Polymer Translocation through Nanometer Pores
Abstract
:1. Introduction
2. Theoretical Model
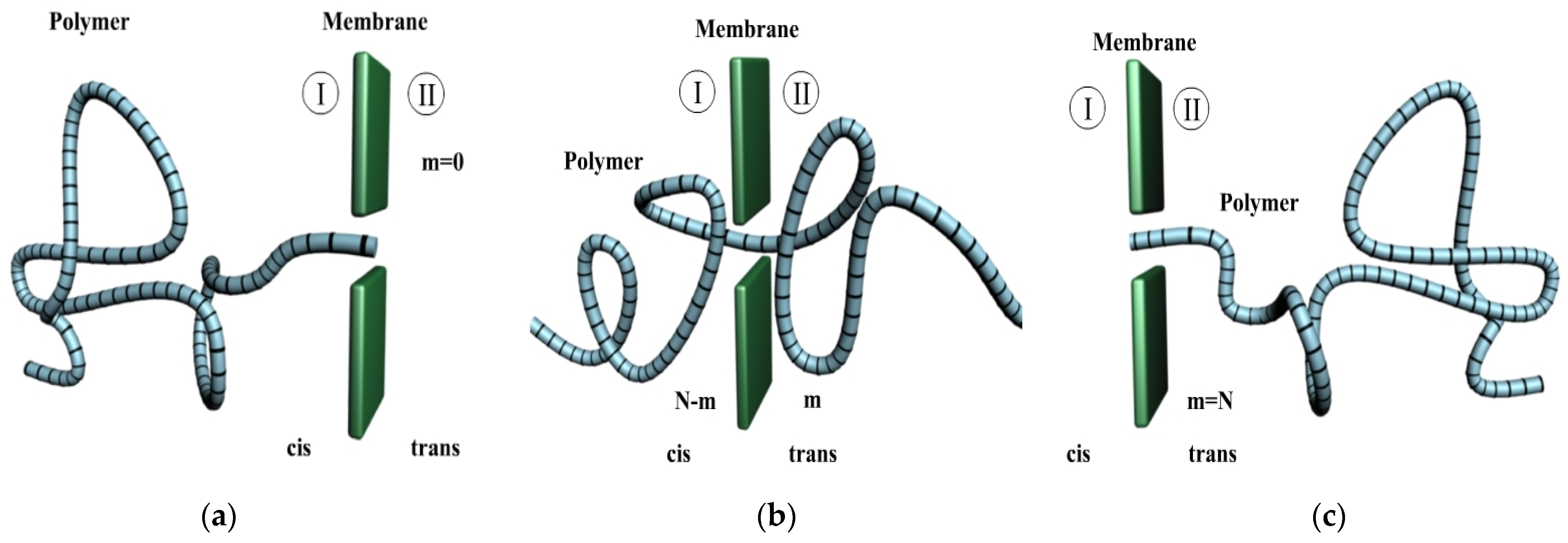
2.1. Charged Polymer Translocation Process
- computation of time-evolution of translocation τ,
- obtaining of the polymer flux through the pore J,
- (I)
- the free energy barrier estimation,
- (II)
- the polymer evolution equations,
- (III)
- the flux probability of translocation across the barrier.
