Abstract
Viruses, the most abundant biological entities on the planet, are capable of infecting organisms from all three branches of life, although the majority infect bacteria where the greatest degree of cellular diversity lies. However, the characterization and assessment of viral diversity in natural environments is only beginning to become a possibility. Through the development of a novel technique for the harvest of viral DNA and the application of 454 pyrosequencing, a snapshot of the diversity of the DNA viruses harvested from a standing pond on a cattle farm has been obtained. A high abundance of viral genotypes (785) were present within the virome. The absolute numbers of lambdoid and Shiga toxin (Stx) encoding phages detected suggested that the depth of sequencing had enabled recovery of only ca. 8% of the total virus population, numbers that agreed within less than an order of magnitude with predictions made by rarefaction analysis. The most abundant viral genotypes in the pond were bacteriophages (93.7%). The predominant viral genotypes infecting higher life forms found in association with the farm were pathogens that cause disease in cattle and humans, e.g. members of the Herpesviridae. The techniques and analysis described here provide a fresh approach to the monitoring of viral populations in the aquatic environment, with the potential to become integral to the development of risk analysis tools for monitoring the dissemination of viral agents of animal, plant and human diseases.
1. Introduction
Viruses, most of which infect microorganisms, are the most abundant biological entities on the planet and are capable of infecting organisms from all three branches of life [1]. Because bacteria represent the greatest portion of microbial life and diversity of earth, bacteriophages form the largest division of the viruses and have the potential to alter the genetic composition, and ultimately the functioning of microbial communities [2]. Viruses are important, possibly the most important, drivers of microbial evolution; resistance to infection can be due to the alteration of surface epitopes or the development of alternative intracellular anti-viral mechanisms [3]. In their role as agents of genetic exchange, they not only alter the genetic makeup of host cells, but also play a pivotal role in the movement of genetic material between ecosystems [4].
However the characterization and assessment of viral diversity in the natural environment is hindered by the limitations of current isolation and recovery methods, and the specificity requirements of individual propagation and identification techniques. The size, nature, rapid evolution and genomic flexibility of viruses make them notoriously difficult to study [5,6]. Propagation of viruses is also likely to be untenable in many cases because their hosts cannot be isolated or cultured in the laboratory [7]. For example, the greatest diversity is found amongst the bacteriophages yet it has been predicted that less than 0.0002% of the global bacteriophage pangenome has been sampled thus far [8]. Studies of viral ecology are further constrained by the absence of single genetic elements that are shared by all viruses [9,10]. Although genome comparisons have shown that conserved genes are present within certain groups such as the lambdoid phages [11], these genes will not enable the identification of novel viral groups [12], and viral diversity cannot be determined by using approaches analogous to the cellular RNA profiling approach that has been so successful in revealing the untold diversity within Bacteria and Archaea [1].
Metagenomics, or community genomics, is an approach aimed at analyzing the genomic content of microbial communities within a particular niche [13]. Early metagenomic projects, for example those of Beja and coworkers [13,14,15], were plagued by potential biases imparted mainly by DNA extraction and cloning methods [16]. The traditional approach to metagenome sequencing involves cloning DNA into BACs, fosmids or cosmids and the use of dideoxy chain-termination sequencing [17]. This approach is not well suited to viral metagenomics because some viruses possess an RNA-based genome, DNA viruses often contain extremely small genomes [18] with modified bases making cloning difficult [19], and they can carry genes or sequences that are toxic to bacterial cloning hosts [20]. An alternative approach is the use of next generation sequencing techniques, which provide unbiased, high volumes of short DNA sequences without the requirement for cloning [21]. Pyrosequencing technology has already been used to analyze viral communities in a range of natural environments, including but not exclusive to the examination of the structural and functional changes in coral microbiota [22], the diversity, composition and adaptations of viral assemblages in hot springs [23], Antarctic lake viral communities [24], marine viral communities [5,25] and fresh water communities [26,27] all of which revealed unexpected genetic richness. Viral metagenomic studies have shown that 1 kg of marine sediment contains over a million different viral genotypes [28,29] and at least 1000 unique novel viruses can be identified in the human gut, though the vast majority (>70%) are completely uncharacterized [30]. Many microbial metagenomic projects based on pyrosequencing are limited by complex assembly issues associated with small sequence reads sampled from a mixed community of large genomes; these limitations are less significant in viral metagenomics, due to the relatively small size range of viral genomes [31]. In addition, the hardiness of the viral capsid can be exploited to concentrate and purify viruses away from contaminating microorganisms, prior to sequencing [32].
This study reports a snapshot examination of the diversity of the DNA viruses present in a standing pond on a cattle farm in Cheshire, UK, selected for study as a result of previous work on Shiga toxin (Stx) encoding bacteriophages in relation to the bovine reservoir of E. coli strains pathogenic to man [11,33,34,35]. This was achieved through the development of a novel technique for harvesting uncontaminated viral DNA [11], coupled with the application of 454 pyrosequencing to the viral metagenome so produced. Freshwater viral diversity and community structure has barely been studied, yet it is an important topic that impacts on human and animal disease with implications for environmental monitoring, agricultural practices and public health policy.
