Molecular Imprinted ZnS Quantum Dots-Based Sensor for Selective Sulfanilamide Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Instrumentation
2.2. Synthesis of MEA-Modified Mn2+: ZnS QDs
2.3. Preparation of MIP@QDs Nanocomposites
2.4. Effect of pH Value of Solutions
2.5. Fluorescence Measurement of SAM
2.6. Selectivity Experiments
2.7. Real Sample Analysis
3. Results
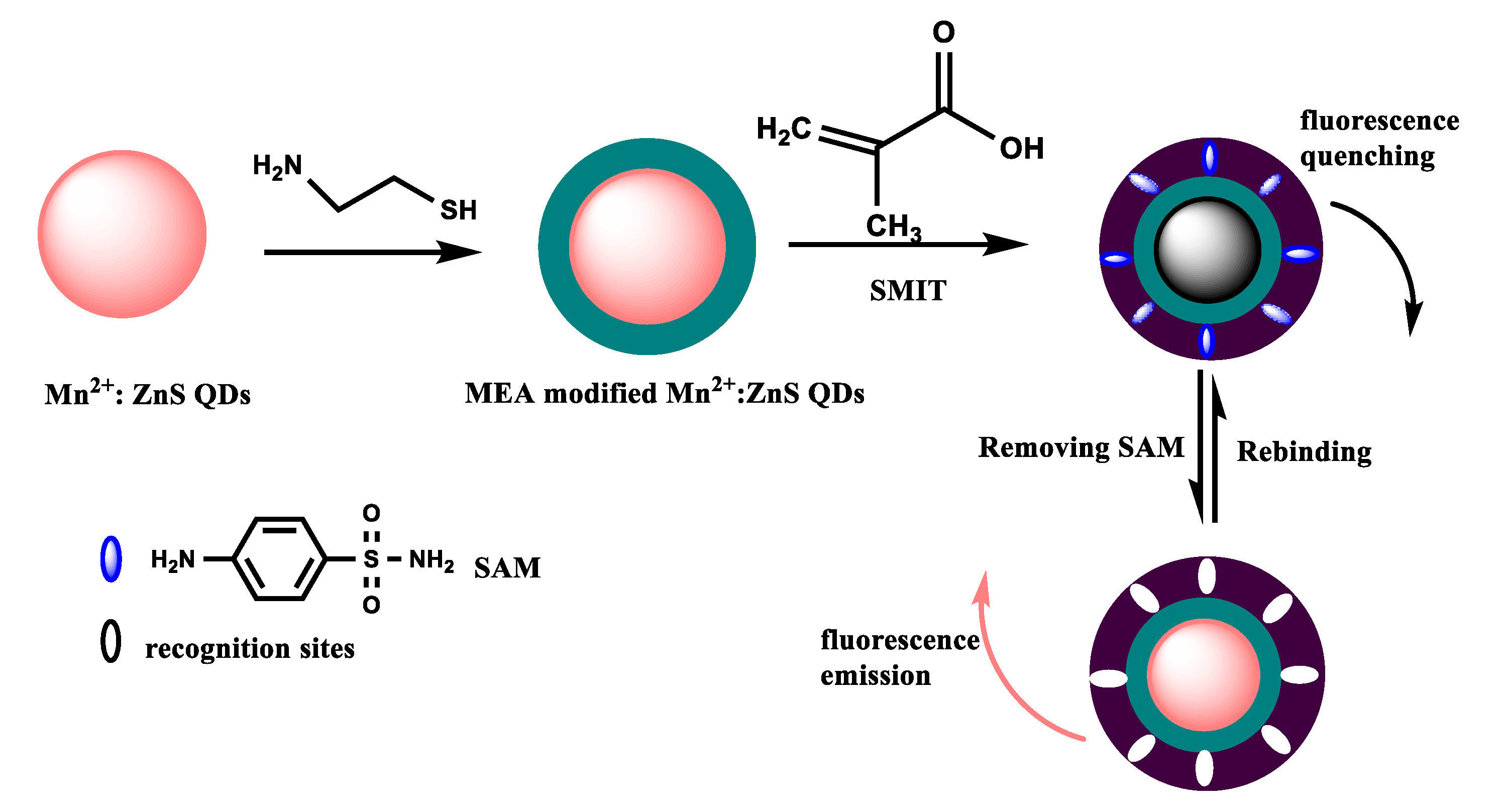
3.1. Fabrication of MIP@QDs-Based Sensor
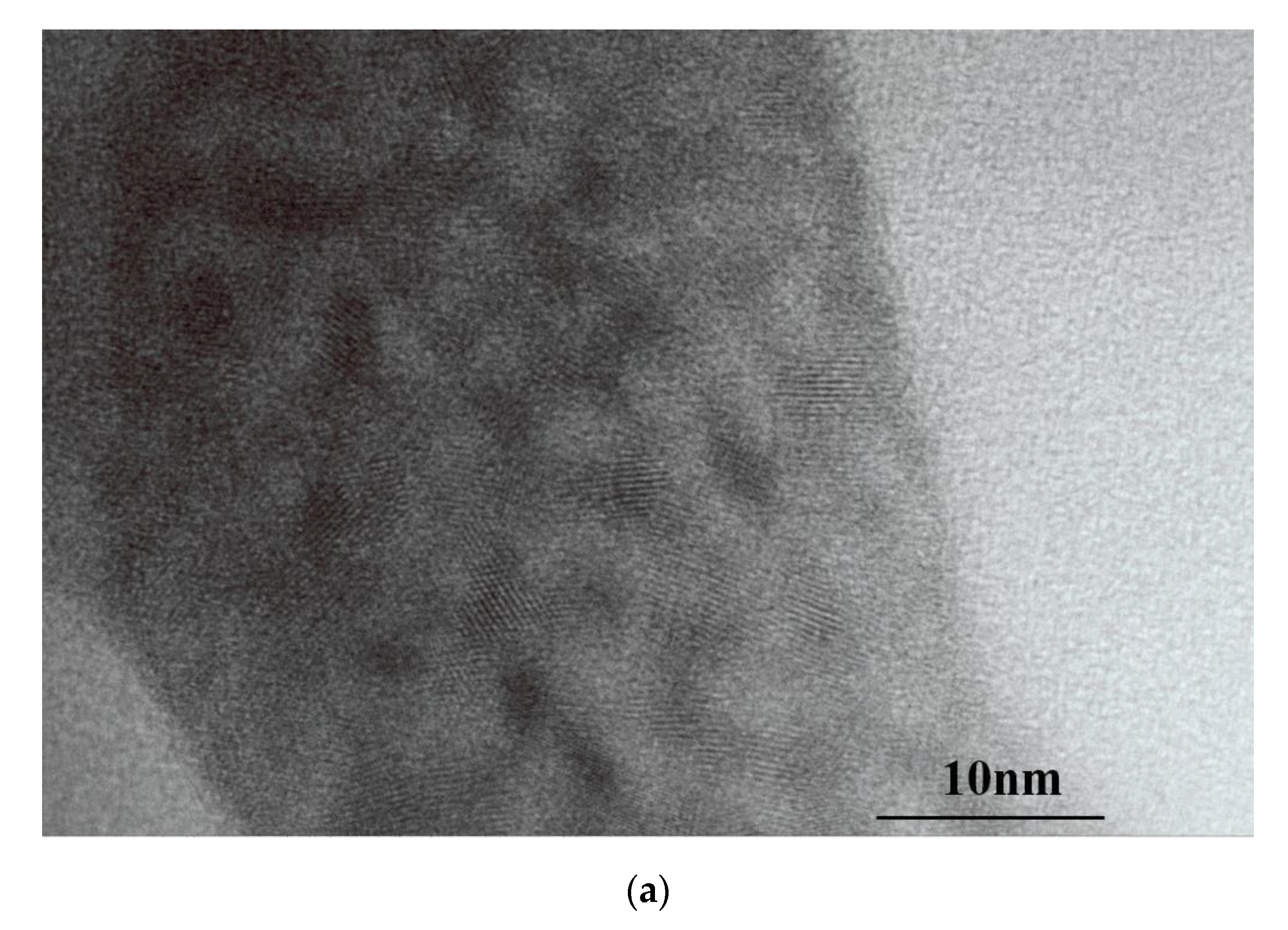
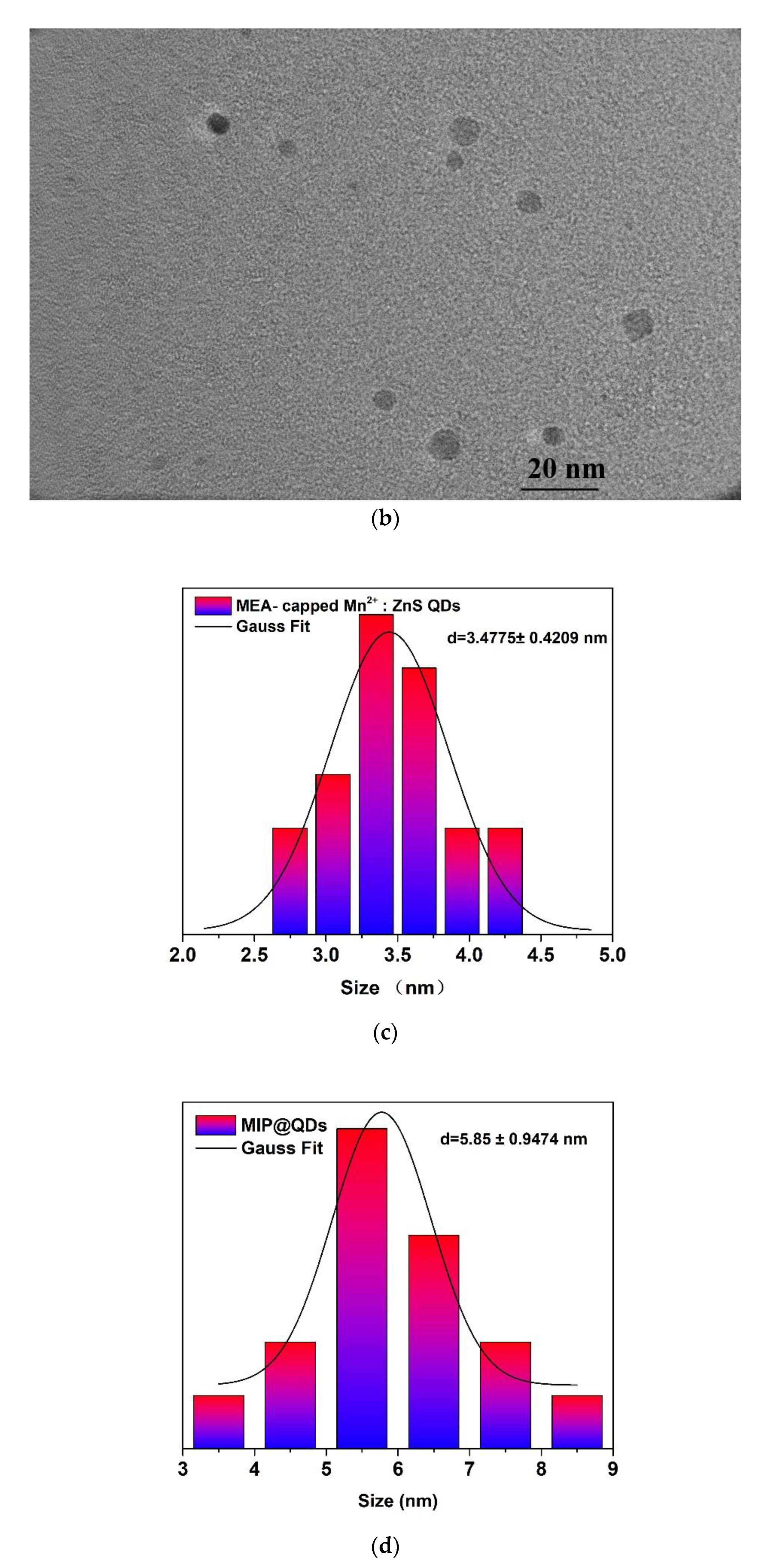
3.2. Characterization
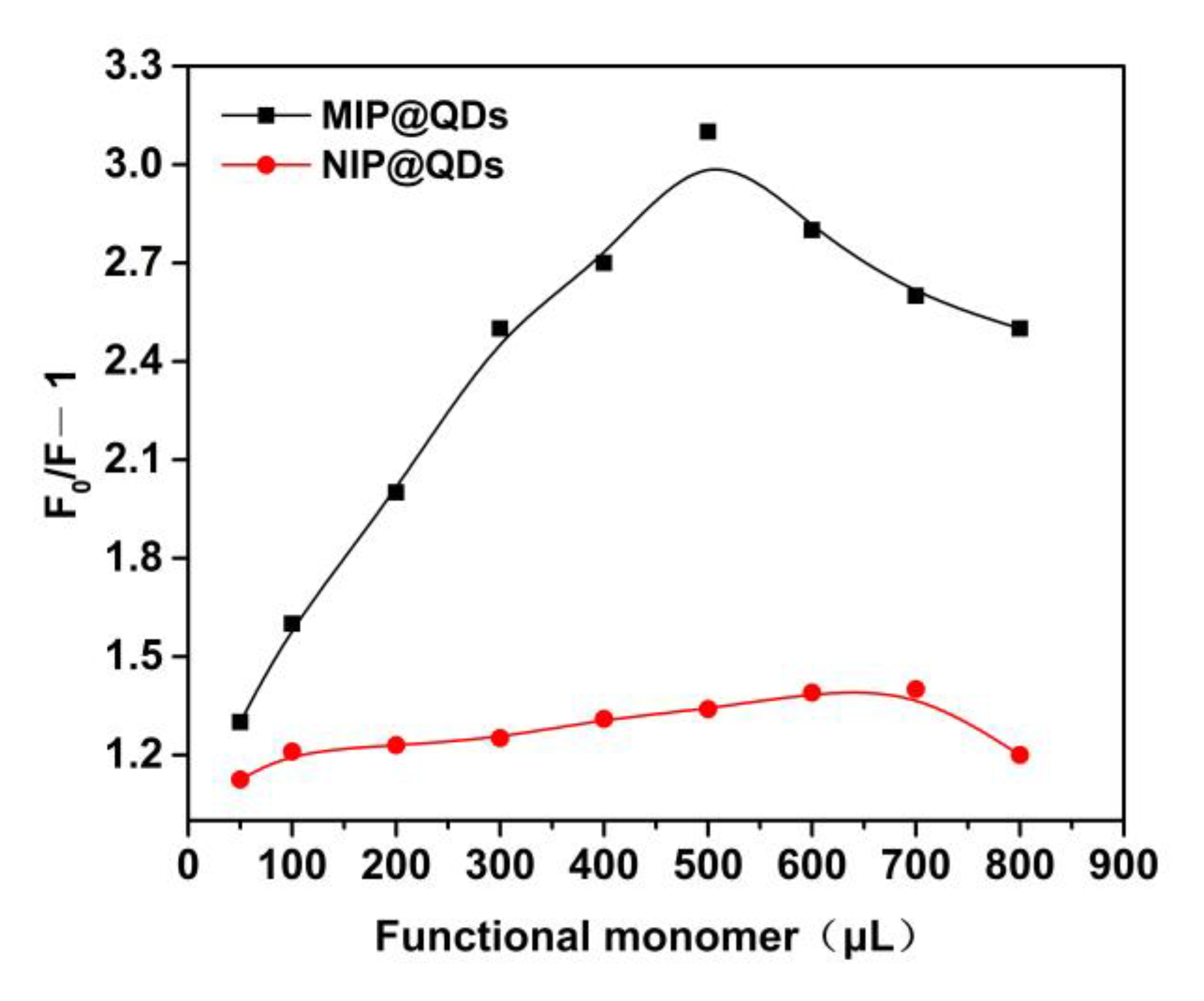
3.3. Effect of the Amount of Functional Monomer
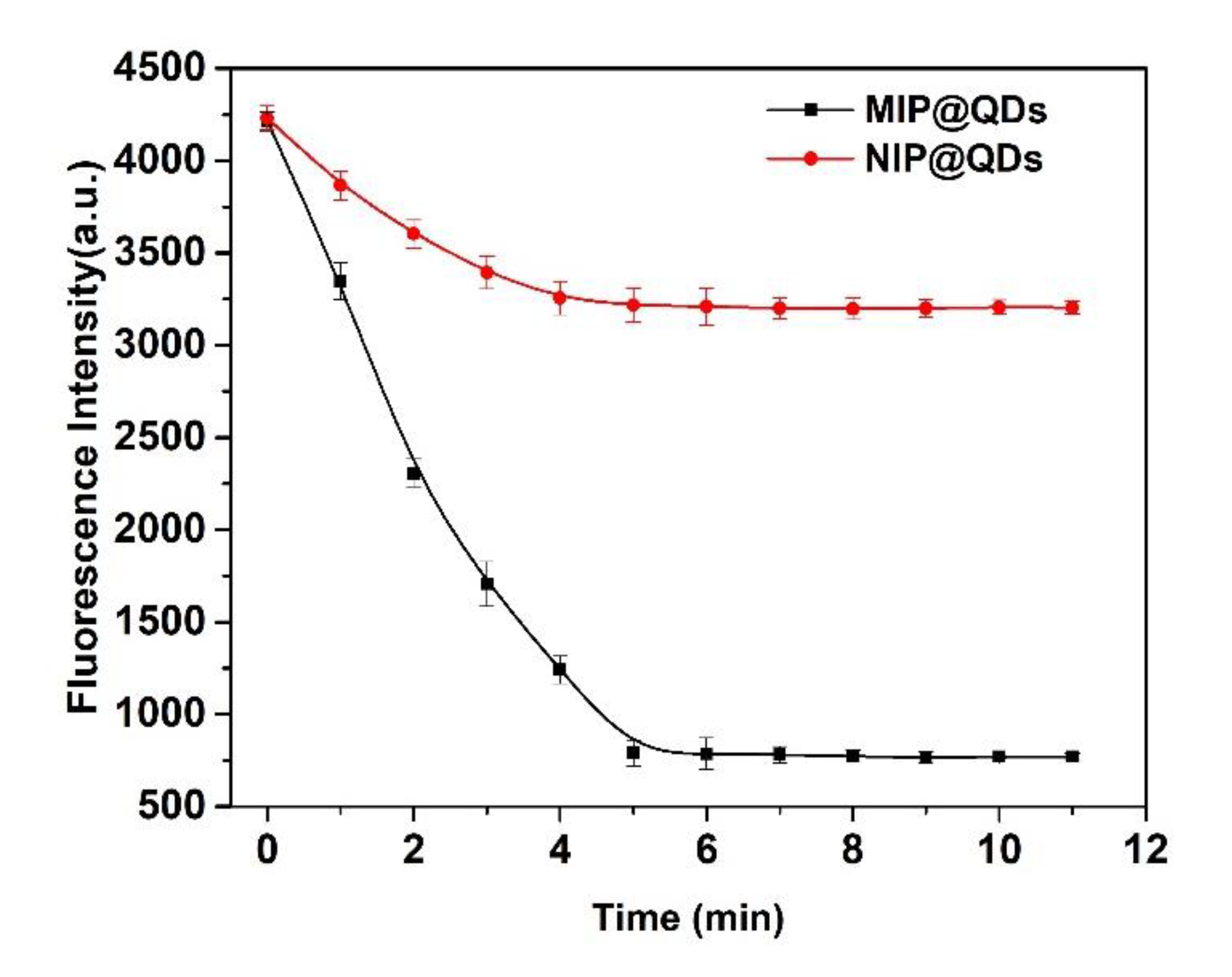
3.4. Response Time
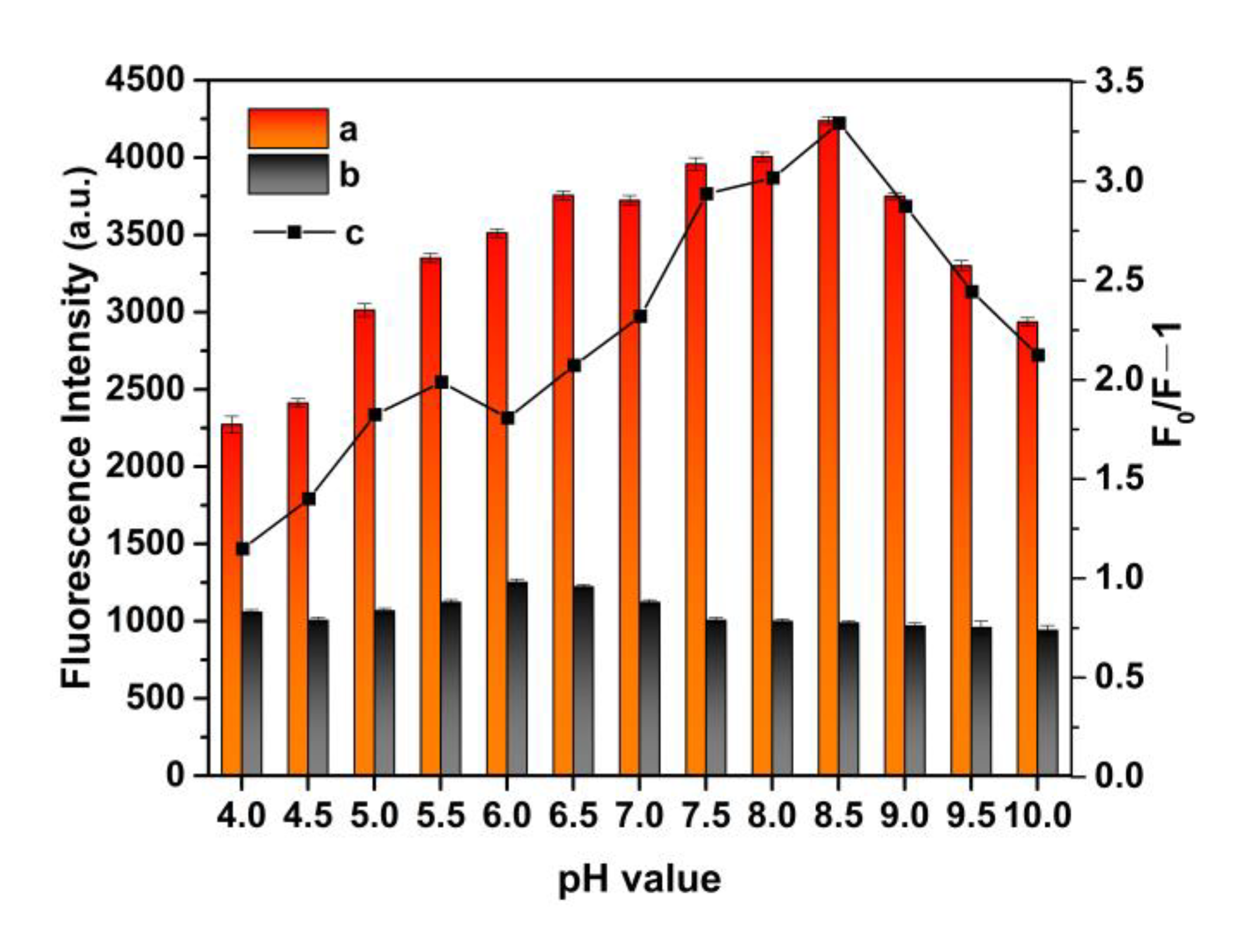
3.5. Effect of pH Value on MIP@QDs
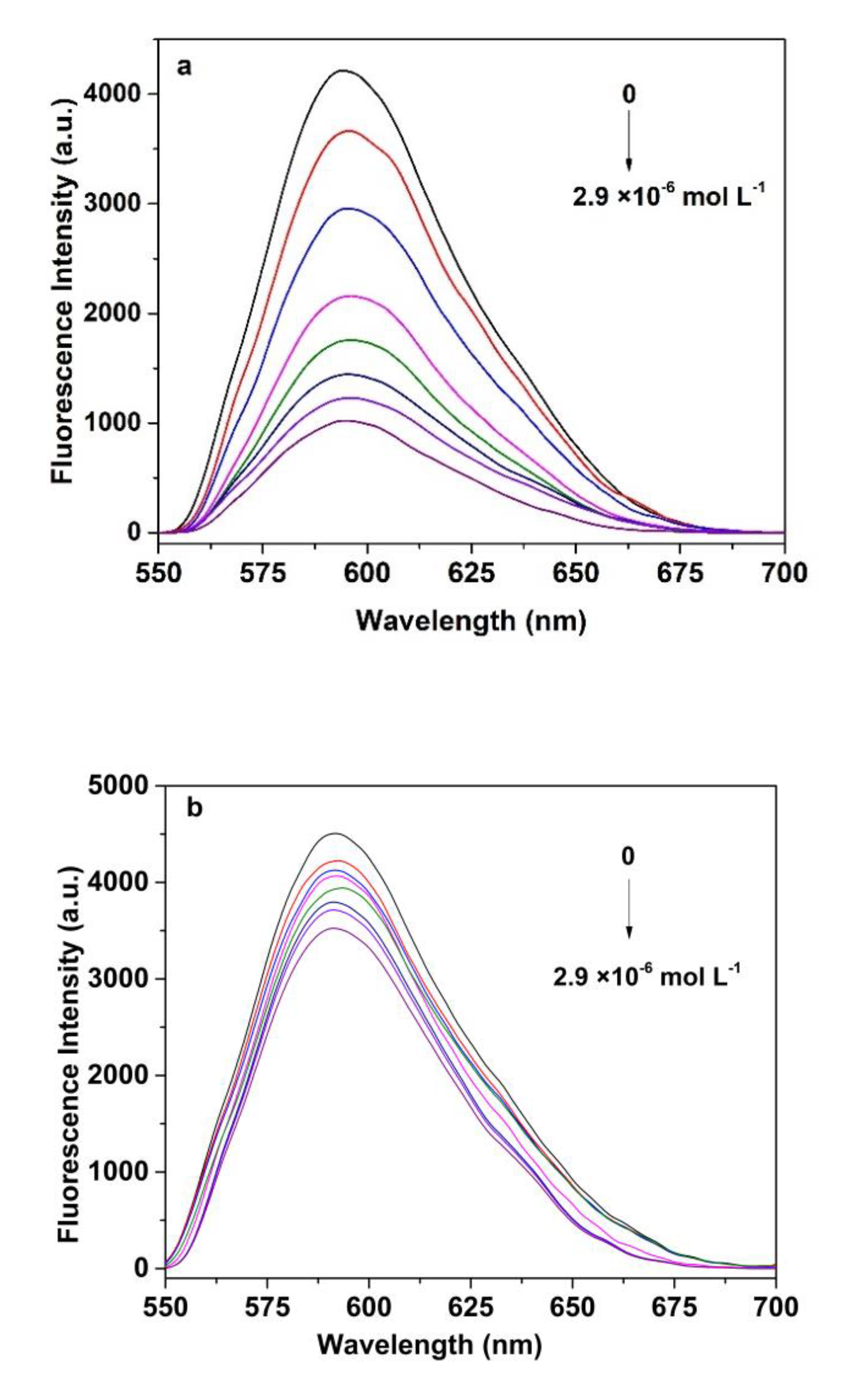
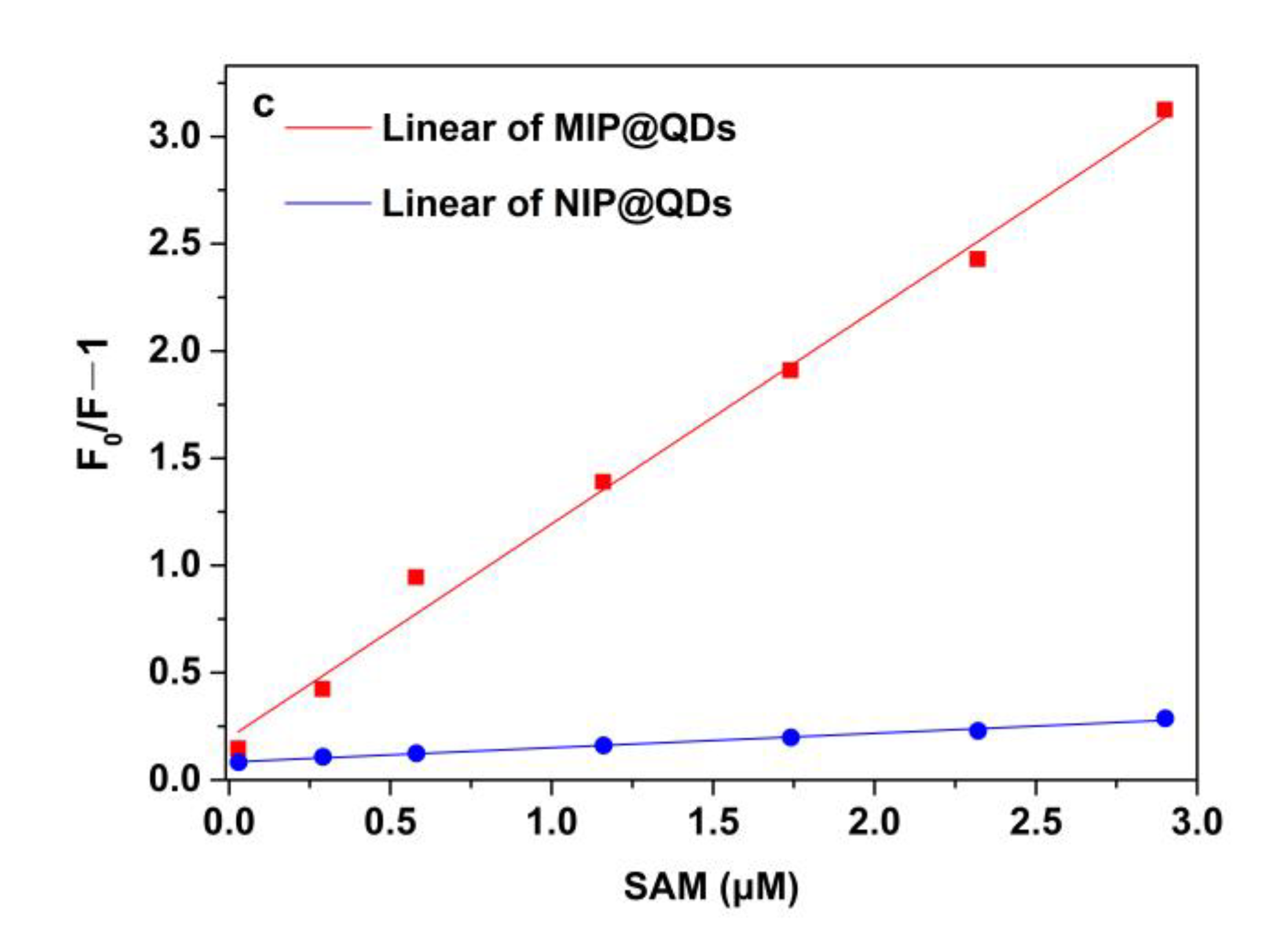
3.6. Fluorescence Measurement of SAM
3.7. Selectivity Experiments
3.8. Application
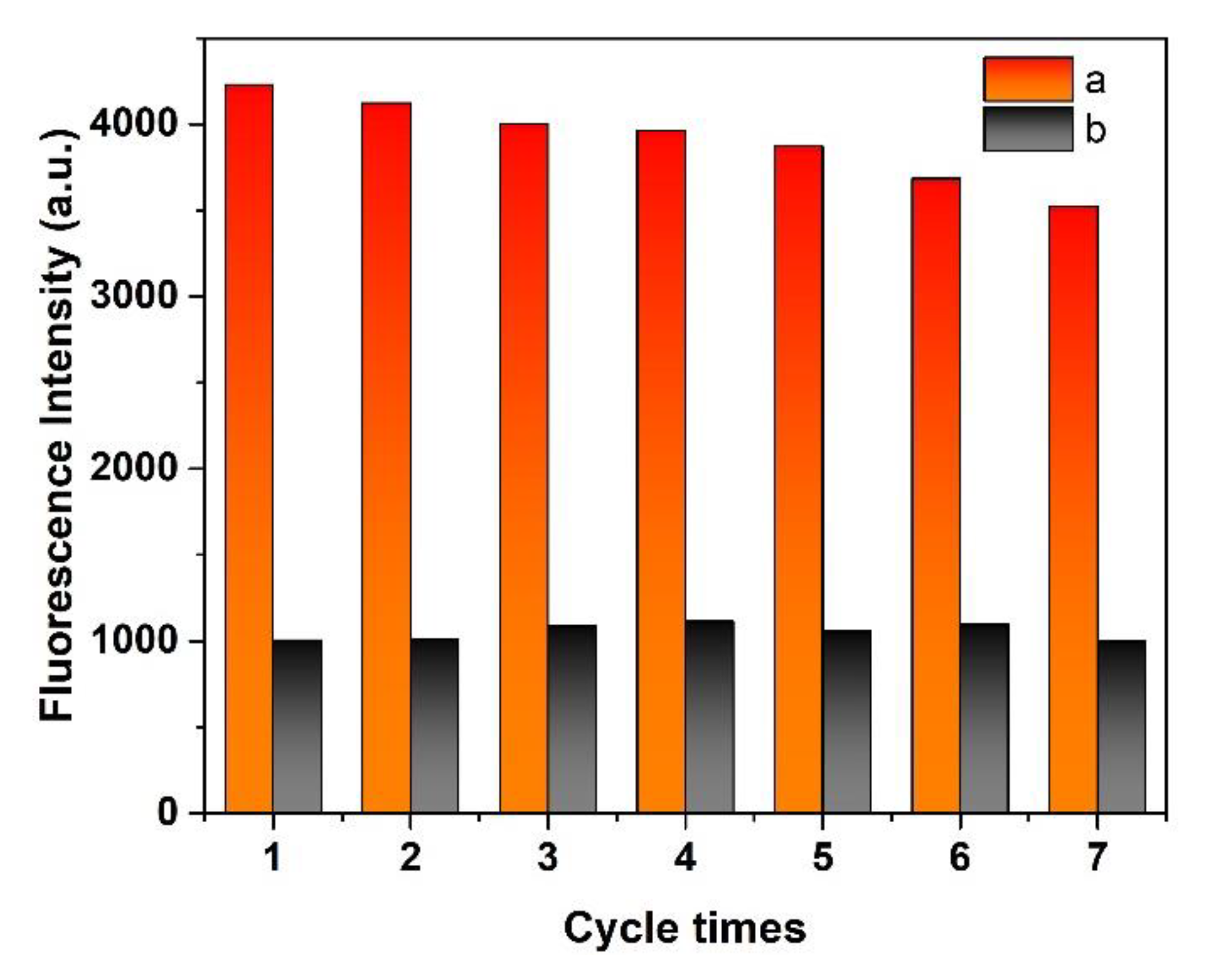
3.9. Recyclability and Stability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wang, S.; Zhang, H.Y.; Wang, L.; Duan, Z.J.; Kennedy, I. Analysis of sulphonamide residues in edible animal products: A review. Food Addit. Contam. 2006, 23, 362–384. [Google Scholar] [CrossRef] [PubMed]
- Dmitrienko, S.G.; Kochuk, E.V.; Apyari, V.V.; Tolmacheva, V.V.; Zolotov, Y.A. Recent advances in sample preparation techniques and methods of sulfonamides detection—A review. Anal. Chim. Acta 2014, 850, 6–25. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, P.S.; Tóth, I.V.; Segundo, M.A.; Lima, J.L.F. Fluoroquinolones and sulfonamides: Features of their determination in water. A review. Int. J. Environ. Anal. Chem. 2016, 96, 182–202. [Google Scholar] [CrossRef]