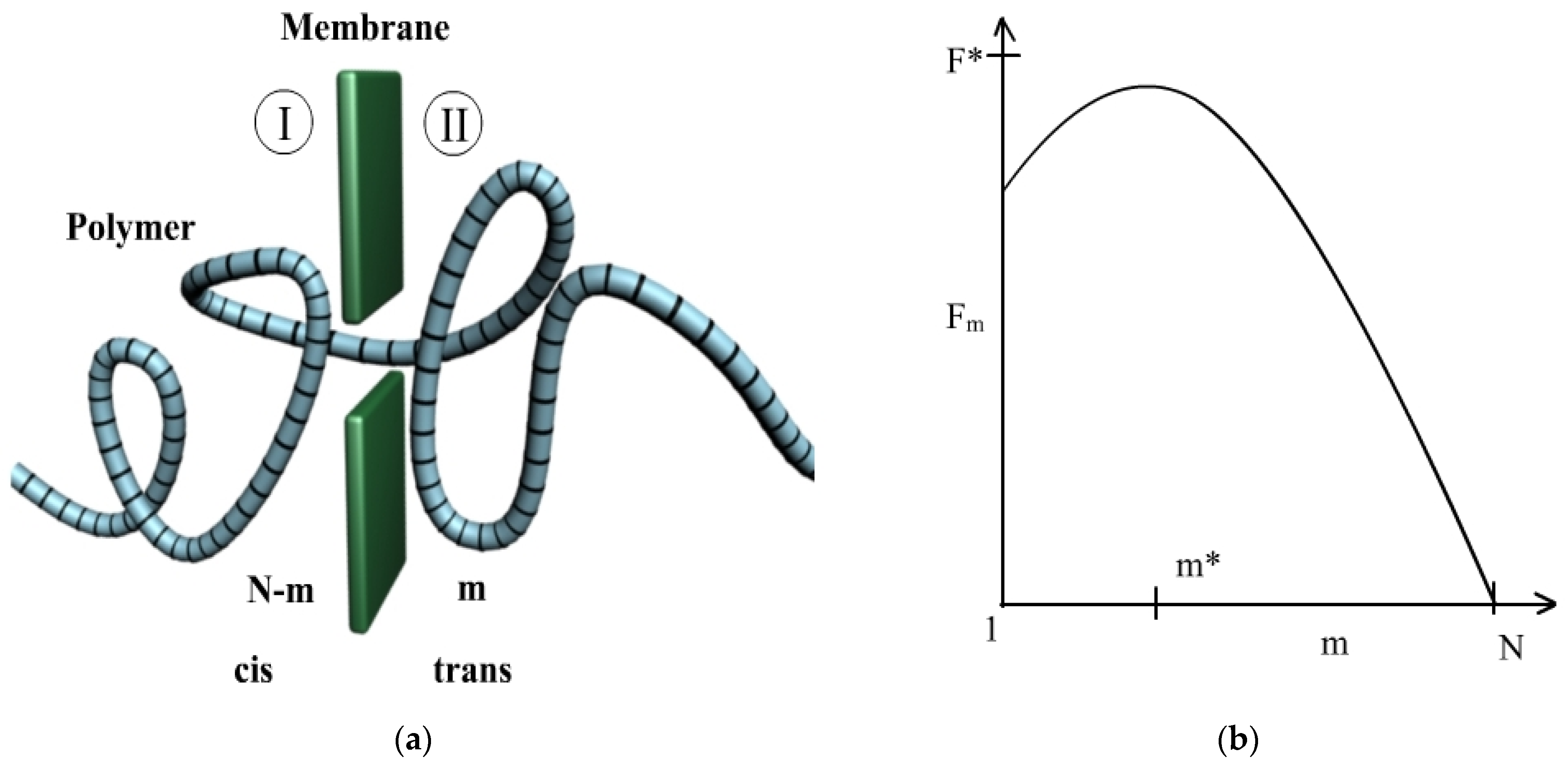
2.1.1. Free Energy
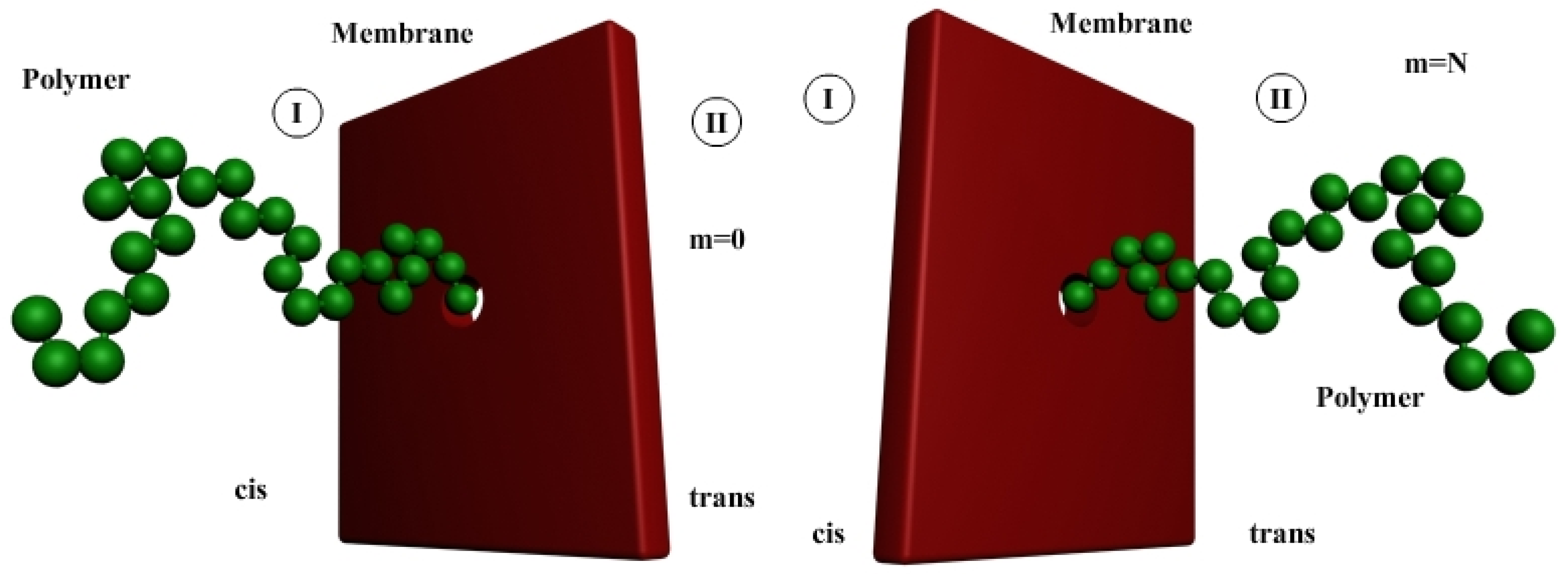
2.1.2. Polymer Translocation through Pores
2.1.3. Flux of Polymers
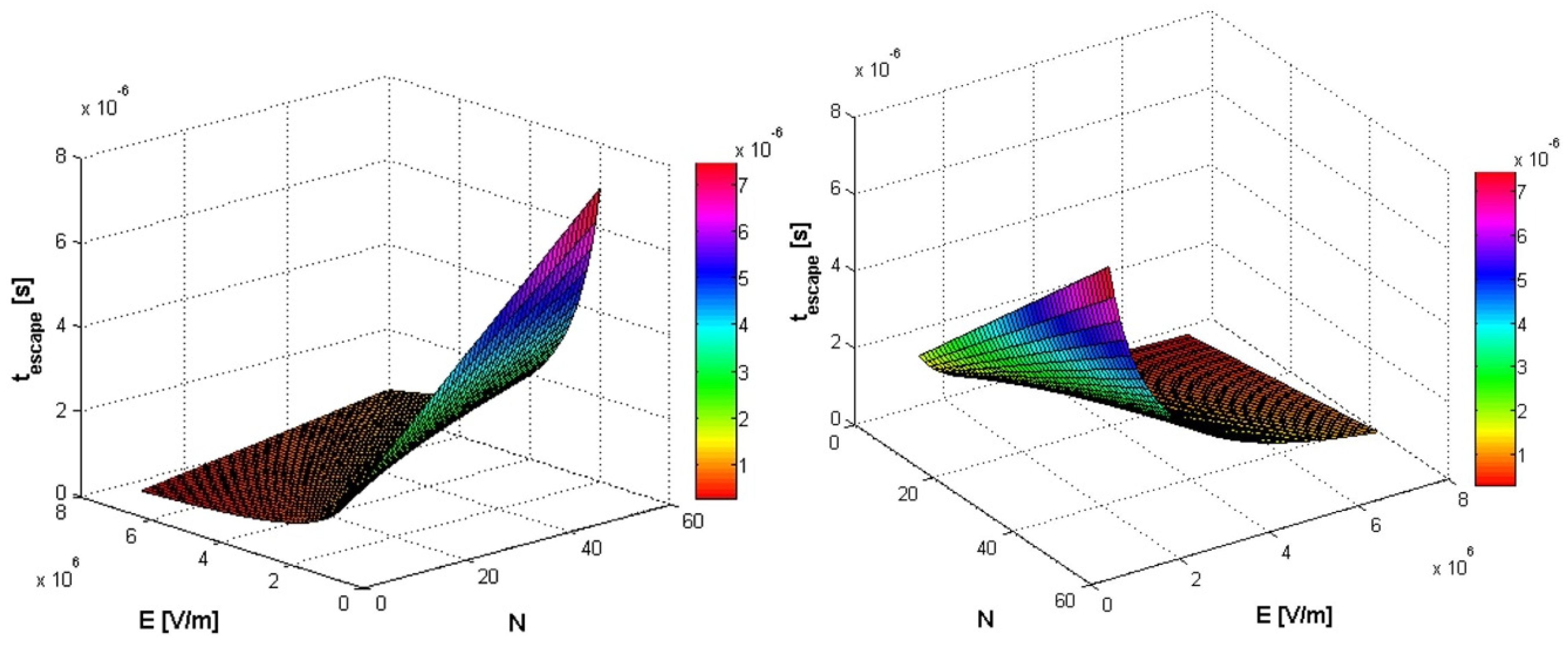
2.1.4. Results and Discussion I
2.2. Linear Polymers Transport under Constant Force
Results and Discussion II
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ambjörnsson, T.; Apell, S.P.; Konkoli, Z.; Di Marzio, E.A.; Kasianowicz, J.J. Charged polymer membrane translocation. J. Chem. Phys. 2002, 117, 4063–4073. [Google Scholar] [CrossRef]
- Sung, W.; Park, P.J. Polymer Translocation through a Pore in a Membrane. Phys. Rev. Lett. 1996, 77, 783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paun, V.P. An estimation of the polymer translocation time through membrane articol. Mater. Plast. 2006, 43, 57–58. [Google Scholar]
- Muthukumar, M. Polymer translocation through a hole. J. Chem. Phys. 1999, 111, 10371–10374. [Google Scholar] [CrossRef]
- Kong, C.Y.; Muthukumar, M. Monte Carlo study of adsorption of a polyelectrolyte onto charged surfaces. J. Chem. Phys. 1998, 109, 1522. [Google Scholar] [CrossRef]
- Muthukumar, M. Translocation of a Confined Polymer through a Hole. Phys. Rev. Lett. 2001, 86, 3188–3191. [Google Scholar] [CrossRef] [PubMed]
- Sofos, F.; Karakasidis, T.E.; Liakopoulos, A. How wall properties control diffusion in grooved nanochannels: A molecular dynamics study. Heat Mass Transf. 2013, 49, 1081–1088. [Google Scholar] [CrossRef]