2. Results and Discussion
2.1. Preparation of DNA
A protocol was developed for the extraction of viral DNA from water samples, with the ultimate goal of minimising the presence of cellular and free DNA. In order to maximize the recovery of viral DNA, removal of bacterial and small eukaryote cells by membrane filtration was avoided as viruses can become immobilized on and trapped within filter membranes [5]. The efficacy of the DNA preparation method was evaluated both pre- and post-sequencing. Pre-sequencing, the samples were subjected to 35 cycles of DNA amplification by PCR using universal oligonucleotide primer sets for the 16S and 18S rRNA genes (Table 1). The inability to amplify bacterial or eukaryotic rRNA genes from the environmental viral DNA sample, as determined by ethidium bromide staining of DNA bands separated by agarose gel electrophoresis with all appropriate positive and negative controls (data not shown), demonstrated that cellular DNA contamination was at a level that could not be detected by PCR amplification and was therefore potentially suitable for use as the template for 454 pyrosequencing.

Table 1.
Oligonucleotide primers used in this study.
| Gene target | Primername | Sequence (5’ -> 3’) | Annealing temperature (°C) | Amplicon size (bp) | Reference |
|---|---|---|---|---|---|
| 16S rRNA | pA | AGAGTTTGATCCTGGCTCAG | 55 | 1534 | [36] |
| pH | aaggaggtgatccsgccgca | ||||
| 18S rRNA | NS1-Euk | ccagtagtcatatgcttgtc | 50 | 1600 | [37] |
| Univ 1390 | gacgggcggtgtgtacaa |
2.2. Metagenomic library output
The 454 pyrosequencing returned 69,162 reads of DNA sequence, which contained 13,669,562 bp of DNA with a fragment size range of 32-404 bp using ¼ of a sequencing picotitre plate (85% of the sequences were >100 bp and > 51% of the sequences were between 200 – 250 bp, Supplementary Table 1). The DNA sequence reads were used directly for downstream analysis, and not assembled as contiguous sequences. The mosaic nature of viruses, particularly bacteriophage genomes [38], is such that subjecting the sequence data to conventional assembly of contigs would be inappropriate. Consequently, pre-packaged bioinformatic tools and computational approaches were applied to unravel the population structure and function of the viral metagenome sample. The methods used here for the prediction of identities from the metagenomic sequence data were largely automated, involving the use of MEGAN [39] and MG-RAST[40] coupled with BLAST [41] for the comparison of this viral metagenome against the non-redundant viral genome [42] and SEED databases (which uses a non-redundant database) [43].
2.3. Post-pyrosequencing analyses
Using MG-RAST, sequences were screened for ribosomal RNA genes using BLASTn against the rRNA gene databases, which were extracted from Greengenes [44] the Ribosomal Database Project (RDP) [45] and the European Ribosomal Database Project (ERDP) [46] using cutoff parameters requiring similarity >5 bp in length and an expect value of less than 1 x 10-5[40]. Of the 69,162 sequences generated, 18,931 (27.37%) were classified (the SEED database assigns a metabolic potential to these sequences) and 50,231 (72.6%) were unclassified (the SEED database fails to assign a potential function to these sequences. Two of the 69,162 sequences possessed homology (Table 2) to SSU rRNA . The first SSU rRNA sequence (254 bp in length) was 97% identical to Dechloromonas, a member of the betaproteobacteria and further BLAST analysis of these sequences only identified organisms isolated from freshwater environments (e.g. accession numbers DQ450182 and DQ22839). The second SSU rRNA sequence (243 bp in length) possessed 95% identity to an unclassified member of the Bacteriodetes. It cannot be ruled out at this point that bacteriophages do not acquire portions or intact copies of SSU rRNA molecules [47], but additionally the low numbers of identified SSU rRNA sequences were below the level of detection by PCR amplification and indicate that the viral metagenomic library was at least heavily enriched for non-cellular DNA.
2.4. Functional analyses of the virome
Gene transfer undoubtedly occurs in the natural environment, however the scale of the process and the implications for the evolution of the virus and its host organism is poorly understood [25]. Horizontal gene transfer of biochemically important genes from host to virus, virus to host and so on, will ultimately shape the microbial biosphere.

Table 2.
MG-RAST sequence classification.
| SSU rRNA homology (%) RDP/ERDP/GREENGENES | Protein Based (%)(SEED database*) | |
|---|---|---|
| Archaea | 0 | 1.99 |
| Bacteria | 0.02 | 41.71 |
| Eukaryota | 0 | 3.10 |
| Virus | 0 | 51.25 |
| Other | 0 | 0.02 |
*Used to determine the metabolic profile of the metagenome.