- Fu, L.; Zhang, X.Y.; Ding, S.; Chen, F.; Lv, Y.F.; Zhang, H.W.; Zhao, S.C. Recent Developments in the Electrochemical Determination of Sulfonamides. Curr. Pharm. Anal. 2022, 18, 4–13. [Google Scholar] [CrossRef]
- Boxall, A.B.A.; Kolpin, D.W.; Holling-Sorensen, B.; Tolls, J. Peer Reviewed: Are Veterinary Medicines Causing Environmental Risks? J. Environ. Sci. Technol. 2003, 37, 286–294. [Google Scholar] [CrossRef]
- Chatzimitakos, T.; Stalikas, C. Zinc ferrite as a magnetic sorbent for the dispersive micro solid-phase extraction of sulfonamides and their determination by HPLC. Microchem. J. 2020, 155, 104670. [Google Scholar] [CrossRef]
- Zhou, Q.X.; Fang, Z. Highly sensitive determination of sulfonamides in environmental water samples by sodium dodecylbenzene sulfonate enhanced micro-solid phase extraction combined with high performance liquid chromatography. Talanta 2015, 141, 170–174. [Google Scholar] [CrossRef]
- Hu, S.P.; Zhao, M.; Xi, Y.Y.; Mao, Q.Q.; Zhou, X.D.; Chen, D.W.; Yan, P.C. Non-targeted screening and determination of sulfonamides: A dispersive micro solid-phase extraction approach to the analysis of milk and honey samples using liquid chromatography- high resolution mass spectrometry. J. Agric. Food Chem. 2017, 65, 1984–1991. [Google Scholar] [CrossRef]
- Turco, A.; Corvaglia, S.; Mazzotta, E.; Pompa, P.P.; Malitesta, C. Preparation and characterization of molecularly imprinted mussel inspired film as antifouling and selective layer for electrochemical detection of sulfamethoxazole. Sens. Actuators B-Chem. 2018, 255, 3374–3383. [Google Scholar] [CrossRef]
