integrative analysis. Medicine 2017, 96, e8261. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.; Quinn, M.C.; et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Wu, L.Y.; Wang, Y.; Zhang, X.S. GOMA: Functional enrichment analysis tool bases on GO modules. Chin. J. Cancer 2013, 32, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Gao, Y.F.; Chen, Y.X.; Wang, Z.B.; Yin, J.Y.; Mao, X.Y.; Li, X.; Zhang, W.; Zhou, H.H.; Liu, Z.Q. Genome-scale analysis identifies GJB2 and ERO1LB as prognosis markers in patients with pancreatic cancer. Oncotarget 2017, 8, 21281–21289. [Google Scholar] [CrossRef] [PubMed]
- Gress, T.M.; Muller-Pillasch, F.; Lerch, M.M.; Friess, H.; Buchler, M.; Adler, G. Expression and in-situ localization of genes coding for extracellular matrix proteins and extracellular matrix degrading proteases in pancreatic cancer. Int. J. Cancer 1995, 62, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Rothman, V.L.; Nicosia, R.F.; Tuszynski, G.P. Expression of thrombospondin-1 in human pancreatic adenocarcinomas: Role in matrix metalloproteinase-9 production. Pathol. Oncol. Res. 2001, 7, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Hirabayashi, K.; Imaizumi, T.; Matsuyama, M.; Yazawa, N.; Dowaki, S.; Tobita, K.; Ohtani, Y.; Tanaka, M.; Inokuchi, S.; et al. Stromal thrombospondin-1 expression is a prognostic indicator and a new marker of invasiveness in intraductal papillary-mucinous neoplasm of the pancreas. Biomed. Res. 2010, 31, 13–19. [Google Scholar] [CrossRef]
- Bocci, G.; Fioravanti, A.; Orlandi, P.; Di Desidero, T.; Natale, G.; Fanelli, G.; Viacava, P.; Naccarato, A.G.; Francia, G.; Danesi, R. Metronomic ceramide analogs inhibit angiogenesis in pancreatic cancer through up-regulation of caveolin-1 and thrombospondin-1 and down-regulation of cyclin D1. Neoplasia 2012, 14, 833–845. [Google Scholar] [CrossRef]