Metagenomics allows the analysis of the complete genetic information contained within the viral community, and in this way the relative abundances of all genes can be determined and used to generate a description of the functional potential of each viral community within a given sample [5,29,30,48,49,50,51]. The metabolic potential of the virome was determined using the SEED database (Table 2, Supplementary Table 2) [43], which comprises all known protein sequences spanning all biological taxonomies [43]. The SEED arranges metabolic pathways into a hierarchical structure in which all of the genes required for a specific task are arranged into subsystems. At the highest level of organization, the subsystems include both catabolic and anabolic functions, for example DNA metabolism, and at the lowest level the subsystems are specific pathways, for example the synthesis pathway for thymidine [26]. It uses a BLASTX algorithm and assigns genes to subsystems, each of which is composed of a group of functionally related proteins where an E-value of <0.001 is considered to be significant [43] so that the genes implicated in specific functional roles are very likely to exist even though they have not yet been identified due to limited amounts of sequence homology, a consequence of short pyrosequencing reads. Here MG-RAST was able to assign 9% of sequences to a subsystem and the relative abundances of sequences assigned to each major subsystem were determined (Figure 1). The remaining sequences were identified as hypothetical proteins. The greatest proportion of sequences that could be assigned to a metabolic function were virulence associated (15%). In light of the fact that viruses infect their host cells it was expected that a variety of virulence associated genes would be identified in any viral metagenome. Virulence genes are important both to viral pathogenesis and to host cells acquiring new pathogenic potential following viral infection. However, the rest of the functional families are slightly more surprising on first inspection. The second most abundant function grouping contains genes associated with carbohydrate and protein metabolism (both 10%). Respiration and phosphate metabolism associated genes made up 5 and 3%, respectively, with motility and chemotaxis associated genes representing 2% of the metabolic profiles. Bacteriophages are known modulators of bacterial virulence [52], but they can also carry genes involved in bacterial phosphate metabolism and photosystems [53], both of which were identified in the functional gene analysis of this virome (Figure 1). Analysis of the viral population structure (Figure 2) demonstrates that phages known to carry these genes are also present. Roseobacter phage SI01 [NC_002519], has previously been shown to encode four predicted proteins involved in phosphate metabolism (RP Pho, RP ribonucleotide reductase, RP Thy1 and RP endodeoxyribonuclease 1), suggesting that phosphate recycling is important to the roseophage SI01 life cycle. The published genome sequence of S-PM2 [NC_006820], a bacteriophage that infects marine Synechococcous strains, encodes the D1 and D2 proteins of PS11, a large protein pigment complex. The presence of this virus in the host may ensure that photo-inhibition does not occur, enabling photosynthesis to proceed during viral infection, providing the energy needed for virus replication [53].
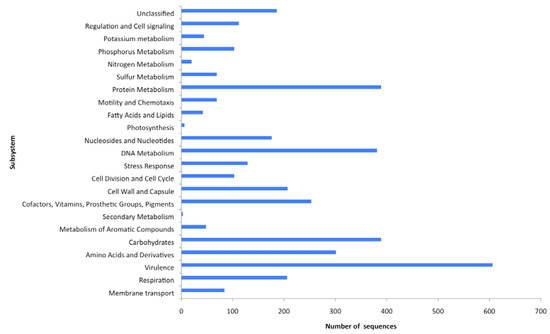
Figure 1.
Functional potential of sequences from the freshwater virome. This was calculated using a BLASTX algorithm against the SEED database. An E-value of <0.001 was considered to be specific.
The presence of motility and chemotaxis genes (17%) (Figure 1) was unexpected as phages and viruses are non-motile, however a functional study of nine viromes by Dinsdale et al., [26] reported a total of 130 SEED annotated motility and chemotaxis proteins present in these metagenomes. The role that they play remains unclear, however, they all carry out functions associated with type 1V pilus and motility, and may be acting as symbiotic relationship facilitators [54]. For example, the pathogenicity of Vibrio cholerae is dependent upon the production of a type 1V pilus, and cholera toxin, and the genes for both are encoded on remnant and infective phages, respectively [55]. In general, the assignment of putative functions to a number of the sequences in this virome suggests that there is little restriction on the types of genes carried by a viral community, and their potential for influencing a wide range of biological processes.
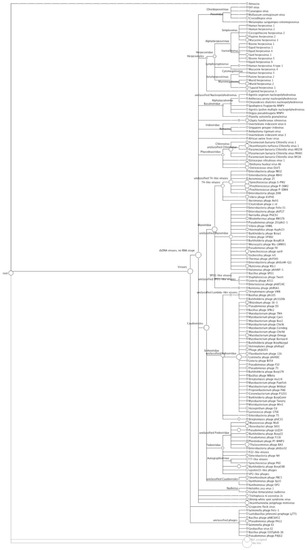
Figure 2.
Phylogenetic diversity of the freshwater metagenome as calculated by MEGAN. Each circle represents a taxon in the NCBI taxonomy, and the size of the circle is scaled logarithmically to represent the number of reads assigned directly to the taxon. This Figure is also available as supplementary file.
2.5. Population Diversity and Structure
In addition to directly causing disease in plants and animals, viruses are important microbial predators that influence global biogeochemical cycling and drive microbial evolution [25]. Much of our knowledge about the roles and diversity of viruses in the natural environment is informed by studies on marine microbial communities. For example 1 kg of marine sediment may contain over a million different viral types, and 200 L of seawater ca. 5,000 viral types [28,29], with the vast majority (> 75 %) of genetic material remaining completely uncharacterized. Other environments, including those more directly impacted by human activity, possess greater viral diversity and novelty [56].
The original BLASTn output, which was generated from the comparison of the dataset against the non-redundant viral genome database, was used to perform a rank abundance analysis of family hits (Figure 3). Rank abundance analysis provides a measure of the total number of organisms (abundance = 42,415 viral genotypes), the number of species within the sample (richness = 785 viral genotypes), and a measure of the relative proportional abundances of the species (evenness) [56]. The T4 like phages dominated the metagenome, as did the cyanophages with 411 and 408 representatives in the virome, respectively. Using regression analyses, and assuming the maximal Y value is calculated at X = 1 and the maximal X value is calculated when Y = 1, the formula y = 5,079.7 (X-1.197) was used to determine from the abundance plot (Figure 3), an estimate of the number of sequences that would have been obtained if 100% of the population had been sampled. This analysis implied that the true diversity of species present was grossly underestimated, as at least 820,000 sequences would be required to ensure recovery of a single sequence fragment from each member of the viral community. A qPCR-based methodology was previously used to quantify the number of lambdoid bacteriophages in the same viral DNA preparation [11] used to generate the virome, thus enabling an independent assessment of the depth of coverage achieved by the 454 pyrosequencing run. BLAST comparisons of the virome dataset revealed 20 lambda- like sequences and 1 Stx phage, and by comparing this with the absolute numbers of lambdoid and Stx phages determined by qPCR [11], it is estimated that the depth of the virome sequencing has resulted in ca. 5% of the viral population in the 2 L of pond water has been sampled. This compares favorably with the theoretical estimate of coverage obtained by the rank abundance analysis (Figure 3) of ca. 8%.