- Liu, C.; Song, D.; Yang, Z.; Wang, Z.; Peng, P.; Liu, J.; Yang, X.; Li, R.; Zhu, Z.; Xue, F. Research on advanced methods of electrochemiluminescence detection combined with optical imaging analysis for the detection of sulfonamides. Analyst 2021, 146, 7611–7617. [Google Scholar] [CrossRef]
- Beitollahi, H.; Tajik, S.; Bartolomeo, A.D. Application of MnO2 Nanorod–Ionic Liquid Modified Carbon Paste Electrode for the Voltammetric Determination of Sulfanilamide. Micromachines 2022, 13, 598. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Kuang, X.; Shen, X.; Zhu, J.; Zhang, B.; Li, H. Improvement of Voltammetric Detection of Sulfanilamide with a Nanodiamond−Modified Glassy Carbon Electrode. Int. J. Electrochem. Sci. 2019, 14, 7858–7870. [Google Scholar] [CrossRef]
- Liang, L.; Li, C.; Zhu, J.; Song, X.; Yu, W.; Zhang, J.; Zhang, S.; Shen, J.; Wang, Z. Dihydropteroate synthase based sensor for screening multi-sulfonamides residue and its comparison with broad-specific antibody based immunoassay by molecular modeling analysis. Anal. Chim. Atca 2019, 1050, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, L.; Xu, L.; Song, S.; Kuang, H.; Cui, G.; Xu, C. Gold immunochromatographic sensor for the rapid detection of twenty-six sulfonamides in foods. Nano Res. 2017, 10, 2833–2844. [Google Scholar] [CrossRef]