- Sofos, F.; Karakasidis, T.E.; Spetsiotis, D. Molecular Dynamics simulations of ion separation in nano-channel water flows using an electric field. Mol. Simul. 2019, 45, 1395–1402. [Google Scholar] [CrossRef]
- Sofos, F.; Karakasidis, T.E.; Liakopoulos, A. Fluid flow at the nanoscale: How fluid properties deviate from the bulk. Nanosci. Nanotechnol. Lett. 2013, 5, 457–460. [Google Scholar] [CrossRef]
- Doi, M.; Edwards, S.F. Theory of Polymer Dynamics; Clarendon Press: Oxford, UK, 1986. [Google Scholar]
- Ma, S.K. Statistical Mechanic; World Scientific: Singapore, 1985. [Google Scholar]
- Kasianowicz, J.J.; Brandin, E.; Branton, D.; Deamer, D.W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA 1996, 93, 13770–13773. [Google Scholar] [CrossRef] [Green Version]
- Meller, A.; Nivon, L.; Branton, D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 2001, 86, 3435–3438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paun, V.P. Theoretical Study of the Polymer Transport through Nanopores. Rev. Chim. 2006, 57, 221–223. [Google Scholar]
- Buyukdagli, S.; Sarabadani, J.; Ala-Nissila, T. Theoretical Modeling of Polymer Translocation: From the Electrohydrodynamics of Short Polymers to the Fluctuating Long. Polymers 2019, 11, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghosh, B.; Chaudhury, S. Translocation Dynamics of an Asymmetrically Charged Polymer through a Pore under the Influence of Different pH Conditions. J. Phys. Chem. B 2019, 123, 4318–4323. [Google Scholar] [CrossRef] [PubMed]
