Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria Strain
2.2. Cellulase Assay
2.3. Screening Fermentation Conditions
2.4. Enzyme Purification
2.5. Enzyme Characterization
2.6. High-Performance Liquid Chromatography and Viscosity Analysis
2.7. Clarification of Apple Juice
3. Results and Discussion
3.1. Screening the Cellulase Biosynthesis Conditions
3.2. Enzyme Purification
3.3. Enzyme Characterization
3.4. Clarification of Apple Juice
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Jakob, M.; Mahendran, A.R.; Gindl-Altmutter, W.; Bliem, P.; Konnerth, J.; Müller, U.; Veigel, S. The strength and stiffness of oriented wood and cellulose-fibre materials: A review. Prog. Mater. Sci. 2022, 125, 100916. [Google Scholar] [CrossRef]
- Li, H.; Zhang, M.; Zhang, Y.; Xu, X.; Zhao, Y.; Jiang, X.; Zhang, R.; Gui, Z. Characterization of cellulose-degrading bacteria isolated from silkworm excrement and optimization of its cellulase production. Polymers 2023, 15, 4142. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Langan, P.; Chanzy, H. Crystal structure and hydrogen-bonding system in cellulose iβ from synchrotron x-ray and neutron fiber diffraction. J. Am. Chem. Soc. 2002, 124, 9074–9082. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Sugiyama, J.; Chanzy, H.; Langan, P. Crystal Structure and Hydrogen Bonding System in Cellulose Iα from Synchrotron X-ray and Neutron Fiber Diffraction. J. Am. Chem. Soc. 2003, 125, 14300–14306. [Google Scholar] [CrossRef] [PubMed]
- Deligey, F.; Frank, M.A.; Cho, S.H.; Kirui, A.; Mentink-Vigier, F.; Swulius, M.T.; Nixon, B.T.; Wang, T. Structure of in vitro-synthesized cellulose fibrils viewed by cryo-electron tomography and 13C natural-abundance dynamic nuclear polarization solid-state NMR. Biomacromolecules 2022, 23, 2290–2301. [Google Scholar] [CrossRef]
- Berruyer, P.; Gericke, M.; Moutzouri, P.; Jakobi, D.; Bardet, M.; Karlson, L.; Schantz, S.; Heinze, T.; Emsley, L. Advanced characterization of regioselectively substituted methylcellulose model compounds by DNP enhanced solid-state NMR spectroscopy. Carbohydr. Polym. 2021, 262, 117944. [Google Scholar] [CrossRef] [PubMed]
- Blasi, A.; Verardi, A.; Lopresto, C.G.; Siciliano, S.; Sangiorgio, P. Lignocellulosic agricultural waste valorization to obtain valuable products: An overview. Recycling 2023, 8, 61. [Google Scholar] [CrossRef]