- Jenkinson, C.; Elliott, V.L.; Evans, A.; Oldfield, L.; Jenkins, R.E.; O’Brien, D.P.; Apostolidou, S.; Gentry-Maharaj, A.; Fourkala, E.O.; Jacobs, I.J.; et al. Decreased Serum Thrombospondin-1 Levels in Pancreatic Cancer Patients Up to 24 Months Prior to Clinical Diagnosis: Association with Diabetes Mellitus. Clin. Cancer Res. 2016, 22, 1734–1743. [Google Scholar] [CrossRef]
- Huang, T.T.; Sun, L.; Yuan, X.L.; Qiu, H. Thrombospondin-1 is a multifaceted player in tumor progression. Oncotarget 2017, 8, 84546–84558. [Google Scholar] [CrossRef]
- Li, L.N.; Leedom, T.A.; Do, J.; Huang, H.H.; Lai, J.Y.; Johnson, K.; Osothprarop, T.F.; Rizzo, J.D.; Doppalapudi, V.R.; Bradshaw, C.W.; et al. Antitumor Efficacy of a Thrombospondin 1 Mimetic CovX-Body. Transl. Oncol. 2011, 4, 239–247. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.S.; Ling, J.; Nye, D.; Papathomas, K.; Dickinson, C. Thrombospondin peptide ABT-898 inhibits inflammation and angiogenesis in a colitis model. World J. Gastroenterol. 2015, 21, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Dossi, R.; Frapolli, R.; Di Giandomenico, S.; Paracchini, L.; Bozzi, F.; Brich, S.; Castiglioni, V.; Borsotti, P.; Belotti, D.; Uboldi, S.; et al. Antiangiogenic activity of trabectedin in myxoid liposarcoma: Involvement of host TIMP-1 and TIMP-2 and tumor thrombospondin-1. Int. J. Cancer 2015, 136, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Conejo, J.R.; Kleeff, J.; Koliopanos, A.; Matsuda, K.; Zhu, Z.W.; Goecke, H.; Bicheng, N.; Zimmermann, A.; Korc, M.; Friess, H.; et al. Syndecan-1 expression is up-regulated in pancreatic but not in other gastrointestinal cancers. Int. J. Cancer 2000, 88, 12–20. [Google Scholar] [CrossRef]
- Szatmari, T.; Otvos, R.; Hjerpe, A.; Dobra, K. Syndecan-1 in Cancer: Implications for Cell Signaling, Differentiation, and Prognostication. Dis. Markers 2015, 2015, 796052. [Google Scholar] [CrossRef] [PubMed]