All sequences (69,162), were compared to the non-redundant viral genome databases, using a BLASTn algorithm, and in accordance with MEGAN recommendations [39], relaxed alignment parameters were used for the BLAST search. Of the 69,162 sequences generated from the 454 output, 42,715 sequences provided hits from the BLASTn search. These results were then loaded into MEGAN and the lowest common ancestor (LCA) algorithm assigned 20,287 sequences to taxa with 22,428 remaining unassigned. The LCA algorithm has an inbuilt threshold that is sufficiently stringent to reduce the chances of generating false identities, hence the assignment of the sequences to taxa is both reliable and conservative. Of the assigned sequence reads, 19,010 (93.7%) were identified as dsDNA viruses (Figure 4), of which 10,525 (55.3%) belonged to the Caudovirales; 3,459 (18.2%) Herpesvirales; 2,034 (10.7%) Phycodnaviridae; 1,197 (6.3%) Baculoviridae; 1,026 (5.4%) Iridoviridae and 1,028 (5.4%) Poxviridae.
The primary viral families in this virome associated with infection of metazoans where the Herpesviridae, a large family of DNA viruses that cause disease in animals and humans worldwide [57]. In this sample of pond water (accessed by cattle for drinking water), this viral family was represented in 18.2% of the classified virome. All four previously described bovine herpes viruses (BHV-1; BHV-2, BHV-4 and BHV-5) were identified (Figure 2) and are known to cause disease in cattle, worldwide [58]. Of these, bovine herpesviruses 1 (BHV-1 [NC_001847]) and 5 (BHV-5 [NC_005261]) are two closely related viruses that infect cattle [59] and cause multiple diseases including infectious bovine rhinotracheitis, abortion, infectious vulvovaginitis and systemic infection in bovine neonates [60]. When animals survive a BHV infection, a life-long latent infection is established in nervous sensory ganglia, providing both a viral reservoir and survival strategy [60]. Viral reactivation of BHV can occur at any time, and the virus can be shed and transmitted to both immune and non-immune hosts [61]. The Phycodnaviridae, are a rapidly expanding collection of large icosahedral dsDNA viruses which infect algae [62] and here comprised 10.7% of the classified virome, mostly represented by different species of the Chloroviruses, ubiquitous in nature and isolated from freshwater throughout the world [62]. 5.4% of the classified virome population was comprised of members of the Iridoviridae and all were members of the group Ranavirus, pathogens which have affected amphibian populations worldwide [63]. Also present in the population are members of the Poxviridae (5.4%), a group of established pathogens that includes zoonotic forms that infect animals and humans [64]. Baculovirdae, a family of large rod shaped viruses, comprised 6.3% of the classified virome; these viruses are extremely species specific, with larval forms of Lepidoptera (moths) the most common hosts [65]. These data suggest that the water in the cattle pond can serve as a reservoir for viruses that directly affect metozoans.
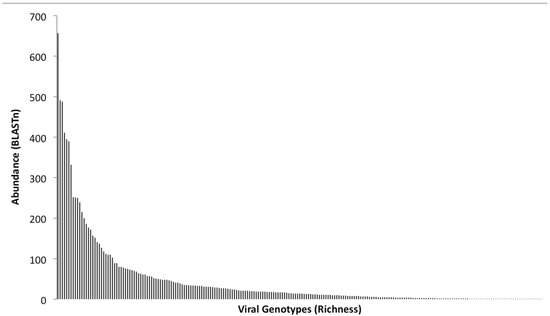
Figure 3.
Rank abundance analysis of BLASTn output. This plot was generated from identifications made by comparison of the 454-sequence dataset against the non-redundant viral genome database. Each bar along the X-axis represents a particular viral genotype and the Y-axis indicates the number of representative sequences identified.
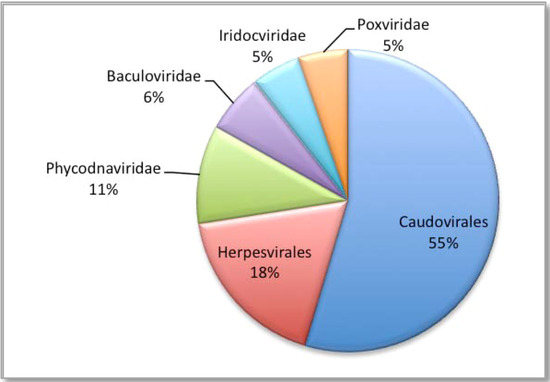
Figure 4.
Percentage of sequences assigned to the dsDNA viruses.
