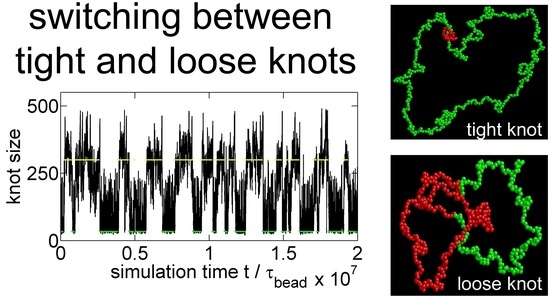
Trapping a Knot into Tight Conformations by Intra-Chain Repulsions
Abstract
:1. Introduction
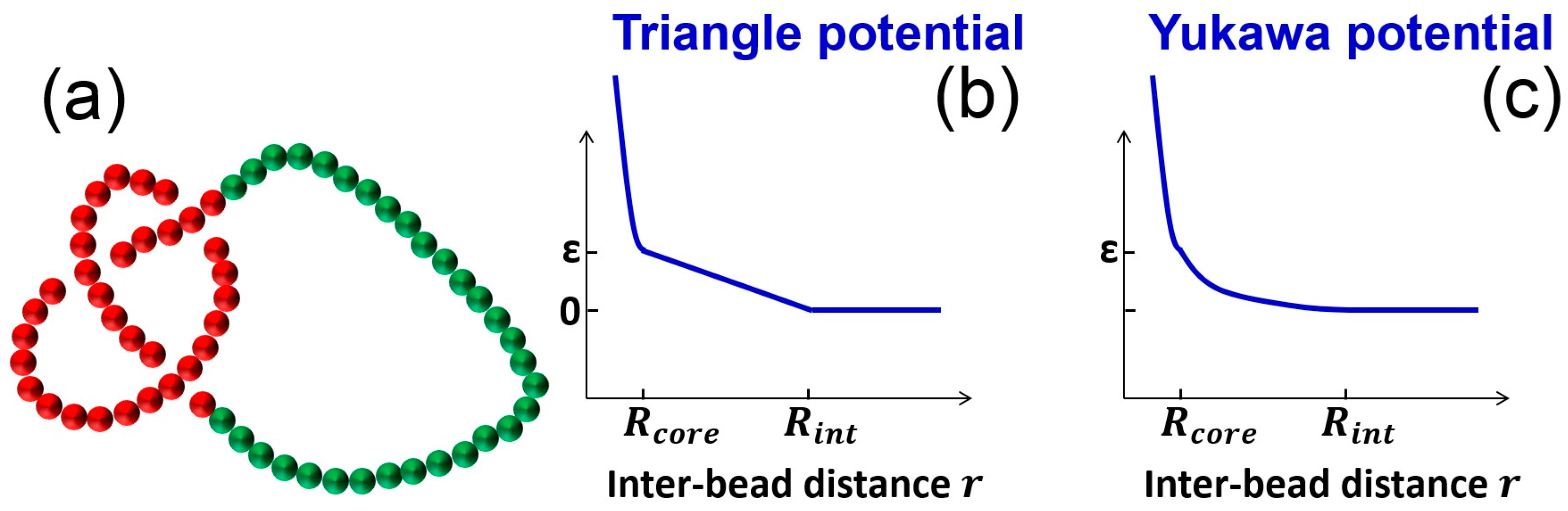
2. Simulation Methods
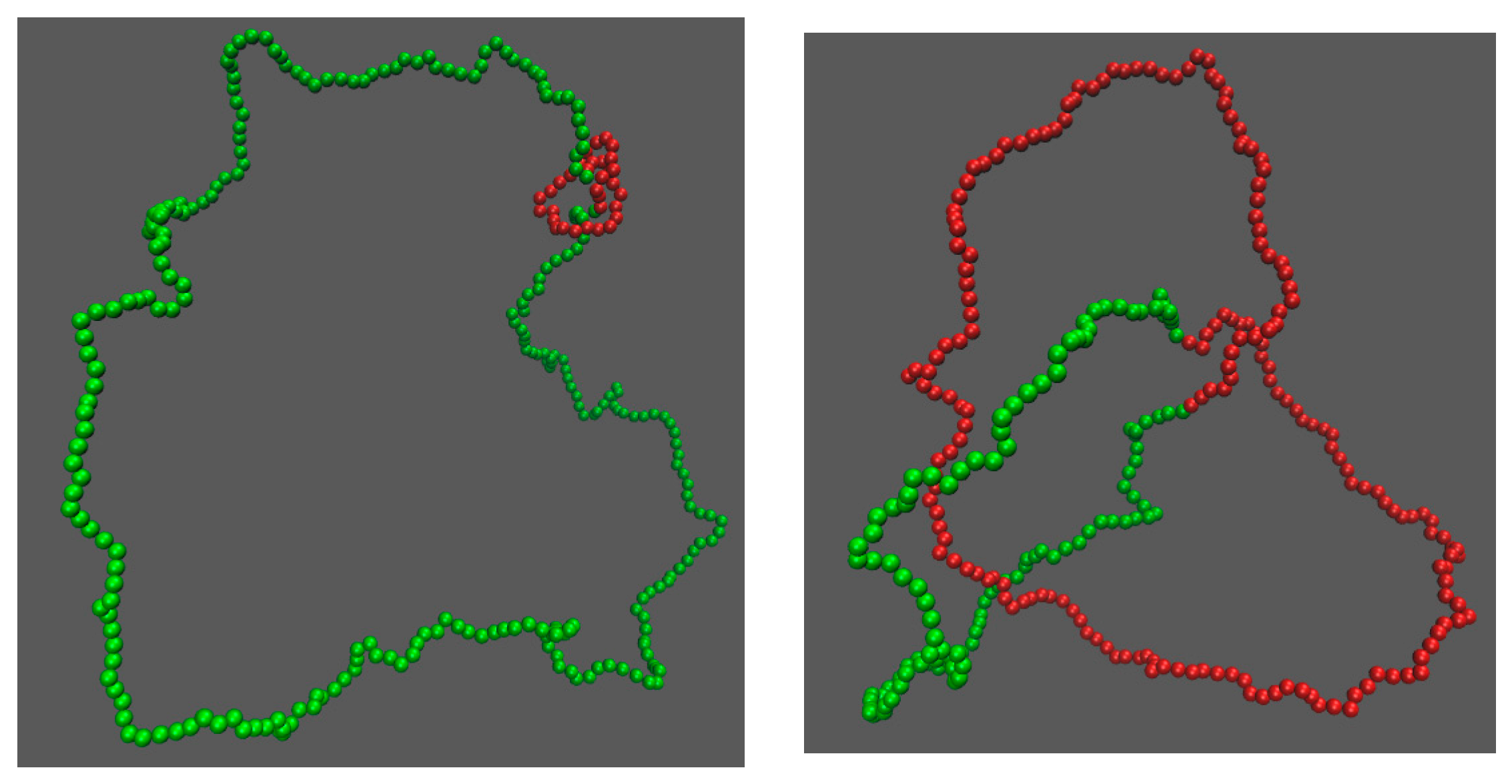
3. Results
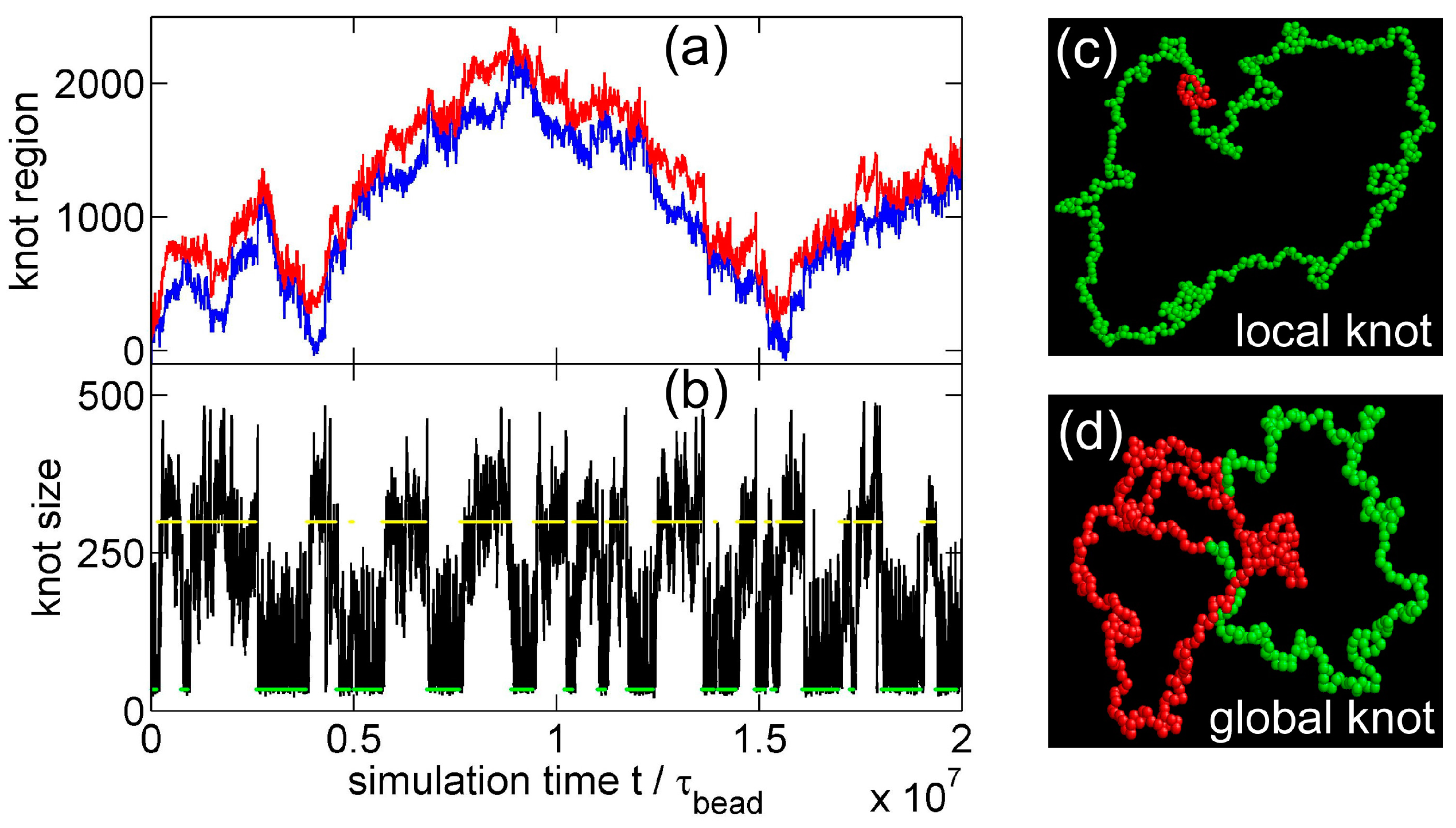
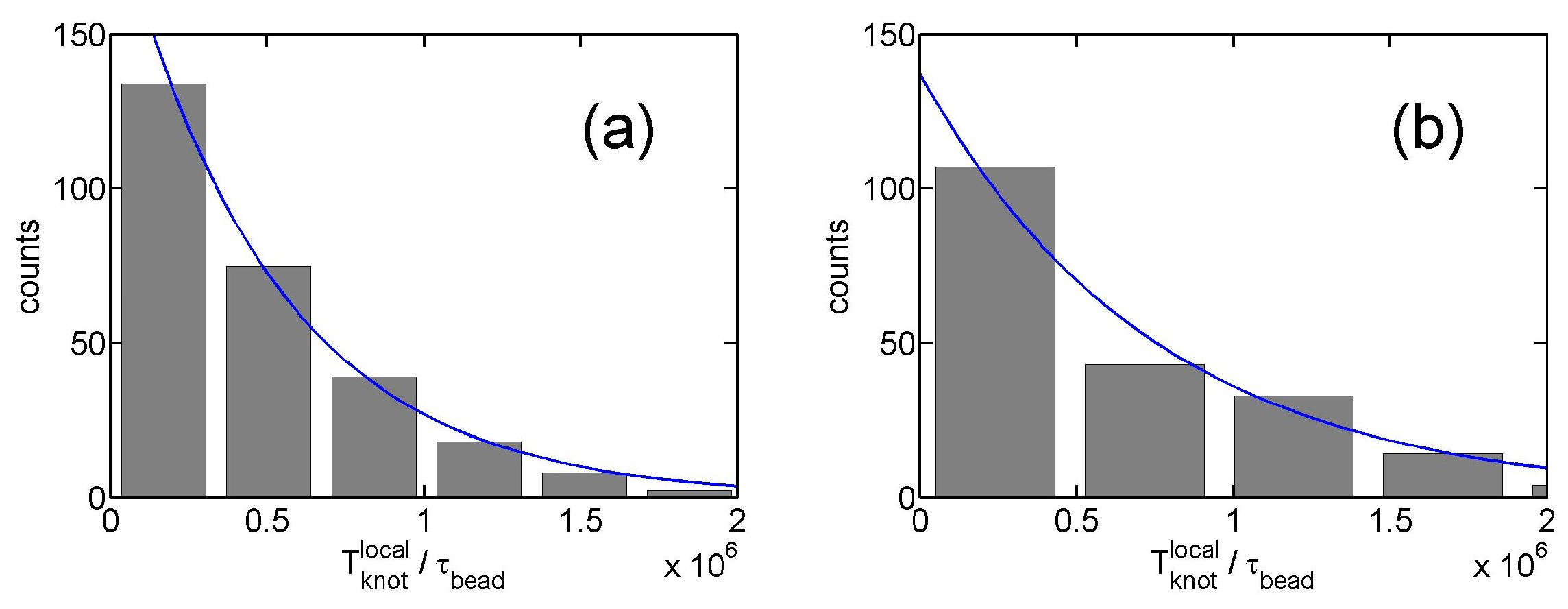
3.1. Knot Breathing in a Flexible Chain with a Triangle Potential
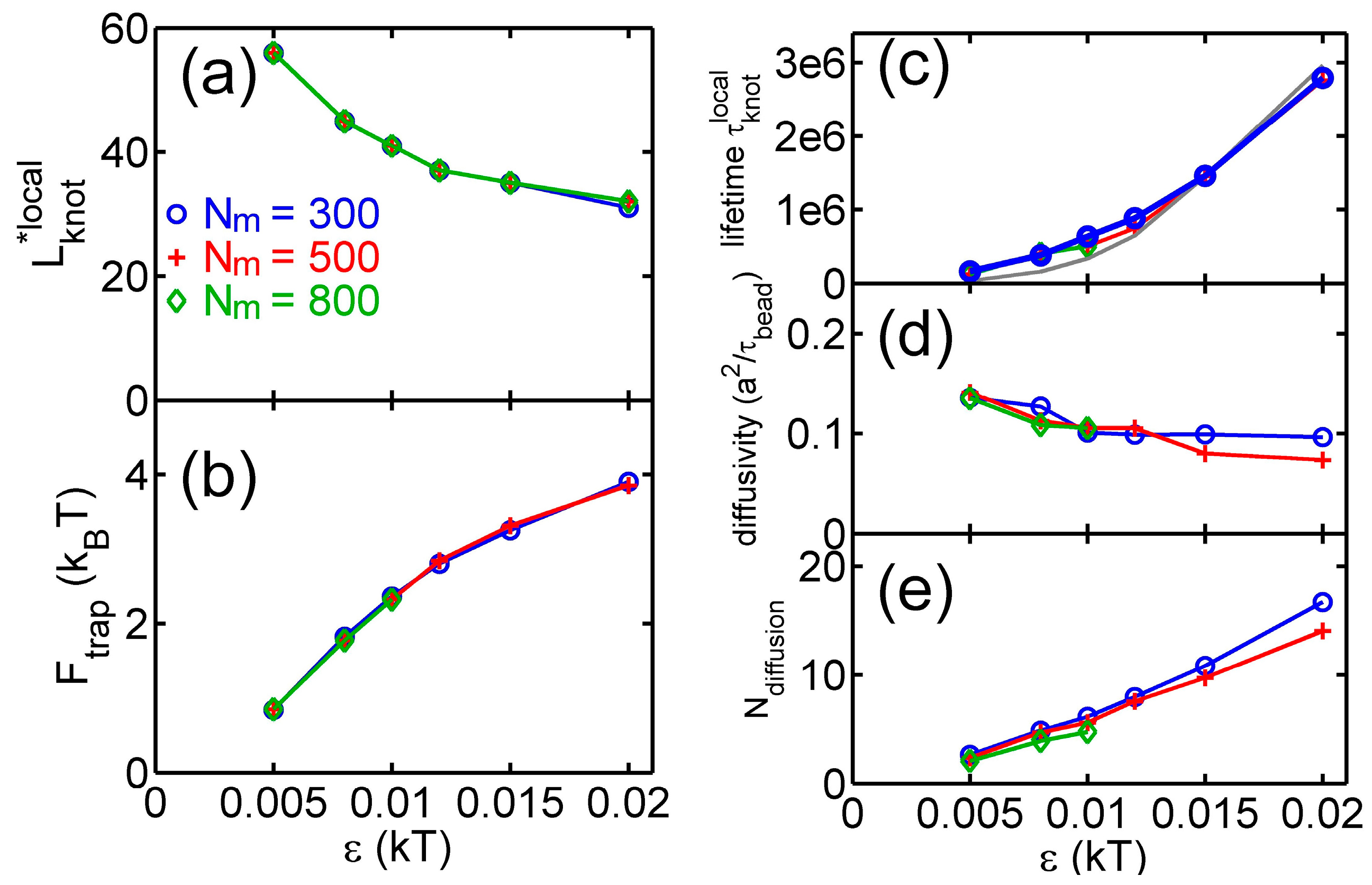
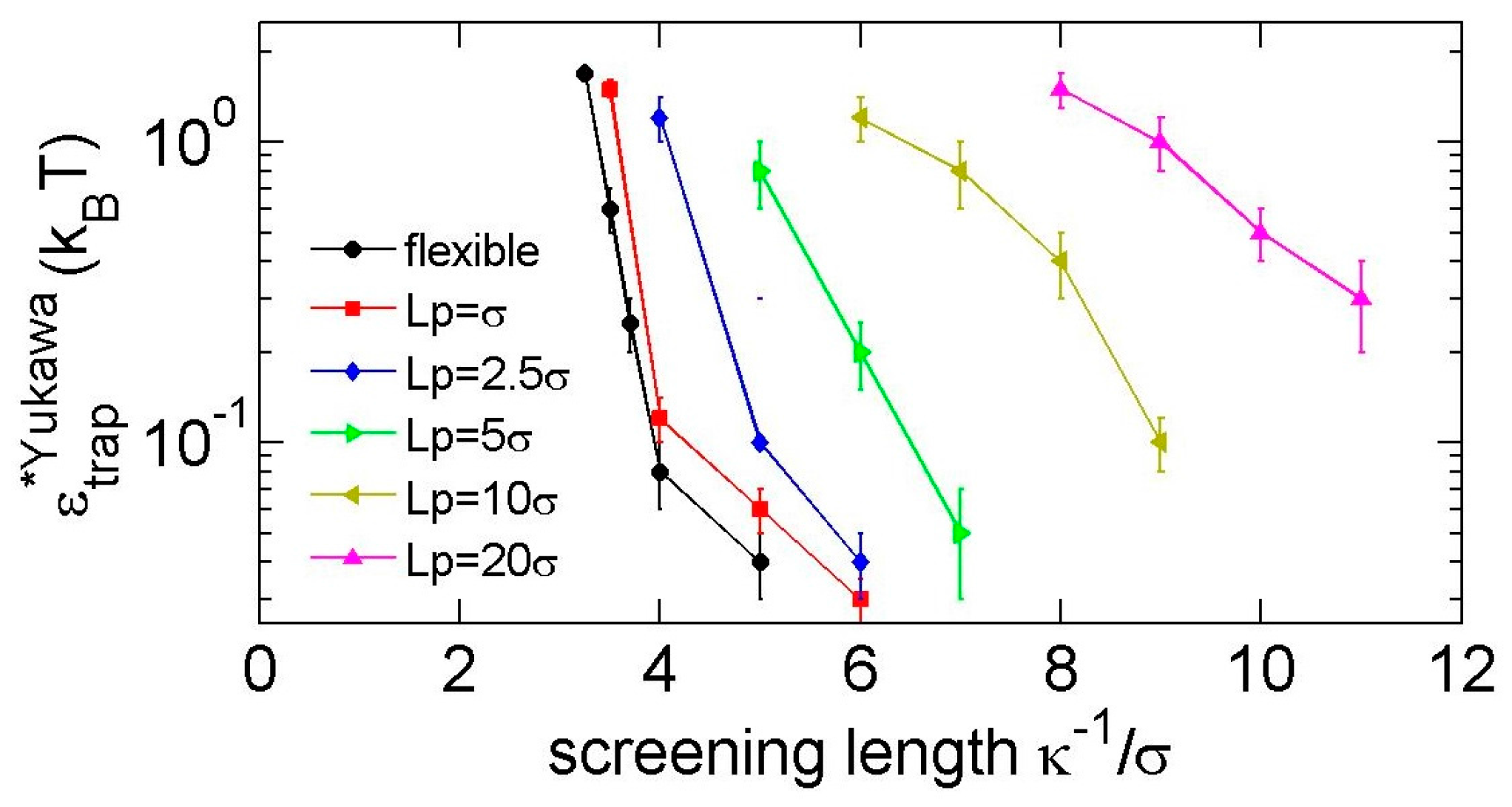
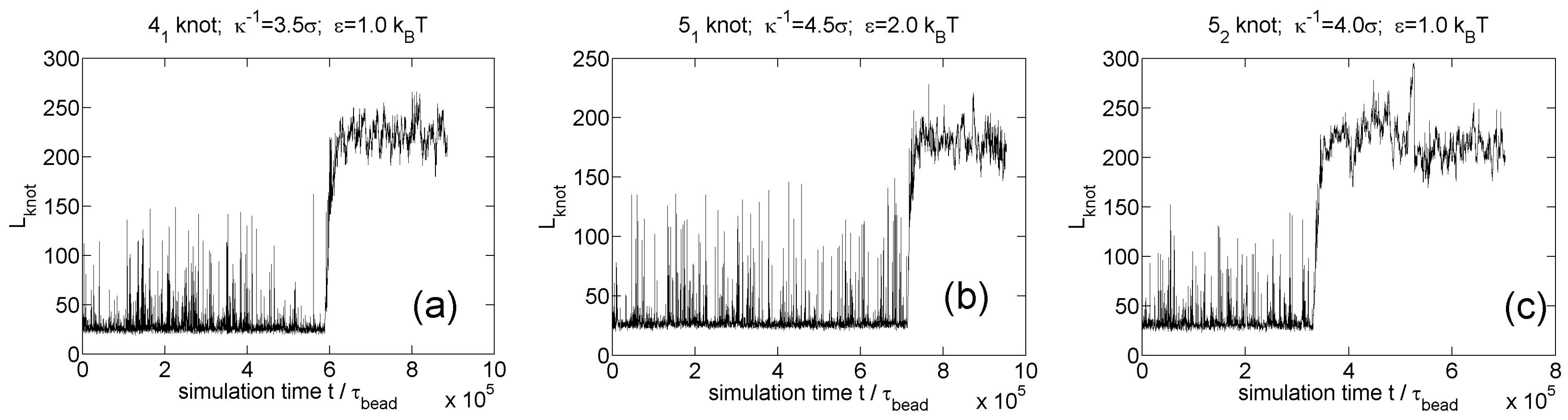
3.2. Knot Breathing in a Semiflexible Chain with a Yukawa Potential
4. Discussion about Length Scales and the Relevance to DNA and Peptides
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rybenkov, V.V.; Cozzarelli, N.R.; Vologodskii, A.V. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl. Acad. Sci. USA 1993, 90, 5307–5311. [Google Scholar] [CrossRef] [PubMed]
- Rybenkov, V.V.; Ullsperger, C.; Vologodskii, A.V.; Cozzarelli, N.R. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science 1997, 277, 690–693. [Google Scholar] [CrossRef] [PubMed]