- Kim, S.J.; Shin, K.C.; Kim, D.W.; Kim, Y.S.; Park, C.S. Cloning and characterization of cellulase from Paenibacillus peoriae MK1 isolated from soil. Fermentation 2023, 9, 873. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, B.K.; Lee, B.H.; Jo, K.I.; Lee, N.K.; Chung, C.H.; Lee, Y.C.; Lee, J.W. Purification and characterization of cellulase produced by Bacillus amyoliquefaciens DL-3 utilizing rice hull. Bioresour. Technol. 2008, 99, 378–386. [Google Scholar] [CrossRef]
- Nisar, K.; Abdullah, R.; Kaleem, A.; Lqtedar, M.; Aftab, M.; Saleem, F. Purification, characterization and thermodynamic analysis of cellulases produced from Thermomyces dupontii and its industrial applications. Saudi J. Biol. Sci. 2022, 29, 103483. [Google Scholar] [CrossRef]
- Ilić, N.; Milić, M.; Beluhan, S.; Dimitrijević-Branković, S. Cellulases: From lignocellulosic biomass to improved production. Energies 2023, 16, 3598. [Google Scholar] [CrossRef]
- Sutaoney, P.; Rai, S.N.; Sinha, S.; Choudhary, R.; Gupta, A.K.; Singh, S.K.; Banerjee, P. Current perspective in research and industrial applications of microbial cellulases. Int. J. Biol. Macromol. 2024, 264, 130639. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.Y.; Liu, Y.S.; Zeng, Y.N.; Himmel, M.E.; Baker, J.O.; Bayer, E.A. How does plant cell wall nanoscale architecture correlate with enzymatic digestibility? Science 2012, 338, 1055–1060. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.O.; Asevedo, E.A.; Souza Filho, P.F.; Santos, E.S.d. Extraction of cellulases produced through solid-state fermentation by Trichoderma reesei CCT-2768 using green coconut fibers pretreated by steam explosion combined with alkali. Biomass 2024, 4, 92–106. [Google Scholar] [CrossRef]
- Singh, A.; Bajar, S.; Devi, A.; Pant, D. An overview on the recent developments in fungal cellulase production and their industrial applications. Bioresour. Technol. Rep. 2021, 14, 100652. [Google Scholar] [CrossRef]
- Benatti, A.L.T.; Polizeli, M.d.L.T.d.M. Lignocellulolytic biocatalysts: The main players involved in multiple biotechnological processes for biomass valorization. Microorganisms 2023, 11, 162. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, G.; Liu, H.; Wu, Z.; Gong, D.; Ru, X.; Gong, X.; Pi, Q.; Yang, Q. Application of Aspergillus niger in practical biotechnology of industrial recovery of potato starch by-products and its flocculation characteristics. Microorganisms 2022, 10, 1847. [Google Scholar] [CrossRef]