- Orecchia, P.; Conte, R.; Balza, E.; Petretto, A.; Mauri, P.; Mingari, M.C.; Carnemolla, B. A novel human anti-syndecan-1 antibody inhibits vascular maturation and tumour growth in melanoma. Eur. J. Cancer 2013, 49, 2022–2033. [Google Scholar] [CrossRef]
- Schonfeld, K.; Zuber, C.; Pinkas, J.; Hader, T.; Bernoster, K.; Uherek, C. Indatuximab ravtansine (BT062) combination treatment in multiple myeloma: Pre-clinical studies. J. Hematol. Oncol. 2017, 10, 13. [Google Scholar] [CrossRef]
- Delgado, J.L.; Hsieh, C.M.; Chan, N.L.; Hiasa, H. Topoisomerases as anticancer targets. Biochem. J. 2018, 475, 373–398. [Google Scholar] [CrossRef]
- Bai, J.; Sata, N.; Nagai, H. Gene expression analysis for predicting gemcitabine sensitivity in pancreatic cancer patients. HPB 2007, 9, 150–155. [Google Scholar] [CrossRef][Green Version]
- Zhou, Z.; Liu, S.; Zhang, M.; Zhou, R.; Liu, J.; Chang, Y.; Zhao, Q. Overexpression of Topoisomerase 2-alpha Confers a Poor Prognosis in Pancreatic Adenocarcinoma Identified by Co-Expression Analysis. Digest. Dis. Sci. 2017, 62, 2790–2800. [Google Scholar] [CrossRef]
- Zhou, H.; Chen, Q.; Tan, W.; Qiu, Z.; Li, S.; Song, Y.; Gao, S. Integrated clinicopathological features and gene microarray analysis of pancreatic neuroendocrine tumors. Gene 2017, 625, 72–77. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Kato, T.; Olugbile, S.; Tamura, K.; Chung, S.; Miyamoto, T.; Matsuo, Y.; Salgia, R.; Nakamura, Y.; Park, J.H. Effective growth-suppressive activity of maternal embryonic leucine-zipper kinase (MELK) inhibitor against small cell lung cancer. Oncotarget 2016, 7, 13621–13633. [Google Scholar] [CrossRef] [PubMed]