- Buyukdagli, S.; Ala-Nissila, T. Controlling polymer capture and translocation by electrostatic polymer-pore interactions. J. Chem. Phys. 2017, 147, 114904. [Google Scholar] [CrossRef]
- Paun, V.P. Polymer dynamics simulation at nanometer scale in a 2D diffusion model. Mater. Plast. 2007, 44, 393–395. [Google Scholar]
- Paun, V.-P.; Chiroiu, V.; Munteanu, L. Polymer Transport Process through Biological Membranes with Nanometric Pores. In “Progress in Nanoscience and Nanotechnologies” Monograph; Kleps, I., Ion, A.C., Dascalu, D., Eds.; Romanian Academy Press: Bucharest, Romanian, 2007; pp. 267–275. [Google Scholar]
- Hamidabad, M.N.; Abdolvahab, R.H. Translocation through a narrow pore under a pulling force. Sci. Rep. 2019, 9, 17885. [Google Scholar] [CrossRef] [Green Version]
- Menais, T. Polymer translocation under a pulling force: Scaling arguments and threshold forces. Phys. Rev. E 2018, 97, 022501. [Google Scholar] [CrossRef] [Green Version]
- Paun, V.P. Two-dimensional diffusion model for the biopolymers dynamics at nanometer scale. Open Phys. 2009, 7, 607–613. [Google Scholar] [CrossRef]
- Ermak, D.L.; Buckholz, H. Numerical integration of the Langevin equation: Monte Carlo simulation. J. Comput. Phys. 1980, 35, 169–182. [Google Scholar] [CrossRef]
- Van Gunsteren, W.F.; Berendsen, H.J.C. Algorithms for Brownian dynamics. Mol. Phys. 1982, 45, 637–647. [Google Scholar] [CrossRef]
- Brünger, A.; Brooks III, C.L.; Karplus, M. Stochastic boundary conditions for molecular dynamics simulations of ST2 water. Chem. Phys. Lett. 1984, 105, 495–500. [Google Scholar] [CrossRef]