- Santos, G.B.; de Sousa Francisco Filho, Á.; Rêgo da Silva Rodrigues, J.; Rodrigues de Souza, R. Cellulase production by Aspergillus niger using urban lignocellulosic waste as substrate: Evaluation of different cultivation strategies. J. Environ. Manag. 2022, 305, 114431. [Google Scholar] [CrossRef]
- Lee, D.S.; Song, Y.; Lee, Y.G.; Bae, H.J. Comparative evaluation of adsorption of major enzymes in a cellulase cocktail obtained from Trichoderma reesei onto different types of lignin. Polymers 2022, 14, 167. [Google Scholar] [CrossRef]
- Asha, B.M.; Revathi, M.; Yadav, A.; Sakthivel, N. Purification and characterization of a thermophilic cellulase from a novel cellulolytic strain, Paenibacillus barcinonensis. J. Microbiol. Biotechnol. 2012, 22, 1501–1509. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Wang, C.L.; Wang, S.L. Microbial conversion of shrimp heads to proteases and chitin as an effective dye adsorbent. Polymers 2020, 12, 2228. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Wang, S.L. Production of thermophilic chitinase by Paenibacillus sp. TKU052 by bioprocessing of chitinous fishery wastes and its application in N-acetyl-D-glucosamine production. Polymers 2021, 13, 3048. [Google Scholar] [CrossRef]
- Doan, C.T.; Tran, T.N.; Tran, T.P.H.; Nguyen, T.T.; Nguyen, H.K.; Tran, T.K.T.; Vu, B.T.; Trinh, T.H.T.; Nguyen, A.D.; Wang, S.L. Chitosanase production from the liquid fermentation of squid pens waste by Paenibacillus elgii. Polymers 2023, 15, 3724. [Google Scholar] [CrossRef] [PubMed]
- Ugwuoji, E.T.; Nwagu, T.N.; Ezeogu, L.I. Detergent-stable amylase production by Paenibacillus lactis strain OPSA3 isolated from soil; optimization by response surface methodology. Biotechnol. Rep. 2023, 39, e00808. [Google Scholar] [CrossRef]
- Lee, T.H.; Lee, Y.E. Cloning, sequencing and expression of the gene encoding a thermostable β -Xylosidase from Paenibacillus sp. DG-22. J. Life Sci. 2007, 17, 1197–1203. [Google Scholar] [CrossRef]
- Mostafa, Y.S.; Alamri, S.A.; Hashem, M.; Nafady, N.A.; Abo-Elyousr, K.A.M.; Mohamed, Z.A. Thermostable cellulase biosynthesis from Paenibacillus alvei and its utilization in lactic acid production by simultaneous saccharification and fermentation. Open Life Sci. 2020, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.D.; Pham, T.P.; Vu, B.T.; Le, N.T.N. Data on annotation and analysis of genome sequence of Paenibacillus elgii YSY-1.2, a promising chitinase-producing, plant-growth-promoting, and biocontrol agent. Data Brief 2024, 54, 110285. [Google Scholar] [CrossRef] [PubMed]