- Hiwatashi, K.; Ueno, S.; Sakoda, M.; Iino, S.; Minami, K.; Yonemori, K.; Nishizono, Y.; Kurahara, H.; Mataki, Y.; Maemura, K.; et al. Expression of Maternal Embryonic Leucine Zipper Kinase (MELK) Correlates to Malignant Potentials in Hepatocellular Carcinoma. Anticancer Res. 2016, 36, 5183–5188. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chung, C.H.; Miller, A.; Panopoulos, A.; Hao, E.; Margolis, R.; Terskikh, A.; Levine, F. Maternal embryonic leucine zipper kinase regulates pancreatic ductal, but not beta-cell, regeneration. Physiol. Rep. 2014, 2, e12131. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, R.; Hong, C.S.; Smith, L.G.F.; Kornblum, H.I.; Nakano, I. Maternal Embryonic Leucine Zipper Kinase: Key Kinase for Stem Cell Phenotype in Glioma and Other Cancers. Mol. Cancer Ther. 2014, 13, 1393–1398. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Suzuki, H.; Miyamoto, T.; Takamatsu, N.; Tatsuguchi, A.; Ueda, K.; Kijima, K.; Nakamura, Y.; Matsuo, Y. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget 2012, 3, 1629–1640. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.B.; Pothula, S.P.; Xu, Z.; Lee, A.K.; Goldstein, D.; Pirola, R.C.; Apte, M.V.; Wilson, J.S. The role of the hepatocyte growth factor/c-MET pathway in pancreatic stellate cell-endothelial cell interactions: Antiangiogenic implications in pancreatic cancer. Carcinogenesis 2014, 35, 1891–1900. [Google Scholar] [CrossRef]
- Neuzillet, C.; Couvelard, A.; Tijeras-Raballand, A.; de Mestier, L.; de Gramont, A.; Bedossa, P.; Paradis, V.; Sauvanet, A.; Bachet, J.B.; Ruszniewski, P.; et al. High c-Met expression in stage I-II pancreatic adenocarcinoma: Proposal for an immunostaining scoring method and correlation with poor prognosis. Histopathology 2015, 67, 664–676. [Google Scholar] [CrossRef]
- Brandes, F.; Schmidt, K.; Wagner, C.; Redekopf, J.; Schlitt, H.J.; Geissler, E.K.; Lang, S.A. Targeting cMET with INC280 impairs tumour growth and improves efficacy of gemcitabine in a pancreatic cancer model. BMC Cancer 2015, 15, 71. [Google Scholar] [CrossRef]