DNA sequences identified with the tailed bacteriophages (Caudovirales), predominate (93.17%) in the cattle pond virome (Figure 2), and comprise the Myoviridae, Siphoviridae and Podoviridae-like viruses. The Myoviridae were the most represented (53%), predominantly T4-like viruses (58% of the Myoviridae sequences) with exemplars such as Synechococcus phage S-PM2 and Prochlorococcus phages P-SSML. Additional members of the Myoviridae whose presence was indicated, included the Spo1-like viruses that infect Bacillus spp. The Siphoviridae, characterized as having long non-contractile tails, made up 30% of the identified Caudovirales, dominated by the mycobacteria-like phages (72%). Members of the Siphoviridae which infect mycobacterial hosts have proven useful for the diagnosis of mycobacterial infections, including bovine tuberculosis, a zoonosis, which can spread to humans through inhalation of infectious droplet nuclei and by ingestion of raw milk [66]. To date, 50 mycobacteriophages have been fully sequenced, and sequences matching 11 of these are represented in the virome (Figure 2) [67,68]. Analysis of the sequenced genomes suggests that these viruses may also play a more important role in human disease than originally thought. CJW1 [NC_004681] and phage omega [NC_004688] (Figure 2) encode close homologs to the leprosy and tuberculosis antigen Lsr2 suggesting a role for phage in Mycobacterial virulence, and phage Bxz2 [NC_004682] (Figure 2) encodes a homolog with ~35% identify to the human Ro protein, which has major involvement in the autoimmune response in Lupus and Sjögrens syndrome [69]. Whether these genes are involved in virulence is unknown at present, but there is precedence for phage encoded genes being important in diseases, for example, the ability to produce Shiga toxin (Stx), ultimately the most important virulence factor of enterohaemorrhagic E. coli, is conferred to E. coli following infection with Stx-bacteriophage(s), lambdoid phages that carry the Shiga toxin operon [52]. These are a heterogeneous group of phages with stx gene carriage as their common property [11,52], but there is evidence that short-tailed phages (Podoviridae) are the most epidemiologically significant [35,70]. The virome contained sequences originating from lambda-like phages (20) and a single stx gene sequence, and as discussed above, estimates of the population of lambdoid and Stx phages obtained by qPCR analysis in this pond water sample [11] were used to determine the depth of sampling achieved in the 454 pyrosequenced virome. The Podoviridae sequences in the virome were dominated by the Roseobacter S101-like, and Thalassomonas BA3-like phages [NC_009990] (Figure 2), perhaps indicating that even in water directly contaminated by cattle faeces (thereby contaminated with bovine-associated gut bacteria and bacteriophages), the bacteriophages infecting indigenous freshwater bacterial species predominate the viral community. Nevertheless, these data indicate that the cattle pond can serve as a reservoir for viruses with an indirect pathogenic phenotype for higher life-forms, e.g. temperate phages that directly alter the pathogenic phenotype of their bacterial hosts. This mechanism, coupled with the ability the cattle farm pond to harbor viruses infecting metazoan life, demonstrates the potential of water sources to serve as reservoirs for viruses, and sites for dissemination to new hosts, directly (via viral pathogens) or indirectly (via temperate phages).
3. Experimental Section
3.1. Viral DNA Extraction
Two litre water samples were collected from a farm in Cheshire, U.K. in August 2008 [9]. NaCl2 (0.5M) was added to each sample and allowed to dissolve to facilitate the dissociation of viral particles from cellular and particulate surfaces [11,71]. Cell and environmental debris were removed by slow speed centrifugation (6000 x g for 5 min). DNase (Ambion) and RNase (Sigma) were added to a final concentration of 5 µg mL-1, and the samples were incubated at 37°C for 30 min. Polyethylene glycol (PEG) with an average molecular weight of 8000 (Sigma) was gradually added to a final concentration of 10% (w/v), and the sample was incubated for 18 h at 4 °C. The sample was centrifuged (10,000 x g for 10 min at 4°C), the supernatants discarded and the pellets suspended in 7.5 mL TBT buffer (100 mM Tris-HCL, pH 7.0; 100 mM NaCl2; 100 mM MgCl2). An equal volume of chloroform was added, the samples were subjected to centrifugation (4000 x g, 20 min), and the top phase was harvested [11]. A second DNase (Ambion) and RNase (Sigma) step was carried out as described above. Viruses were precipitated by incubation with 33% (w/v) PEG on ice for 60 min and harvested by centrifugation (10,000 x g; 10 min at 4°C). A third DNase and RNase digestion was performed, followed by extraction using an equal volume of equilibrated phenol:chloroform:isoamyl alcohol (25:24:1, pH8.0). The mixture was centrifuged (13,000 x g for 5 min), and the aqueous phase harvested and subjected to a further three rounds of phenol:chloroform:isoamyl alcohol extraction. Viral DNA was precipitated by the addition of equal volumes isopropanol, 10% (v/v) 3 M sodium acetate (pH 5.2.) and glycogen (20 mg mL-1) followed by incubation at -20°C for 18 h. The sample was centrifuged (10,000 x g for 30 min at 4°C). The resultant viral DNA pellet was washed with ice-cold 70% ethanol, air-dried and suspended in 15 µL of sterile dH20.
3.2. End-Point polymerase chain reaction
To demonstrate the removal of contaminating prokaryotic or eukaryotic genomic DNA from the viral DNA sample, PCR amplification was performed using universal primer sets specific to bacterial 16S and eukaryotic 18S rRNA genes (Table 1) and Phusion high fidelity DNA polymerase (Finzymes). Bacillus subtilis genomic DNA, and DNA extracted from bovine rumen fluid were used as controls for the 16S and 18S amplifications respectively. Each reaction (50 µL) contained forward and reverse primers (0.5 µM, each), dNTP’s (200 µM), HF buffer (Finzymes), and 0.02 U µL-1 Phusion high fidelity polymerase. Cycling conditions were 98°C for 30 s, 35 cycles (98°C for 5 s, 1 min at the specific annealing temp for each primer (Table 1), 72°C for 1.5 min) and a final extension step of 72°C for 10 min. Amplification products were detected on a 1% TAE agarose gel (40 mM Tris base, 20 mM glacial acetic acid, 1 mM EDTA, 1.5% (w/v) agarose) run for 1 h at 100V.