- Arsuaga, J.; Vazquez, M.; Trigueros, S.; Sumners, D.; Roca, J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc. Natl. Acad. Sci. USA 2002, 99, 5373–5377. [Google Scholar] [CrossRef] [PubMed]
- Arsuaga, J.; Vazquez, M.; McGuirk, P.; Trigueros, S.; Sumners, D.; Roca, J. DNA knots reveal a chiral organization of DNA in phage capsids. Proc Natl. Acad. Sci. USA 2005, 102, 9165–9169. [Google Scholar] [CrossRef] [PubMed]
- Marenduzzo, D.; Orlandini, E.; Stasiak, A.; Sumners, D.; Tubiana, L.; Micheletti, C. DNA-DNA interactions in bacteriophage capsids are responsible for the observed DNA knotting. Proc Natl. Acad. Sci. USA 2009, 106, 22269–22274. [Google Scholar] [CrossRef] [PubMed]
- Taylor, W.R. A deeply knotted protein structure and how it might fold. Nature 2000, 406, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Virnau, P.; Mirny, L.A.; Kardar, M. Intricate knots in proteins: Function and evolution. PLoS. Comput. Biol. 2006, 2, e122. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.L.; Yen, S.C.; Yu, S.H.; Hwang, J.K. pKNOT: The protein KNOT web server. Nucleic Acids Res. 2007, 35, 420–424. [Google Scholar]