- Rizwani, W.; Allen, A.E.; Trevino, J.G. Hepatocyte Growth Factor from a Clinical Perspective: A Pancreatic Cancer Challenge. Cancers 2015, 7, 1785–1805. [Google Scholar] [CrossRef]
- Pothula, S.P.; Xu, Z.; Goldstein, D.; Biankin, A.V.; Pirola, R.C.; Wilson, J.S.; Apte, M.V. Hepatocyte growth factor inhibition: A novel therapeutic approach in pancreatic cancer. Br. J. Cancer 2016, 114, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Takiguchi, S.; Inoue, K.; Matsusue, K.; Furukawa, M.; Teramoto, N.; Iguchi, H. Crizotinib, a MET inhibitor, prevents peritoneal dissemination in pancreatic cancer. Int. J. Oncol. 2017, 51, 184–192. [Google Scholar] [CrossRef] [PubMed]
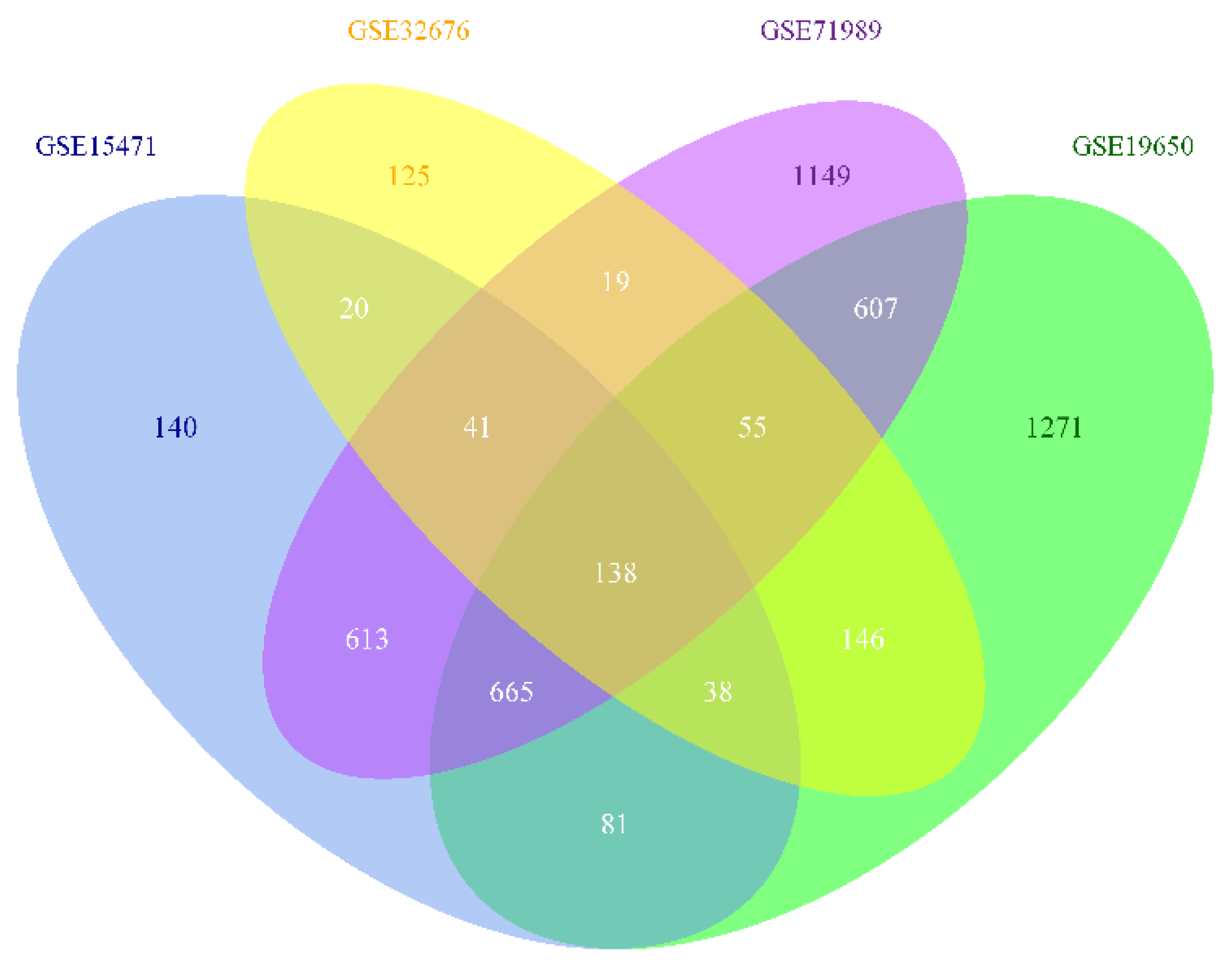
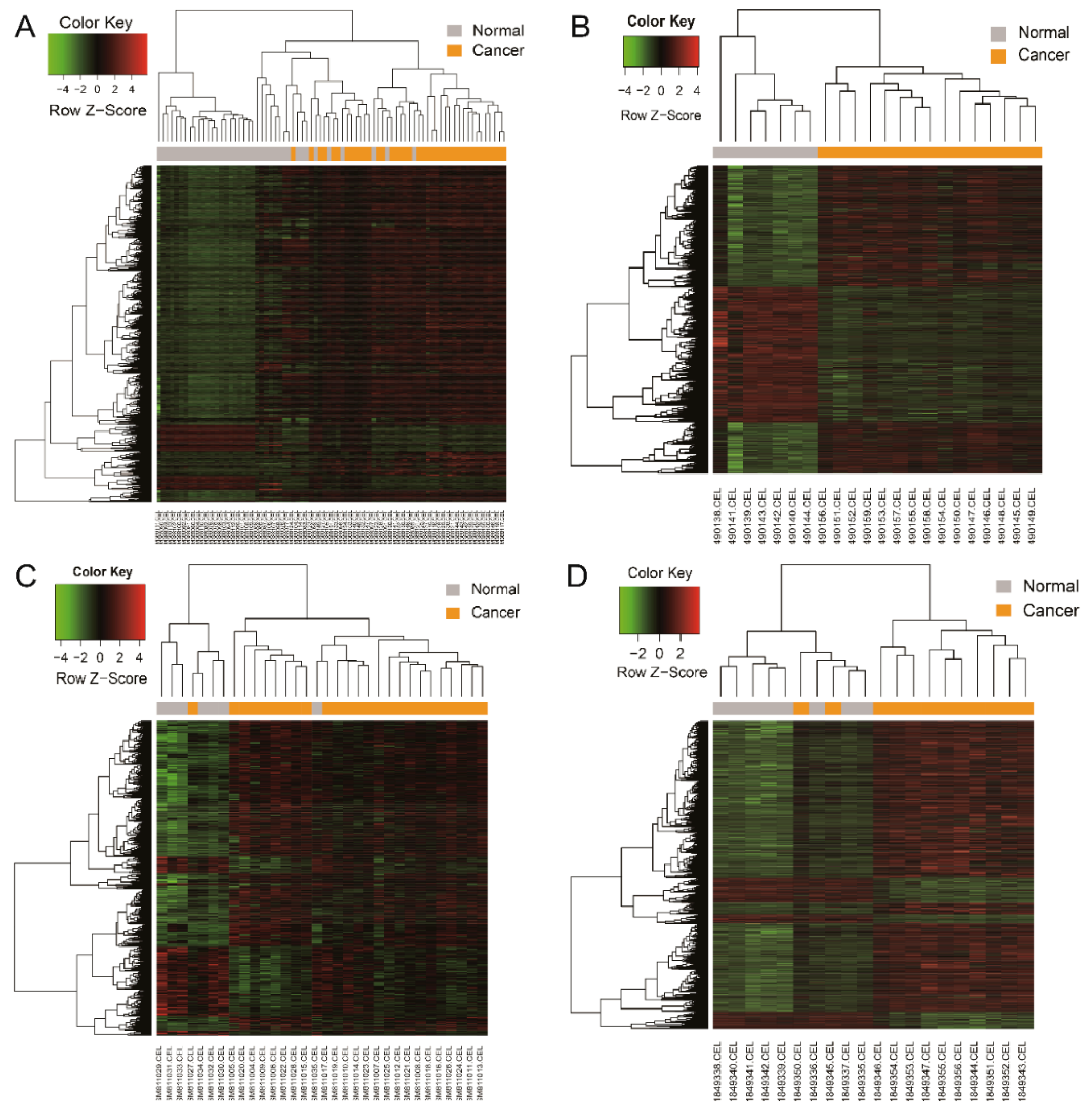
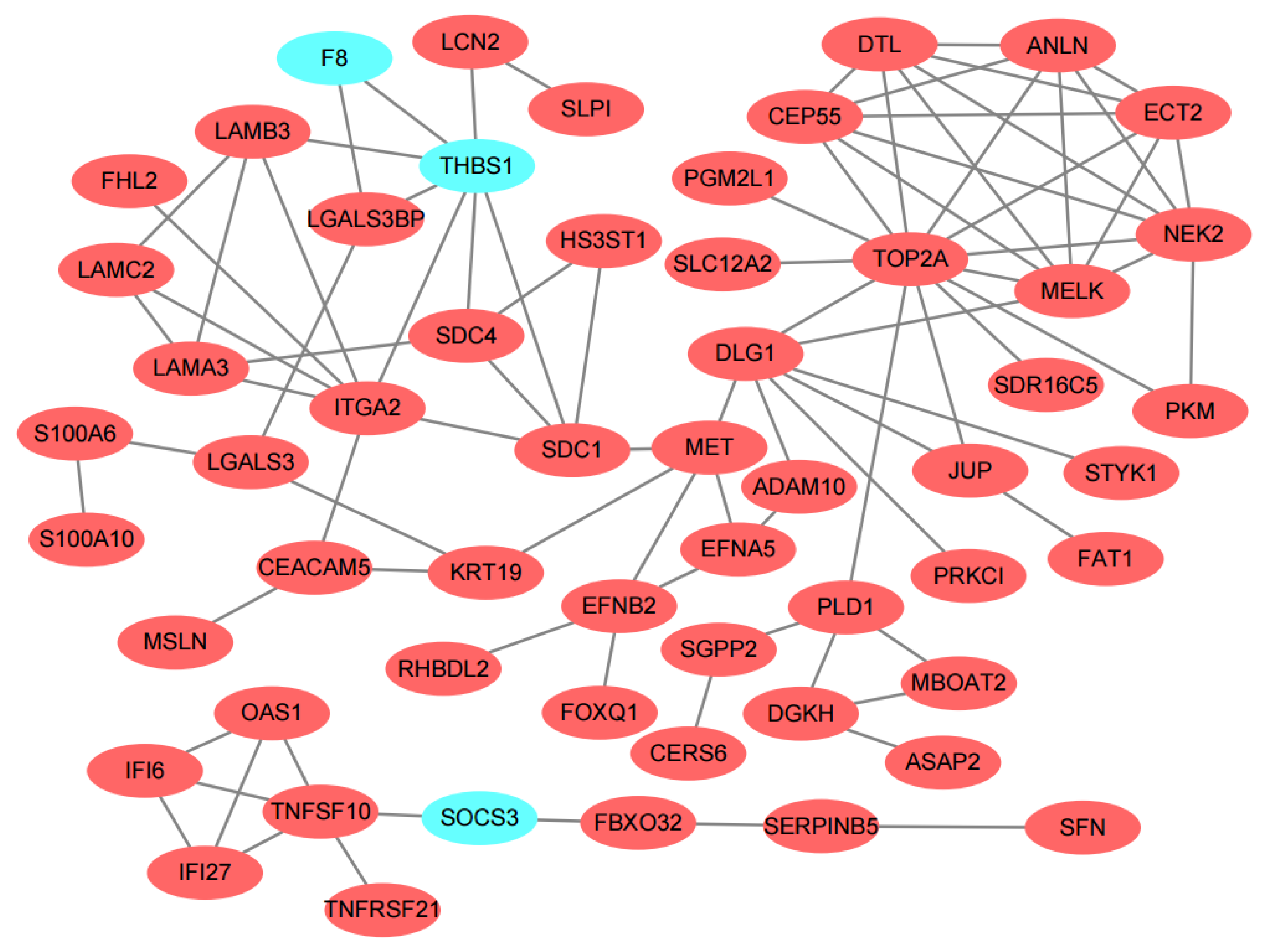
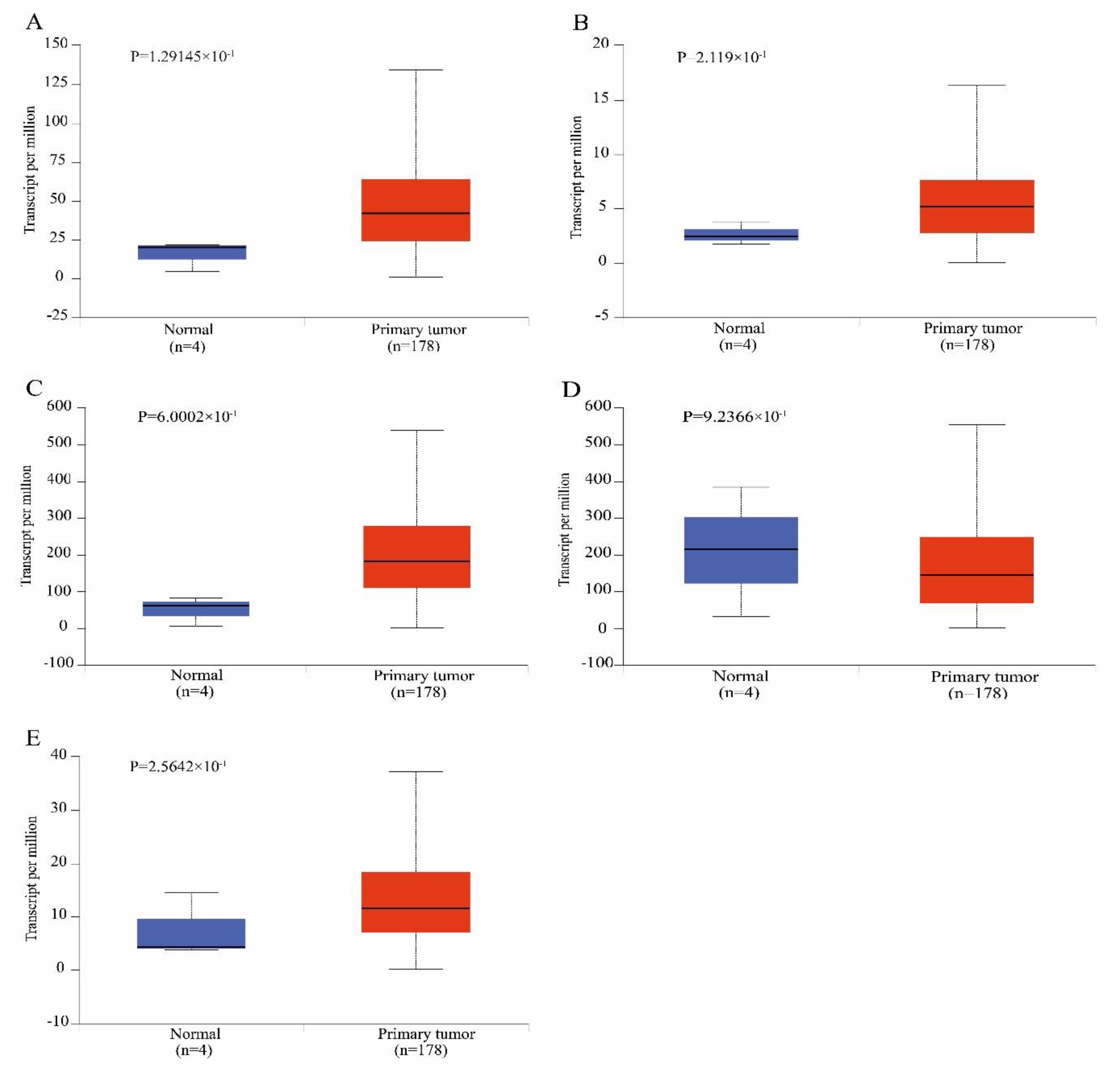
| Category | Term | Count | p-Value | Genes |
|---|---|---|---|---|
| BP | GO:0016477 ~ cell migration | 10 | 6.34 × 106 | JUP, STYK1, SDC1, TSPAN1, FAT1, PRKCI, THBS1, SDC4, CEACAM1, ADAM9 |
| BP | GO:0007155 ~ cell adhesion | 13 | 1.89 × 104 | EFNB2, FERMT1, ITGA2, JUP, LGALS3BP, LAMB3, LAMA3, FAT1, MSLN, LAMC2, THBS1, CEACAM1, ADAM9 |
| BP | GO:0098609 ~ cell-cell adhesion | 9 | 1.06 × 103 | PKM, FLRT3, S100P, RPL14, CAPG, ANLN, SFN, GPRC5A, CEACAM1 |
| BP | GO:0022617 ~ extracellular matrix disassembly | 5 | 2.63 × 103 | LAMB3, LAMA3, ADAM10, CAPG, LAMC2 |
| BP | GO:0031581 ~ hemidesmosome assembly | 3 | 3.56 × 103 | LAMB3, LAMA3, LAMC2 |
| CC | GO:0070062 ~ extracellular exosome | 46 | 1.35 × 107 | S100A6, TSPO, FXYD3, SLC44A1, TSPAN1, RPL14, MAL2, MARCKSL1, MLPH, SFN, SDC4, GPRC5A, ZG16B, PKM, LGALS3BP, FAT1, CEACAM5, NQO1, THBS1, MYOF, CEACAM1, DLG1, ADAM9, S100P, ADAM10, SLC12A2, LGALS3, TMC5, PRKCI, SLC6A14, S100A10, S100A14, ANXA3, LCN2, JUP, MTMR11, SDC1, KRT19, TNFSF10, LAMA3, C1ORF116, SERPINB5, CAPG, CTSE, SLPI, AOC1 |