- Li, C.; Luo, X.; Li, Y.; Yang, H.; Liang, X.; Wen, K.; Cao, Y.; Li, C.; Wang, W.; Shi, W.; et al. A Class-selective immunoassay for sulfonamides residue detection in milk using a superior polyclonal antibody with broad specificity and highly uniform affinity. Molecules 2019, 24, 443. [Google Scholar] [CrossRef]
- Dai, T.; Duan, J.; Li, X.; Xu, X.; Shi, H.; Kang, W. determination of sulfonamide residues in food by capillary zone electrophoresis with on-line chemiluminescence detection based on an ag(iii) complex. Int. J. Mol. Sci. 2017, 18, 1286. [Google Scholar] [CrossRef]
- Yang, L.; Ni, H.J.; Li, X.Z.; Zhang, X.Y.; Wen, K.; Ke, K.; Yang, H.; Shi, W.M.; Zhang, S.; Shen, J.Z.; et al. Development of a highly specific chemiluminescence aptasensor for sulfamethazine detection in milk based on in vitro selected aptamers. Sens. Actuators B-Chem. 2019, 281, 801–811. [Google Scholar] [CrossRef]
- Li, X.; Li, Q.; Xue, A.; Chen, H.; Li, S. Dispersive liquid–liquid microextraction coupled with single-drop microextraction for the fast determination of sulfonamides in environmental water samples by high performance liquid chromatography-ultraviolet detection. Anal. Methods 2016, 8, 517–525. [Google Scholar] [CrossRef]
- Dmitrienko, S.G.; Kochuk, E.V.; Tolmacheva, V.V.; Apyari, V.V.; Zolotov, Y.A. Determination of the total content of some sulfonamides in milk using solid-phase extraction coupled with off-line derivatization and spectrophotometric detection. Food Chem. 2015, 188, 51–56. [Google Scholar] [CrossRef]
- Nasir, A.N.M.; Yahaya, N.; Zain, N.N.M.; Lim, V.; Kamaruzaman, S.; Saad, B.; Nishiyama, N.; Yoshida, N.; Hirota, Y. Thiol-functionalized magnetic carbon nanotubes for magnetic micro-solid phase extraction of sulfonamide antibiotics from milks and commercial chicken meat products. Food Chem. 2019, 144, 458–466. [Google Scholar] [CrossRef]