- Dai, L.; Zhou, Y. Characterizing the existing and potential structural space of proteins by large-scale multiple loop permutations. J. Mol. Biol. 2011, 408, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, F.; Lim, N.C.; Mandal, S.S.; Pelz, B.; Ng, W.P.; Schlierf, M.; Jackson, S.E.; Rief, M. Knotting and unknotting of a protein in single molecule experiments. Proc. Natl. Acad. Sci. USA 2016, 113, 7533–7538. [Google Scholar] [CrossRef] [PubMed]
- Orlandini, E.; Whittington, S.G. Statistical topology of closed curves: Some applications in polymer physics. Rev. Mod. Phys. 2007, 79, 611–642. [Google Scholar] [CrossRef]
- Micheletti, C.; Marenduzzo, D.; Orlandini, E. Polymers with spatial or topological constraints: Theoretical and computational results. Phys. Rep. 2011, 504, 1–73. [Google Scholar] [CrossRef]
- Micheletti, C.; Marenduzzo, D.; Orlandini, E.; Summers, D. Knotting of random ring polymers in confined spaces. J. Chem. Phys. 2006, 124, 064903. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; van der Maarel, J.R.C.; Doyle, P.S. Effect of nanoslit confinement on the knotting probability of circular DNA. ACS Macro. Lett. 2012, 1, 732–736. [Google Scholar] [CrossRef]