- Sanghi, A.; Garg, N.; Gupta, V.K.; Mittal, A.; Kuhad, R.C. One-step purification and characterization of cellulase-free xylanase produced by alkalophilic Bacillus subtilis ash. Braz. J. Microbiol. 2010, 41, 467–476. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, D.H.; Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, C.L.; Wang, S.L. Proteases production and chitin preparation from the liquid fermentation of chitinous fishery by-products by Paenibacillus elgii. Mar. Drugs 2021, 19, 477. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Dinh, T.K.L.; Duong, T.H.N.; Phan, T.T.U.; Le, T.T.L.; Tran, T.D.; Hoang, P.H.Q.; Nguyen, A.D.; Wang, S.L. Optimization production of an endo-β-1,4-xylanase from Streptomyces thermocarboxydus using wheat bran as sole carbon source. Recycling 2024, 9, 50. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Tran, T.N.; Doan, C.T.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Antioxidant and anti-diabetes potential of water-soluble chitosan–glucose derivatives produced by Maillard reaction. Polymers 2019, 11, 1714. [Google Scholar] [CrossRef] [PubMed]
- Islam, F.; Roy, N. Screening, purification and characterization of cellulase from cellulase producing bacteria in molasses. BMC Res. Notes 2018, 11, 445. [Google Scholar] [CrossRef]
- Datsomor, O.; Yan, Q.; Opoku-Mensah, L.; Zhao, G.; Miao, L. Effect of different inducer sources on cellulase enzyme production by white-rot basidiomycetes Pleurotus ostreatus and Phanerochaete chrysosporium under submerged fermentation. Fermentation 2022, 8, 561. [Google Scholar] [CrossRef]
- Lugani, Y.; Singla, R.; Sooch, B.S. Optimization of cellulase production from newly isolated Bacillus sp. Y3. J. Bioproc. Biotech. 2015, 5, 1000264. [Google Scholar] [CrossRef]
- Malik, W.A.; Javed, S. Enhancement of cellulase production by cellulolytic bacteria SB125 in submerged fermentation medium and biochemical characterization of the enzyme. Int. J. Biol. Macromol. 2024, 263, 130415. [Google Scholar] [CrossRef] [PubMed]
- Maravi, P.; Kumar, A. Optimization and statistical modeling of microbial cellulase production using submerged culture. J. Appl. Biol. Biotechnol. 2021, 9, 142–152. [Google Scholar]