3.3. pyrosequencing
Sequencing was performed using the Roche 454 GS-FLX instrument at the NERC funded Advanced Genomics Facility at the University of Liverpool, according to the method for pyrosequencing by Marguilies et al. [72].
3.4. Bioinformatical analyses
A BLAST comparison was performed using the BLASTn algorithm [41], against the non-redundant viral genome database [42]. The output of this analysis was used for two different analyses. First, the rank abundance data was generated using the top 15 hits (e< 0.1) of each sequence fragment to identify viral genotypes. Secondly, the output file was uploaded into a windows version of the MEGAN program [39], which was used to estimate and interactively explore the taxonomical distribution of the dataset. The program uses a simple algorithm that assigns each read to the lowest common ancestor (LCA) of the set of taxa with matches in the comparison. The LCA parameters used were: min support, 5; min score, 35.0; top percent, 10.0; win score, 0.0.
3.5. MG-RAST
All freshwater DNA 454 sequences were uploaded in a FASTA format to the MG-RAST server at the SEED [43]. The raw data presented here can be found on the MG-RAST (Public: MGPhage (4442702.3) from project PhageMG).
4. Conclusions
This paper provides a snapshot examination of the viral diversity present in a single sample of DNA harvested from a freshwater pond on a cattle farm in Cheshire, UK. A broad range of viruses were found that are directly and indirectly associated with diseases affecting plants, animals and man, including Stx phages, Herpesviridae, and Poxviridae. These data support the concept that the aquatic environment plays a more important role as a reservoir of viral pathogens than may have been previously been appreciated. In addition to directly causing disease, viruses are also important predators, controlling microbial population size and structure, influencing global biogeochemical cycling and driving microbial evolution through gene transfer. Analysis of functional potential of genetic information within the virome identified homology to genes associated with virulence, photosystems, metabolism and even motility. Freshwater viral diversity and community structure has barely been studied and even though this virome possessed a high diversity of viral genotypes (785), rank-abundance analyses indicated that the depth of sequencing employed enabled only ca. 8% of the virus population in the 2 L freshwater sample to be examined. Less than 62% of the 69,162 sequences (42,715 sequence reads) were assigned a BLASTn hit using the viral databases. Less than 30% of 69,162 reads could be assigned to a function/genotype via SEED (18,931 sequence reads) and MEGAN (20,287 sequence reads). Techniques like those reported here, represent a fundamental breakthrough in our ability to detect and monitor viral populations. Further work on determining abundances linked with disease monitoring and viral dynamics may be integral to the development of risk analysis tools. Delivery of this information to the end users, such as government advisory bodies for environment, human health, water safety, control of recreational water and agriculture will enable us to pre-emptively deal with disease affecting and impacting upon animals, plants and humankind.
Acknowledgments
This research was funded by the Natural Environment Research Council (UK), and the Veterinary Laboratory Agency (UK).
Supplementary Files
References and Notes
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Microbiol. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Weinbauer, M.G.; Rassoulzadegan, F. Are viruses driving microbial diversification and diversity? Environ. Microbiol. 2004, 6, 1–11. [Google Scholar] [CrossRef]
- Thingstad, E.V. Elements of a theory for the mechanisms controlling abundance, diversity and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol. Oceanogr. 2000, 45, 1320–1328. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, 368. [Google Scholar] [CrossRef]
- Brussow, H.; Hendrix, R.W. Phage genomics: small is beautiful. Cell 2002, 108, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.-W.; DuBow, M.S. Viruses of prokaryotes, Volume 1: General properties of bacteriophages; 1987; CRC Press: Boca Raton, FL, USA. [Google Scholar]
- Rohwer, F. Global phage diversity. Cell 2003, 113, 141. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Edwards, R. The Phage Proteomic Tree: a genome-based taxonomy for phage. J. Bacteriol. 2002, 184, 4529–4535. [Google Scholar] [CrossRef] [PubMed]
- Thurber, R.V. Current insights into phage biodiversity and biogeography. Curr. Opin. Microbiol. 2009, 12, 582–587. [Google Scholar] [CrossRef] [PubMed]
- Rooks, D.J.; Yan, Y.; McDonald, J.E.; Woodward, M.J.; McCarthy, A.J.; Allison, H.E. Development and validation of a qPCR-based method for quantifying Shiga toxin-encoding and other lambdoid bacteriophages. Environ. Microbiol. 2010, 12, 1194–1204. [Google Scholar] [CrossRef] [PubMed]
- Short, C.M.; Suttle, C.A. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 2005, 71, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Hall, N. Advanced sequencing technologies and their wider impact in microbiology. J. Exp. Biol. 2007, 210, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Beja, O.; Spudich, E.N.; Spudich, J.L.; Leclerc, M.; DeLong, E.F. Proteorhodopsin phototrophy in the ocean. Nature 2001, 411, 786–789. [Google Scholar] [CrossRef] [PubMed]