- Dai, L.; Renner, C.B.; Doyle, P.S. Metastable knots in confined semiflexible chains. Macromolecules 2015, 48, 2812–2818. [Google Scholar] [CrossRef]
- Vologodskii, A. Brownian dynamics simulation of knot diffusion along a stretched DNA molecule. Biophys. J. 2006, 90, 1594–1597. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Makarov, D.E. Langevin dynamics simulations of the diffusion of molecular knots in tensioned polymer chains. J. Phys. Chem. A. 2007, 111, 10338–10344. [Google Scholar] [CrossRef] [PubMed]
- Virnau, P.; Kantor, Y.; Kardar, M. Knots in globule and coil phases of a model polyethylene. J. Am. Chem. Soc. 2005, 127, 15102–15106. [Google Scholar] [CrossRef] [PubMed]
- D’Adamo, G.; Micheletti, C. Molecular crowding increases knots abundance in linear polymers. Macromolecules 2015, 48, 6337–6346. [Google Scholar] [CrossRef]
- Poier, P.; Likos, C.N.; Matthews, R. Influence of rigidity and knot complexity on the knotting of confined polymers. Macromolecules 2014, 47, 3394–3400. [Google Scholar] [CrossRef] [PubMed]
- Matthews, R.; Louis, A.A.; Likos, C.N. Effect of bending rigidity on the knotting of a polymer under tension. ACS Macro. Lett. 2012, 1, 1352–1356. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Di Ventra, M.; Micheletti, C. Topological jamming of spontaneously knotted polyelectrolyte chains driven through a nanopore. Phys. Rev. Lett. 2012, 109, 118301. [Google Scholar] [CrossRef] [PubMed]
- Narsimhan, V.; Renner, C.B.; Doyle, P.S. Translocation dynamics of knotted polymers under a constant or periodic external field. Soft Matter 2016, 12, 5041–5049. [Google Scholar] [CrossRef] [PubMed]
- Grosberg, A.Y.; Rabin, Y. Metastable tight knots in a wormlike polymer. Phys. Rev. Lett. 2007, 99, 217801. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Renner, C.B.; Doyle, P.S. Metastable tight knots in semiflexible chains. Macromolecules 2014, 47, 6135–6140. [Google Scholar] [CrossRef]
- Dai, L.; Renner, C.B.; Doyle, P.S. Origin of metastable knots in single flexible chains. Phys. Rev. Lett. 2015, 114, 037801. [Google Scholar] [CrossRef] [PubMed]