- Boondaeng, A.; Keabpimai, J.; Trakunjae, C.; Vaithanomsat, P.; Srichola, P.; Niyomvong, N. Cellulase production under solid-state fermentation by Aspergillus sp. IN5: Parameter optimization and application. Heliyon 2024, 10, e26601. [Google Scholar] [CrossRef] [PubMed]
- Sohail, M.; Ahmad, A.; Khan, S.A. Production of cellulase from Aspergillus terreus MS105 on crude and commercially purified substrates. 3 Biotech 2016, 6, 103. [Google Scholar] [CrossRef]
- Dar, M.A.; Pawar, K.D.; Rajput, B.P.; Rahi, P.; Pandit, R.S. Purification of a cellulase from cellulolytic gut bacterium, Bacillus tequilensis G9 and its evaluation for valorization of agro-wastes into added value byproducts. Biocatal. Agric. Biotechnol. 2019, 20, 101219. [Google Scholar] [CrossRef]
- Elsababty, Z.E.; Abdel-Aziz, S.H.; Ibrahim, A.M.; Guirgis, A.A.; Dawwam, G.E. Purification, biochemical characterization, and molecular cloning of cellulase from Bacillus licheniformis strain Z9 isolated from soil. J. Genet. Eng. Biotechnol. 2022, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.L.; Zhang, Z.; Wu, M.; Wu, Y.; Feng, J.X. Isolation, screening, and identification of cellulolytic bacteria from natural reserves in the subtropical region of China and optimization of cellulase production by Paenibacillus terrae ME27-1. BioMed Res. Int. 2014, 2014, 512497. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.M.; Hong, S.J.; Math, R.K.; Islam, S.M.; Kim, J.O.; Lee, Y.H.; Kim, H.; Yun, H.D. Cloning of two cellulase genes from endophytic Paenibacillus polymyxa GS01 and comparison with cel 44C-man 26A. J. Basic Microbiol. 2008, 48, 464–472. [Google Scholar] [CrossRef] [PubMed]
- Park, I.H.; Chang, J.; Lee, Y.S.; Fang, S.J.; Choi, Y.L. Gene cloning of endoglucanase Cel5A from cellulose-degrading Paenibacillus xylanilyticus KJ-03 and purification and characterization of the recombinant enzyme. Protein J. 2012, 31, 238–245. [Google Scholar] [CrossRef]
- Dhar, H.; Kasana, R.C.; Dutt, S.; Gulati, A. Cloning and expression of low temperature active endoglucanase EG5C from Paenibacillus sp. IHB B 3084. Int. J. Biol. Macromol. 2015, 81, 259–566. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.H.; Tsai, C.H.; Lin, P.H.; Chang, K.C.; Tu, J.; Wang, Y.N.; Yang, C.Y. Characterization and pulp refining activity of a Paenibacillus campinasensis cellulase expressed in Escherichia coli. Bioresour. Technol. 