- Bou-Rabee, N. Time Integrators for Molecular Dynamics. Entropy 2014, 16, 138–162. [Google Scholar] [CrossRef] [Green Version]
- Hamidabad, M.N.; Asgari, S.; Abdolvahab, R.H. Nanoparticle-assisted polymer translocation through a nanopore. Polymer 2020, 204, 122847. [Google Scholar] [CrossRef]
- Meller, A.; Nivon, L.; Brandin, E.; Golovchenko, J.A.; Branton, D. Rapid nanopore discrimination between single polynucleotide molecules. Proc. Natl. Acad. Sci. USA 2000, 97, 1079. [Google Scholar] [CrossRef] [Green Version]
- Meller, A.; Branton, D. Single Molecule Measurements of DNA Transport through a Nanopore. Electrophoresis 2002, 23, 2583. [Google Scholar] [CrossRef]
- Luo, K.F.; Ala-Nissila, T.; Ying, S.C.; Bhattacharya, A. Sequence dependence of DNA translocation through a nanopore. Phys. Rev. Lett. 2008, 100, 058101. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paun, M.-A.; Paun, V.-A.; Paun, V.-P. Polymer Translocation through Nanometer Pores. Polymers 2022, 14, 1166. https://doi.org/10.3390/polym14061166
Paun M-A, Paun V-A, Paun V-P. Polymer Translocation through Nanometer Pores. Polymers. 2022; 14(6):1166. https://doi.org/10.3390/polym14061166
Chicago/Turabian StylePaun, Maria-Alexandra, Vladimir-Alexandru Paun, and Viorel-Puiu Paun. 2022. "Polymer Translocation through Nanometer Pores" Polymers 14, no. 6: 1166. https://doi.org/10.3390/polym14061166
APA StylePaun, M.-A., Paun, V.-A., & Paun, V.-P. (2022). Polymer Translocation through Nanometer Pores. Polymers, 14(6), 1166. https://doi.org/10.3390/polym14061166