- Grosberg, A.Y. Do knots self-tighten for entropic reasons? Polym. Sci. Ser. A 2016, 58, 864–872. [Google Scholar] [CrossRef]
- Arai, Y.; Yasuda, R.; Akashi, K.; Harada, Y.; Miyata, H.; Kinosita, K.; Itoh, H. Tying a molecular knot with optical tweezers. Nature 1999, 399, 446–448. [Google Scholar] [PubMed]
- Bao, X.R.; Lee, H.J.; Quake, S.R. Behavior of complex knots in single DNA molecules. Phys. Rev. Lett. 2003, 91, 265506. [Google Scholar] [CrossRef] [PubMed]
- Metzler, R.; Reisner, W.; Riehn, R.; Austin, R.; Tegenfeldt, J.O.; Sokolov, I.M. Diffusion mechanisms of localised knots along a polymer. Eur. Phys. Lett. 2006, 76, 696. [Google Scholar] [CrossRef]
- Tang, J.; Du, N.; Doyle, P.S. Compression and self-entanglement of single DNA molecules under uniform electric field. Proc. Natl. Acad. Sci. USA 2011, 108, 16153–16158. [Google Scholar] [CrossRef] [PubMed]
- Renner, C.B.; Doyle, P.S. Stretching self-entangled DNA molecules in elongational fields. Soft Matter 2015, 11, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, E.; Valle, F.; Adamcik, J.; Witz, G.; Metzler, R.; De Los Rios, P.; Roca, J.; Dietler, G. Fractal dimension and localization of DNA knots. Phys. Rev. Lett. 2007, 98, 058102. [Google Scholar] [CrossRef] [PubMed]