2010, 101, 7882–7888. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.H.T.; Kim, J.; Shim, J.; Chang, S.; Chung, W. Coconut mesocarp-based lignocellulosic waste as a substrate for cellulase production from high promising multienzyme-producing Bacillus amyloliquefaciens FW2 without pretreatments. Microorganisms 2022, 10, 327. [Google Scholar] [CrossRef]
- Kim, D.; Ku, S. Bacillus cellulase molecular cloning, expression, and surface display on the outer membrane of Escherichia coli. Molecules 2018, 23, 503. [Google Scholar] [CrossRef]
- Malik, W.A.; Javed, S. Biochemical characterization of cellulase from Bacillus subtilis strain and its effect on digestibility and structural modifications of lignocellulose rich biomass. Front. Bioeng. Biotechnol. 2021, 9, 800265. [Google Scholar] [CrossRef]
- Wu, B.; Guddat, L.W.; Chang, S.; He, B.; Schenk, G. Processivity and enzymatic mechanism of a multifunctional family 5 endoglucanase from Bacillus subtilis BS-5 with potential applications in the saccharification of cellulosic substrates. Biotechnol. Biofuels 2018, 11, 20. [Google Scholar] [CrossRef]
- Wu, S.; Wu, S. Processivity and the mechanisms of processive endoglucanases. Appl. Biochem. Biotechnol. 2020, 190, 448–463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wang, Y.; Li, Z.; Zhou, X.; Zhang, W.; Zhao, Y.; Lu, X. Characterization of a multi-function processive endoglucanase CHU_2103 from Cytophaga hutchinsonii. Appl. Microbiol. Biotechnol. 2014, 98, 6679–6687. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Kumar, A.; Luo, X.; Shi, H.; Liu, Z.; Wu, G. Highly alkali-stable and cellulase-free xylanases from Fusarium sp. 21 and their application in clarification of orange juice. Int. J. Biol. Macromol. 2020, 155, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Ozyilmaz, G.; Gunay, E. Clarification of apple, grape and pear juices by co-immobilized amylase, pectinase and cellulase. Food Chem. 2023, 398, 133900. [Google Scholar] [CrossRef] [PubMed]
- Aleksandra, D.C.; Tomasz, T.; Tadeusz, T. Antioxidant activity of apples-an impact of maturity stage and fruit part. Acta Sci. Pol. Technol. Aliment. 2011, 10, 443–454. [Google Scholar]
- Asale, Y.; Dessalegn, E.; Assefa, D.; Abdisa, M. Phytochemicals and antioxidant activity of different apple cultivars grown in South Ethiopia: Case of the wolayta zone. Int. J. Food Prop. 2020, 24, 354–363. [Google Scholar] [CrossRef]