- Shen, F.; Xu, Y.J.; Wang, Y.; Chen, J.; Wang, S. Rapid and ultra-trace levels analysis of 33 antibiotics in water by on-line solid-phase extraction with ultra-performance liquid chromatography-tandem mass spectrometry. J. Chromatogr. A 2022, 1677, 463304. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Wang, X.; Ming, M.; Li, J.; Ye, N. Determination of sulfonamides in milk by capillary electrophoresis with PEG@MoS2 as a dispersive solid-phase extraction sorbent. R. Soc. Open Sci. 2018, 5, 172104. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lv, Y.; Chen, Y.; Li, Y.; Wang, Y.; Zhao, H. Trimetallic Ag@Pt-Rh core-shell nanocubes modified anode for voltammetric sensing of dopamine and sulfanilamide. Chem. Eng. Sci. 2022, 249, 117326. [Google Scholar] [CrossRef]
- Vanoni, C.R.; Winiarski, J.P.; Nagurniak, G.R.; Magosso, H.A.; Jost, C.L. A novel electrochemical sensor based on silsesquioxane/nickel (ii) phthalocyanine for the determination of sulfanilamide in clinical and drug samples. Electroanalysis 2019, 31, 867–875. [Google Scholar] [CrossRef]
- He, B.; Chen, W. Carboxyl multiwalled carbon nanotubes through ultrasonic dispersing in dimethylfomamide modified electrode as a sensitive amperometric sensor for detection of sulfonamide. Int. J. Electrochem. Sci. 2015, 10, 4335–4345. [Google Scholar]