- Plesa, C.; Verschueren, D.; Pud, S.; van der Torre, J.; Ruitenberg, J.W.; Witteveen, M.J.; Jonsson, M.P.; Grosberg, A.Y.; Rabin, Y.; Dekker, C. Direct observation of DNA knots using a solid-state nanopore. Nat. Nanotechnol. 2016, 11, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Doyle, P.S. Effects of intrachain interactions on the knot size of a polymer. Macromolecules 2016, 49, 7581–7587. [Google Scholar] [CrossRef]
- Orlandini, E.; Baiesi, M.; Zonta, F. How local flexibility affects knot positioning in ring polymers. Macromolecules 2016, 49, 4656–4662. [Google Scholar] [CrossRef]
- Plimpton, S. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995, 117, 1–19. [Google Scholar] [CrossRef]
- Kremer, K.; Grest, G.S. Dynamics of entangled linear polymer melts: A molecular-dynamics simulation. J. Chem. Phys. 1990, 92, 5057–5086. [Google Scholar] [CrossRef]
- Frankkamenetskii, M.D.; Vologodskii, A.V. Topological aspects of polymer physics—Theory and its biophysical applications. Sov. Phys. Usp. 1981, 24, 679–696. [Google Scholar] [CrossRef]
- Narsimhan, V.; Renner, C.B.; Doyle, P.S. Jamming of knots along a tensioned chain. ACS Macro. Lett. 2016, 5, 123–127. [Google Scholar] [CrossRef]
- Murphy, M.; Rasnik, I.; Cheng, W.; Lohman, T.M.; Ha, T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004, 86, 2530–2537. [Google Scholar] [CrossRef]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, L.; Doyle, P.S. Trapping a Knot into Tight Conformations by Intra-Chain Repulsions. Polymers 2017, 9, 57. https://doi.org/10.3390/polym9020057
Dai L, Doyle PS. Trapping a Knot into Tight Conformations by Intra-Chain Repulsions. Polymers. 2017; 9(2):57. https://doi.org/10.3390/polym9020057
Chicago/Turabian StyleDai, Liang, and Patrick S. Doyle. 2017. "Trapping a Knot into Tight Conformations by Intra-Chain Repulsions" Polymers 9, no. 2: 57. https://doi.org/10.3390/polym9020057
APA StyleDai, L., & Doyle, P. S. (2017). Trapping a Knot into Tight Conformations by Intra-Chain Repulsions. Polymers, 9(2), 57. https://doi.org/10.3390/polym9020057