- Hosseini, S.S.; Khodaiyan, F.; Mousavi, S.M.; Azimi, S.Z. Clarification of the pomegranate juice in a bioreactor packed by pectinase enzymes immobilized on the glass bead activated with polyaldehyde polysaccharides. LWT-Food Sci. Technol. 2021, 137, 110500. [Google Scholar] [CrossRef]
- Chen, X.; Xu, Y.; Wu, J.; Yu, Y.; Zou, B.; Li, L. Effects of pectinase pre-treatment on the physicochemical properties, bioactive compounds, and volatile components of juices from different cultivars of guava. Foods 2023, 12, 330. [Google Scholar] [CrossRef]

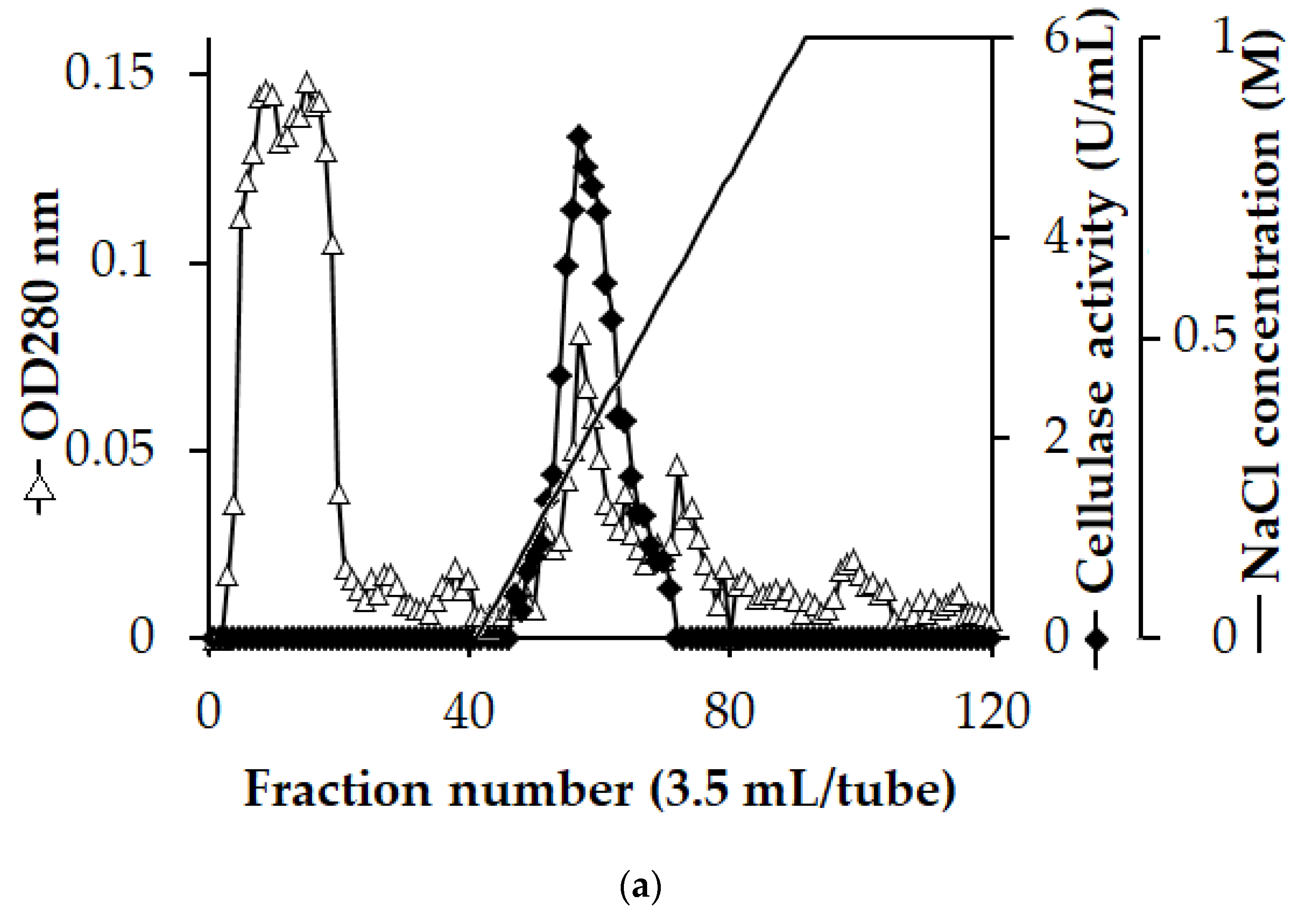

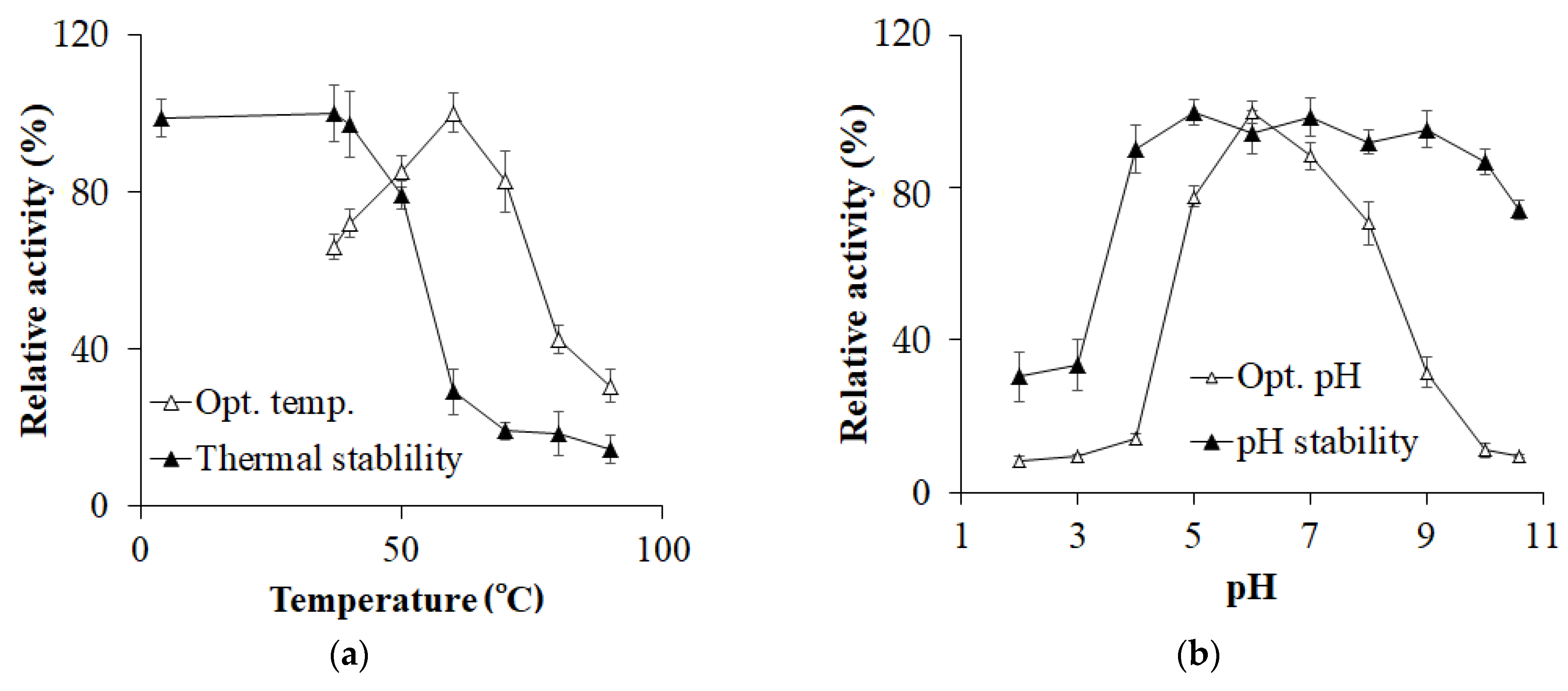
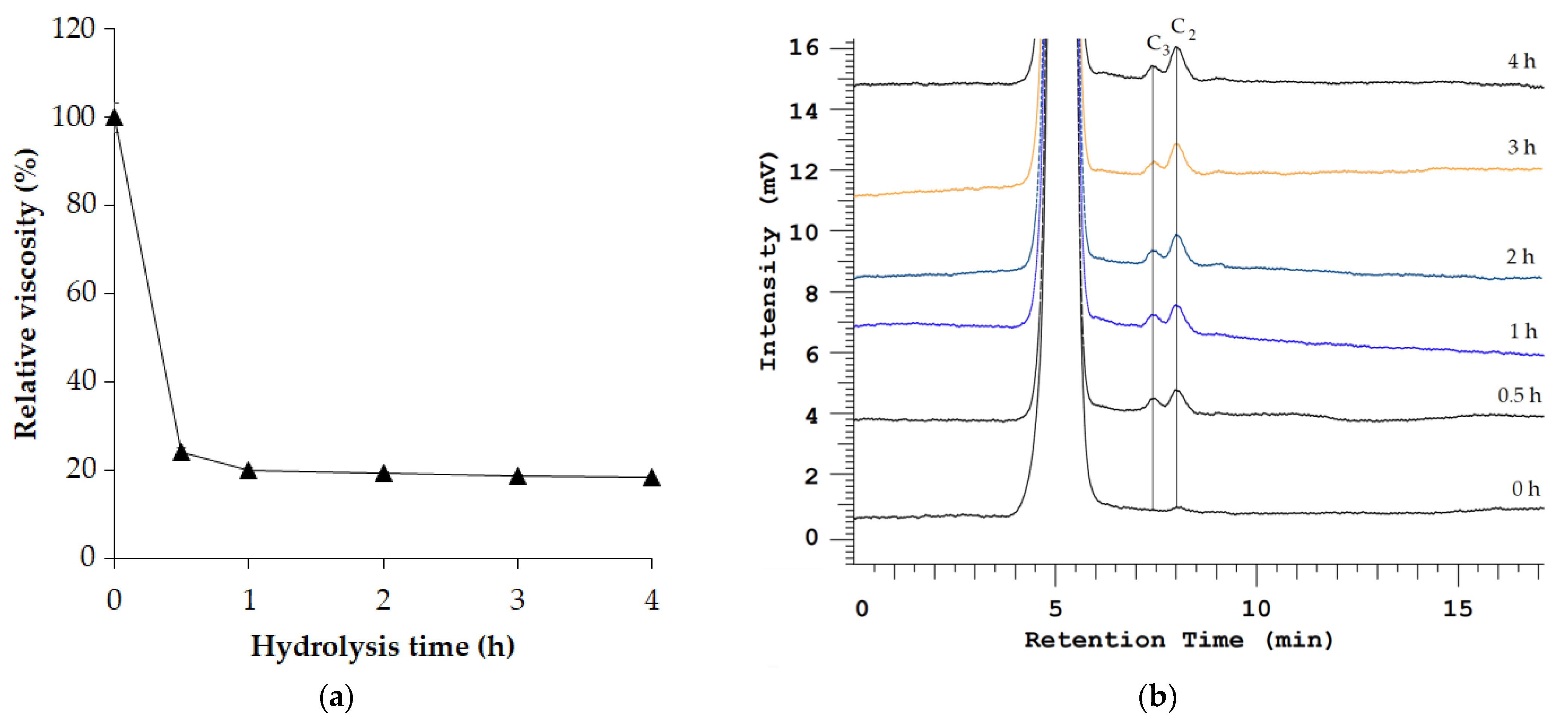
| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture supernatant | 519.5 | 4419.3 | 8.5 | 100 | 1.0 |
| EtOH precipitation | 262.6 | 3652.1 | 13.9 | 83 | 1.6 |
| High Q column | 4.3 | 1554.5 | 358.7 | 35 | 42.2 |
| Cellulase-Producing Strain | MW (kDa) | Opt. pH | Opt. Temp. (°C) | Metal Ion | Ref. | |
|---|---|---|---|---|---|---|
| Inhibitor | Activator | |||||
| Paenibacillus elgii TKU051 | 45 | 6.0 | 60 | Cu2+ | Mn2+ | This study |
| Paenibacillus peoriae MK1 | 65 | 5.0 | 40 | Mn2+ | Cu2+, Ba2+, Mg2+, and Fe2+ | [8] |
| Paenibacillus terrae ME27-1 | 5.5 | 50 | [42] | |||
| Paenibacillus polymyxa GS01 | 33 | 7.0 | 50 | [43] | ||
| 60 | 6.0 | 60 | ||||
| Paenibacillus xylanilyticus KJ-03 | 64 | 5.0 | 40 | Fe3+, Mn2+, Zn2+, and Hg2+ | Ca2+ and Cu2+ | [44] |
| Paenibacillus sp. IHB B 3084 | 63.5 | 40 | [45] | |||
| Paenibacillus barcinonensis | 58.6 | 6.0 | 65 | Hg2+ | Mg2+, Fe2+, Mn2+, and Zn2+ | [20] |
| Paenibacillus campinasensis BL11 | 38 | 7.0 | 60 | Hg2+, Cu2+, and Zn2+ | Mn2+ and Co2+ | [46] |
| Paenibacillus sp. | 67 | 7.0 | 40 | [33] | ||
| Bacillus licheniformis Z9 | 54.4 | 7.4 | 30 | Mg2+ and Na+ | Fe3+, Cu2+, and Ca2+ | [41] |
| Bacillus amyloliquefaciens FW2 | 55 | Fe2+ and Zn2+ | Mg2+ and Ca2+ | [47] | ||
| B. licheniformis ATCC 14580 | 61.7 | 7.0 | 60 | [48] | ||
| B. tequilensis G9 | 43 | 5.0 | 40 | Pb2+ | Zn2+, Ca2+, and Co2+ | [40] |
| Substrate | Relative Activity |
|---|---|
| CMC * | 100.00 ± 4.65 |
| Cellulose powder | 29.47 ± 0.58 |
| Chitin | 12.44 ± 0.50 |
| Chitosan | 23.96 ± 3.06 |
| Xylan | 20.26 ± 0.75 |
| Alginate | 19.44 ± 1.19 |