- Faria, L.V.; Lisboa, T.P.; Matias, T.A.; Sousa, R.A.; Matos, M.A.C.; MunoZ, R.A.A.; Matos, R.C. Use of reduced graphene oxide for sensitive determination of sulfanilamide in synthetic biological fluids and environmental samples by batch injection analysis. J. Electroanal. Chem. 2021, 892, 115298. [Google Scholar] [CrossRef]
- Alivisatos, A.P. Semiconductor Clusters, Nanocrystals, and Quantum Dots. Science 1996, 271, 933–937. [Google Scholar] [CrossRef]
- Chen, H.; Lin, L.; Li, H.; Lin, J.M. Quantum dots-enhanced chemiluminescence: Mechanism and application. Coord. Chem. Rev. 2013, 263, 86–100. [Google Scholar] [CrossRef]
- Zhang, Y.; Hou, D.; Wang, Z.; Cai, N.; Au, C. Nanomaterial-based dual-emission ratiometric fluorescent sensors for biosensing and cell imaging. Polymers 2021, 13, 2540. [Google Scholar] [CrossRef]
- Chen, P.; Qu, R.; Peng, W.; Wang, X.; Huang, K.; He, Y.; Zhang, X.; Meng, Y.; Liu, T.; Chen, J.; et al. Visual and dual-fluorescence homogeneous sensor for the detection of pyrophosphatase in clinical hyperthyroidism samples based on selective recognition of CdTe QDs and coordination polymerization of Ce. J. Mater. Chem. C 2021, 9, 41414149. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, S.; Chen, W.; Li, Y.; Wei, Y.P.; Luo, A.Q. Magnetic fluorescence molecularly imprinted polymer based on feox/zns nanocomposites for highly selective sensing of bisphenol A. Polymers 2019, 11, 1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, K.; Yu, X.; Qin, L.; Wu, Y. dS QDs: Facile synthesis, design and application as an “on–off” sensor for sensitive and selective monitoring Cu2+, Hg2+ and Mg2+ in foods. Food Chem. 2022, 390, 133116. [Google Scholar] [CrossRef] [PubMed]