| CC | GO:0005886 ~ plasma membrane | 52 | 3.79 × 105 | SLC44A1, TSPAN1, MARCKSL1, SDC4, PKM, MGLL, CEACAM1, DLG1, NET1, ADAM10, EFNB2, F8, PRKCI, JUP, STYK1, KRT19, SDC1, EFNA5, AOC1, MELK, TNFRSF21, FXYD3, CLDN18, ASAP2, GPRC5A, FAT1, MSLN, AHNAK2, MYOF, FLRT3, KLF5, OSBPL3, SLC12A2, LGALS3, KLB, MET, SLC6A14, ITGA2, DGKH, ITPR3, KCNK1, DOCK5, S100A14, ANXA3, CLDN23, GJB2, KCNN4, P2RX1, PON2, SYTL2, RHBDL2, IFI6 |
| CC | GO:0005923 ~ bicellular tight junction | 7 | 1.87 × 104 | CLDN18, EPPK1, PRKCI, ECT2, AOC1, CLDN23, DLG1 |
| CC | GO:0005925 ~ focal adhesion | 11 | 6.35 × 104 | FLRT3, JUP, SDC1, ADAM10, FAT1, EFNB2, FERMT1, FHL2, ITGA2, SDC4, ADAM9 |
| CC | GO:0005913 ~ cell-cell adherens junction | 9 | 2.70 × 103 | PKM, JUP, S100P, RPL14, CAPG, ANLN, SFN, GPRC5A, DLG1 |
| MF | GO:0043236 ~ laminin binding | 4 | 7.91 × 104 | LGALS3, ITGA2, THBS1, ADAM9 |
| MF | GO:0042803 ~ protein homodimerization activity | 15 | 9.42 × 104 | FLRT3, S100A6, ADAM10, S100A10, ECT2, JUP, LCN2, CTSE, MGLL, CEACAM5, AOC1, AGR2, TOP2A, CEACAM1, MTUS2 |
| MF | GO:0019903 ~ protein phosphatase binding | 5 | 1.18 × 103 | JUP, KCNN4, NEK2, MET, CEACAM1 |
| MF | GO:0098641 ~ cadherin binding involved in cell-cell adhesion | 9 | 1.35 × 103 | PKM, JUP, S100P, RPL14, CAPG, ANLN, SFN, GPRC5A, DLG1 |
| MF | GO:0005509 ~ calcium ion binding | 13 | 6.50 × 103 | S100A6, S100P, CAPN8, S100A10, MMP28, ITPR3, S100A14, ANXA3, FAT1, SYTL2, THBS1, AOC1, MELK |
| KEGG | hsa04512: ECM-receptor interaction | 7 | 5.33 × 103 | SDC1, LAMB3, LAMA3, ITGA2, LAMC2, THBS1, SDC4 |
| KEGG | hsa05205: Proteoglycans in cancer | 6 | 1.93 × 102 | SDC1, MET, ITGA2, THBS1, SDC4, ITPR3 |
| KEGG | hsa04510: Focal adhesion | 6 | 2.17 × 102 | LAMB3, LAMA3, MET, ITGA2, LAMC2, THBS1 |