- Beja, O.; Suzuki, M.T.; Koonin, E.V.; Aravind, L.; Hadd, A.; Nguyen, L.P.; Villacorta, R.; Amjadi, M.; Garrigues, C.; Jovanovich, S.B.; et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2000, 2, 516–529. [Google Scholar] [CrossRef]
- Martiny, J.B.; Bohannan, B.J.; Brown, J.H.; Colwell, R.K.; Fuhrman, J.A.; Green, J.L.; Horner-Devine, M.C.; Kane, M.; Krumins, J.A.; Kuske, C.R.; et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006, 4, 102–112. [Google Scholar] [CrossRef]
- Venter, J.C.; Remington, K.; Heidelberg, J.F.; Halpern, A.L.; Rusch, D.; Eisen, J.A.; Wu, D.; Paulsen, I.; Nelson, K.E.; Nelson, W.; et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 2004, 304, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Steward, G.F.; Montiel, J.L.; Azam, F. Genome size distributions indicate variability and similarities among marine viral assemblages from diverse environments. Limnol. Oceanogr. 2000, 45, 1697–1706. [Google Scholar] [CrossRef]
- Warren, R.A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol 1980, 34, 137–158. [Google Scholar] [PubMed]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Delwart, E.L. Viral metagenomics. Rev. Med. Virol. 2007, 17, 115–131. [Google Scholar] [CrossRef] [PubMed]
- Vega Thurber, R.; Willner-Hall, D.; Rodriguez-Mueller, B.; Desnues, C.; Edwards, R.A.; Angly, F.; Dinsdale, E.; Kelly, L.; Rohwer, F. Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 2009, 11, 2148–2163. [Google Scholar] [CrossRef] [PubMed]
- Schoenfeld, T.; Patterson, M.; Richardson, P.M.; Wommack, K.E.; Young, M.; Mead, D. Assembly of viral metagenomes from yellowstone hot springs. Appl. Environ. Microbiol. 2008, 74, 4164–4174. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Bueno, A.; Tamames, J.; Velazquez, D.; Moya, A.; Quesada, A.; Alcami, A. High diversity of the viral community from an Antarctic lake. Science 2009, 326, 858–861. [Google Scholar] [CrossRef] [PubMed]
- Rohwer, F.; Prangishvili, D.; Lindell, D. Roles of viruses in the environment. Environ. Microbiol. 2009, 11, 2771–2774. [Google Scholar] [CrossRef] [PubMed]
- Dinsdale, E.A.; Edwards, R.A.; Hall, D.; Angly, F.; Breitbart, M.; Brulc, J.M.; Furlan, M.; Desnues, C.; Haynes, M.; Li, L.; et al. Functional metagenomic profiling of nine biomes. Nature 2008, 452, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Rosario, K.; Nilsson, C.; Lim, Y.W.; Ruan, Y.; Breitbart, M. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 2009, 11, 2806–2820. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Felts, B.; Kelley, S.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 2004, 271, 565–574. [Google Scholar] [CrossRef]
- Breitbart, M.; Salamon, P.; Andresen, B.; Mahaffy, J.M.; Segall, A.M.; Mead, D.; Azam, F.; Rohwer, F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U.S.A. 2002, 99, 14250–14255. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Hewson, I.; Felts, B.; Mahaffy, J.M.; Nulton, J.; Salamon, P.; Rohwer, F. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 2003, 185, 6220–6223. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.; Wilson, W. Aquatic virus diversity accessed through omic techniques: a route map to function. Curr. Opin. Microbiol. 2008, 11, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Thurber, R.V.; Haynes, M.; Breitbart, M.; Wegley, L.; Rohwer, F. Laboratory procedures to generate viral metagenomes. Nat. Protoc. 2009, 4, 470–483. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.E.; Smith, D.L.; Fogg, P.C.; McCarthy, A.J.; Allison, H.E. High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga toxin-encoding Escherichia coli strains. Appl. Environ. Microbiol. 2010, 76, 2360–2365. [Google Scholar] [CrossRef] [PubMed]
- Robinson, S.E.; Wright, E.J.; Hart, C.A.; Bennett, M.; French, N.P. Intermittent and persistent shedding of Escherichia coli O157 in cohorts of naturally infected calves. J. Appl. Microbiol. 2004, 97, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; Wareing, B.M.; Fogg, P.C.; Riley, L.M.; Spencer, M.; Cox, M.J.; Saunders, J.R.; McCarthy, A.J.; Allison, H.E. Multilocus characterization scheme for Shiga toxin-encoding bacteriophages. Appl. Environ. Microbiol. 2007, 73, 8032–8040. [Google Scholar] [CrossRef] [PubMed]
- Edwards, U.; Rogall, T.; Blocker, H.; Emde, M.; Bottger, E.C. Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989, 17, 7843–7853. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics . In PCR protocols: a guide to methods and applications; Innis, M.A., Gelfand, D.H., Sninsky, J.I., White, T.J., Eds.; 1990; Academic Press: San Diego, CA, USA. [Google Scholar]
- Martinsohn, J.T.; Radman, M.; Petit, M.A. The lambda red proteins promote efficient recombination between diverged sequences: implications for bacteriophage genome mosaicism. PLoS Genet. 2008, 4, 1000065. [Google Scholar] [CrossRef]
- Huson, D.H.; Auch, A.F.; Qi, J.; Schuster, S.C. MEGAN analysis of metagenomic data. Genome Res. 2007, 17, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Paarmann, D.; D'Souza, M.; Olson, R.; Glass, E.M.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 2008, 9, 386. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Pruitt, K.D.; Tatusova, T.; Maglott, D.R. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005, 33, D501–D504. [Google Scholar] [CrossRef]