- Ou, P.; Wu, J.; Lin, Y.; Tan, X.; Wu, Y.; Chen, Z.; Wei, F.; Huang, K. Flexible photoelectrochemical sensor for highly sensitive chloramphenicol detection based on M-TiO2-CdTe QDs/CdS QDs composite. Anal. Bioanal. Chem. 2022, 414, 2065–2078. [Google Scholar] [CrossRef]
- Haupt, K.; Linares, A.V.; Bompart, M.; Bui, B.T.S. Molecular Imprinting; Springer: New York, NY, USA, 2012; pp. 1–28. [Google Scholar]
- Haupt, K.; Mosbach, K. Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, B.; Li, Y.; Liu, C.B.; Jiao, P.F.; Wei, Y.P. Molecularly imprinted magnetic fluorescent nanocomposite-based sensor for selective detection of lysozyme. Nanomaterials 2021, 11, 1575. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Gálvez, A.; Ait-Lahcen, A.; Mercante, L.A.; Morales-Narváez, E.; Amine, A.; MerkoçI, A. Molecularly imprinted polymer-decorated magnetite nanoparticles for selective sulfonamide detection. Anal. Chem. 2016, 88, 3578–3584. [Google Scholar] [CrossRef]
- Lin, Z.Z.; Li, L.; Fu, G.Y.; Lai, Z.Z.; Peng, A.H.; Huang, Z.Y. Molecularly imprinted polymer-based photonic crystal sensor array for the discrimination of sulfonamides. Anal. Chim. Acta 2020, 1101, 32–40. [Google Scholar] [CrossRef]
- Buensuceso, C.E.; Tiu, B.D.B.; Lee, L.P.; Sabido, P.M.G.; Nuesca, G.M.; Caldona, E.B.; Mundo, F.R.; Advincula, R.C. Electropolymerized-molecularly imprinted polymers (E-MIPS) as sensing elements for the detection of dengue infection. Anal. Bioanal. Chem. 2022, 414, 1347–1357. [Google Scholar] [CrossRef]
- Fan, Y.; Zeng, G.; Ma, X. Effects of prepolymerization on surface molecularly imprinted polymer for rapid separation and analysis of sulfonamides in water. J. Colloid Interface Sci. 2020, 571, 21–29. [Google Scholar] [CrossRef]