| KEGG | hsa00512: Mucin type O-Glycan biosynthesis | 3 | 2.41 × 102 | GALNT5, C1GALT1, ST6GALNAC1 |
| KEGG | hsa04151: PI3K-Akt signaling pathway | 7 | 5.10 × 102 | LAMB3, LAMA3, MET, ITGA2, EFNA5, LAMC2, THBS1 |
| Topological Algorithm | Top 30 Genes Were Ranked by Score |
|---|---|
| Degree | TOP2A, THBS1, NEK2, DLG1, ITGA2, MELK, CEP55, ANLN, ECT2, DTL, SDC1, MET, LAMB3, LAMA3, SDC4, PLD1, EFNB2, DGKH, LAMC2, LGALS3, EFNA5, JUP, KRT19, CEACAM5, LGALS3BP, PKM, SGPP2, ADAM10, HS3ST1, MBOAT2 |
| Edge Percolated Component | TOP2A, MELK, NEK2, ANLN, DTL, CEP55, ECT2, DLG1, MET, SDC1, THBS1, ITGA2, JUP, SDC4, LAMB3, LAMA3, PKM, EFNA5, LAMC2, KRT19, ADAM10, EFNB2, LGALS3BP, HS3ST1, CEACAM5, PLD1, F8, LGALS3, SLC12A2, PRKCI |
| Maximum Neighborhood Component | TOP2A, NEK2, MELK, CEP55, ANLN, ECT2, DTL, ITGA2, LAMB3, THBS1, SDC1, LAMC2, LAMA3, SDC4, DLG1, PKM, DGKH, EFNA5, MET, JUP, HS3ST1, PLD1, MBOAT2, F8, EFNB2, LGALS3BP, PGM2L1, SDR16C5, PRKCI, FHL2 |
| Density of Maximum Neighborhood Component | CEP55, ANLN, ECT2, DTL, NEK2, MELK, LAMC2, LAMA3, TOP2A, LAMB3, ITGA2, SDC4, DLG1, PKM, DGKH, EFNA5, MET, JUP, HS3ST1, PLD1, MBOAT2, F8, EFNB2, LGALS3BP, THBS1, SDC1, PGM2L1, SDR16C5, PRKCI, FHL2 |
| Clustering Coefficient | PKM, CEP55, ANLN, LAMC2, ECT2, DTL, HS3ST1, MBOAT2, F8, NEK2, MELK, LAMB3, LAMA3, DGKH, EFNA5, SDC4, JUP, LGALS3BP, SDC1, ITGA2, TOP2A, THBS1, PLD1, EFNB2, MET, DLG1, PGM2L1, SDR16C5, PRKCI, FHL2 |
| Maximal Clique Centrality | TOP2A, NEK2, MELK, CEP55, ANLN, ECT2, DTL, ITGA2, THBS1, DLG1, TNFSF10, LAMB3, LAMA3, SDC1, OAS1, IFI6, IFI27, LAMC2, MET, SDC4, PLD1, EFNB2, DGKH, LGALS3, JUP, KRT19, CEACAM5, EFNA5, LGALS3BP, GALNT5 |
| Bottleneck | MET, DLG1, TOP2A, SDC1, KRT19, PLD1, MELK, THBS1, LGALS3, ITGA2, EFNB2, DGKH, SGPP2, SDC4, JUP, S100A6, CEACAM5, LCN2, PKM, CEP55, ANLN, PGM2L1, SDR16C5, PRKCI, LAMB3, LAMC2, ECT2, LAMA3, DTL, NEK2 |