- Overbeek, R.; Begley, T.; Butler, R.M.; Choudhuri, J.V.; Chuang, H.Y.; Cohoon, M.; de Crecy-Lagard, V.; Diaz, N.; Disz, T.; Edwards, R.; et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005, 33, 5691–5702. [Google Scholar] [CrossRef] [PubMed]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Chai, B.; Farris, R.J.; Wang, Q.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Bandela, A.M.; Cardenas, E.; Garrity, G.M.; Tiedje, J.M. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007, 35, D169–D172. [Google Scholar] [CrossRef]
- Wuyts, J.; Perriere, G.; Van De Peer, Y. The European ribosomal RNA database. Nucleic Acids Res. 2004, 32, D101–D103. [Google Scholar] [CrossRef]
- Beumer, A.; Robinson, J.B. A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria. Appl. Environ. Microbiol. 2005, 71, 8301–8304. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Rohwer, F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 2005, 39, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Rodriguez-Brito, B.; Wegley, L.; Haynes, M.; Breitbart, M.; Peterson, D.M.; Saar, M.O.; Alexander, S.; Alexander Jr., E.C.; Rohwer, F. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 2006, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Breitbart, M.; Nulton, J.; Salamon, P.; Lozupone, C.; Jones, R.; Robeson, M.; Edwards, R.A.; Felts, B.; Rayhawk, S.; et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 2007, 73, 7059–7066. [Google Scholar] [CrossRef] [PubMed]
- Wegley, L.; Edwards, R.; Rodriguez-Brito, B.; Liu, H.; Rohwer, F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ. Microbiol. 2007, 9, 2707–2719. [Google Scholar] [CrossRef]
- Allison, H. Stx-phages: drivers and mediators of the evolution of STEC and STEC-like pathogens. Future Microbiol. 2007, 2, 165–174. [Google Scholar] [CrossRef]
- Mann, N.H.; Clokie, M.R.; Millard, A.; Cook, A.; Wilson, W.H.; Wheatley, P.J.; Letarov, A.; Krisch, H.M. The genome of S-PM2, a "photosynthetic" T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 2005, 187, 3188–3200. [Google Scholar] [CrossRef] [PubMed]
- Stachowicz, J.J. Mutualism, facilitation, and the structure of ecological communities. Bioscience 2001, 51, 235–246. [Google Scholar] [CrossRef]
- Davis, B.M.; Waldor, M.K. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 2003, 6, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Foster, S.D.; Dunstan, P.K. The analysis of biodiversity using rank abundance distributions . Biometrics 2009. [Google Scholar]
- Ryan, K.; Ray, C. Sherris Medical Microbiology2004, 4th ed; McGraw Hill: London, UK. [Google Scholar]
- Demirkan, I.; Williams, H.F.; Dhawi, A.; Carter, S.D.; Winstanley, C.; Bruce, K.D.; Hart, C.A. Characterization of a spirochaete isolated from a case of bovine digital dermatitis. J. Appl. Microbiol. 2006, 101, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Del Médico Zajac, M.P.; Ladelfa, M.F.; Kotsias, F.; Muylkens, B.; Thiry, J.; Thiry, E.; Romera, S.A. Biology of bovine herpesvirus 5. Vet. J. 2010, 184, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Muylkens, B.; Thiry, J.; Kirten, P.; Schynts, F.; Thiry, E. Bovine herpesvirus 1 infection and infectious bovine rhinotracheitis. Vet. Res. 2007, 38, 181–209. [Google Scholar] [CrossRef] [PubMed]
- Engels, M.; Ackermann, M. Pathogenesis of ruminant herpesvirus infections. Vet. Microbiol. 1996, 53, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Wilson, W.H.; Van Etten, J.L.; Allen, M.J. The Phycodnaviridae: the story of how tiny giants rule the world. Curr. Top. Microbiol. Immunol. 2009, 328, 1–42. [Google Scholar] [PubMed]
- Jancovich, J.K.; Mao, J.; Chinchar, V.G.; Wyatt, C.; Case, S.T.; Kumar, S.; Valente, G.; Subramanian, S.; Davidson, E.W.; Collins, J. P.; et al. Genomic sequence of a ranavirus (family Iridoviridae) associated with salamander mortalities in North America. Virology 2003, 316, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Essbauer, S.; Pfeffer, M.; Meyer, H. Zoonotic poxviruses. Vet. Microbiol. 2010, 140, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, R.D.; Robles, M.; Lopez de Haro, M.; Santos, A. Virial series for fluids of hard hyperspheres in odd dimensions. J. Chem. Phys. 2008, 129, 014510. [Google Scholar] [CrossRef] [PubMed]
- Thoen, C.; LoBue, P.; de Kantor, I. The importance of Mycobacterium bovis as a zoonosis. Vet. Microbiol. 2006, 112, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F. Bacteriophage genomics. Curr. Opin. Microbiol. 2008, 11, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Hatfull, G.F.; Cresawn, S.G.; Hendrix, R.W. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 2008, 159, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Pedulla, M.L.; Ford, M.E.; Houtz, J.M.; Karthikeyan, T.; Wadsworth, C.; Lewis, J.A.; Jacobs-Sera, D.; Falbo, J.; Gross, J.; Pannunzio, N.R.; et al. Origins of highly mosaic mycobacteriophage genomes. Cell 2003, 113, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.L.; James, C.E.; Sergeant, M.J.; Yaxian, Y.; Saunders, J.R.; McCarthy, A.J.; Allison, H.E. Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J. Bacteriol. 2007, 189, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: a Laboratory Manual, 2nd ed; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Margulies, M.; Egholm, M.; Altman, W.E.; Attiya, S.; Bader, J.S.; Bemben, L.A.; Berka, J.; Braverman, M.S.; Chen, Y.J.; Chen, Z.; et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature 2005, 437, 376–380. [Google Scholar] [PubMed]
© 2010 by the authors; licensee MDPI, Basel, Switzerland This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.