| Dextran | 19.82 ± 1.05 |
| Gum arabic | 17.04 ± 0.48 |
| Pectin | 17.83 ± 1.60 |
| β-1,3-Glucan | 11.51 ± 1.02 |
| Starch | 5.69 ± 0.56 |
| 2-Nitrophenyl β-D-galactopyranoside | N.D. |
| 4-Nitrophenyl N-acetyl-β-D-glucosaminide | N.D. |
| 4-Nitrophenyl-β-D-glucopyranoside | N.D. |
| 4-Nitrophenyl-α-D-glucopyranoside | N.D. |
| Chemical | Relative Activity |
|---|---|
| Control | 100 ± 3.06 |
| Na+ | 97.57 ± 8.22 |
| K+ | 96.81 ± 2.45 |
| Zn2+ | 91.82 ± 6.87 |
| Fe2+ | 90.23 ± 2.05 |
| Ca2+ | 97.44 ± 5.18 |
| Ba2+ | 101.46 ± 6.25 |
| Mg2+ | 101.18 ± 3.39 |
| Mn2+ | 140.00 ± 0.27 |
| Cu2+ | 35.88 ± 2.64 |
| E64 | 82.16 ± 5.30 |
| 2-ME | 88.89 ± 7.04 |
| EDTA | 86.36 ± 2.31 |
| 1,10-Phenanthroline | 82.68 ± 5.22 |
| PMSF | 88.56 ± 8.24 |
| SDS | 15.39 ±5.50 |
| CTAB | 86.13 ± 3.14 |
| Triton X-100 | 117.98 ± 5.19 |
| Tween 20 | 104.34 ± 3.33 |
| Tween 40 | 90.38 ± 5.20 |
| Enzyme Concentration | ||||
|---|---|---|---|---|
| 0 U | 1 U | 5 U | 10 U | |
| Clarification (T%) | 34.54 ± 0.62 | 62.52 ± 1.54 | 68.19 ± 1.85 | 71.33 ± 1.92 |
| Reducing sugar (mM) | 0.82 ± 0.08 | 1.02 ± 0.09 | 1.31 ± 0.07 | 1.51 ± 0.10 |
| DPPH radical scavenging activity (%) | 72.78 ± 1.83 | 71.57 ± 1.45 | 70.92 ± 1.30 | 70.02 ± 1.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, C.T.; Tran, T.N.; Pham, T.P.; Tran, T.T.T.; Truong, B.P.; Nguyen, T.T.; Nguyen, T.M.; Bui, T.Q.H.; Nguyen, A.D.; Wang, S.-L. Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii. Polymers 2024, 16, 2037. https://doi.org/10.3390/polym16142037
Doan CT, Tran TN, Pham TP, Tran TTT, Truong BP, Nguyen TT, Nguyen TM, Bui TQH, Nguyen AD, Wang S-L. Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii. Polymers. 2024; 16(14):2037. https://doi.org/10.3390/polym16142037
Chicago/Turabian StyleDoan, Chien Thang, Thi Ngoc Tran, Thi Phuong Pham, Thi Thanh Thao Tran, Ba Phong Truong, Thi Tinh Nguyen, The Manh Nguyen, Thi Quynh Hoa Bui, Anh Dzung Nguyen, and San-Lang Wang. 2024. "Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii" Polymers 16, no. 14: 2037. https://doi.org/10.3390/polym16142037
APA StyleDoan, C. T., Tran, T. N., Pham, T. P., Tran, T. T. T., Truong, B. P., Nguyen, T. T., Nguyen, T. M., Bui, T. Q. H., Nguyen, A. D., & Wang, S.-L. (2024). Production, Purification, and Characterization of a Cellulase from Paenibacillus elgii. Polymers, 16(14), 2037. https://doi.org/10.3390/polym16142037