| EcCentricity | MET, DLG1, PRKCI, SDC1, TOP2A, EFNA5, ADAM10, JUP, STYK1, KRT19, EFNB2, MELK, PKM, CEP55, ANLN, PGM2L1, SDR16C5, ECT2, THBS1, DTL, NEK2, LGALS3, SLC12A2, RHBDL2, SDC4, FAT1, HS3ST1, PLD1, FOXQ1, ITGA2 |
| Closeness | TOP2A, DLG1, MET, MELK, SDC1, THBS1, ITGA2, NEK2, JUP, CEP55, ANLN, ECT2, DTL, PLD1, KRT19, EFNB2, SDC4, EFNA5, CEACAM5, ADAM10, PKM, LAMB3, LGALS3, PGM2L1, SDR16C5, SLC12A2, PRKCI, STYK1, LGALS3BP, LAMA3 |
| Radiality | MET, DLG1, TOP2A, SDC1, MELK, KRT19, JUP, EFNB2, EFNA5, THBS1, ADAM10, ITGA2, PRKCI, STYK1, PLD1, CEACAM5, SDC4, NEK2, CEP55, ANLN, ECT2, DTL, LGALS3, PKM, PGM2L1, SDR16C5, SLC12A2, HS3ST1, FAT1, LAMB3 |
| Stress | MET, DLG1, TOP2A, SDC1, THBS1, PLD1, KRT19, ITGA2, LGALS3, MELK, EFNB2, CEACAM5, S100A6, LCN2, SDC4, LGALS3BP, JUP, DGKH, SGPP2, ADAM10, EFNA5, LAMB3, LAMA3, NEK2, PKM, CEP55, ANLN, PGM2L1, SDR16C5, PRKCI |
| Betweenness | MET, DLG1, TOP2A, SDC1, PLD1, THBS1, KRT19, ITGA2, LGALS3, EFNB2, MELK, CEACAM5, DGKH, SGPP2, JUP, S100A6, LCN2, LGALS3BP, SDC4, EFNA5, ADAM10, LAMB3, LAMA3, NEK2, PKM, CEP55, ANLN, PGM2L1, SDR16C5, PRKCI |
| Common genes of 12 topological algorithms | MET, MELK, SDC1, THBS1, TOP2A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, W.; Li, N.; Liao, F. Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis. Genes 2019, 10, 612. https://doi.org/10.3390/genes10080612
Lu W, Li N, Liao F. Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis. Genes. 2019; 10(8):612. https://doi.org/10.3390/genes10080612
Chicago/Turabian StyleLu, Wenzong, Ning Li, and Fuyuan Liao. 2019. "Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis" Genes 10, no. 8: 612. https://doi.org/10.3390/genes10080612
APA StyleLu, W., Li, N., & Liao, F. (2019). Identification of Key Genes and Pathways in Pancreatic Cancer Gene Expression Profile by Integrative Analysis. Genes, 10(8), 612. https://doi.org/10.3390/genes10080612