- Zhang, R.R.; Gan, X.T.; Xu, J.J.; Pan, Q.F.; Liu, H.; Sun, A.L.; Shi, X.Z.; Zhang, Z.M. Ultrasensitive electrochemiluminescence sensor based on perovskite quantum dots coated with molecularly imprinted polymer for prometryn determination. Food Chem. 2022, 370, 131353. [Google Scholar] [CrossRef]
- Wei, X.; Xu, G.D.; Gong, C.C.; Qin, F.; Gong, X.H.; Li, C. Fabrication and evaluation of sulfanilamide-imprinted composite sensors by developing a custom-tailored strategy. Sens. Actuators B-Chem. 2018, 255, 2697–2703. [Google Scholar] [CrossRef]
- Ali, G.K.; Omer, K.M. Molecular imprinted polymer combined with aptamer (MIP-aptamer) as a hybrid dual recognition element for bio(chemical) sensing applications. review. Talanta 2022, 236, 122878. [Google Scholar] [CrossRef] [PubMed]
- Sobiech, M.; Luliński, P.; Wieczorek, P.P.; Marć, M. Quantum and carbon dots conjugated molecularly imprinted polymers as advanced nanomaterials for selective recognition of analytes in environmental, food and biomedical applications. TrAC Trends Anal. Chem. 2021, 142, 116306. [Google Scholar] [CrossRef]
- Tan, L.; Kang, C.; Xu, S.; Tang, Y.W. Selective room temperature phosphorescence sensing of target protein using Mn-doped ZnS QDs-embedded molecularly imprinted polymer. Biosens. Bioelectron. 2013, 48, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Ji, Y.; Yan, X.P. Surface Molecular imprinting on Mn-Doped ZnS quantum dots for room-temperature phosphorescence optosensing of pentachlorophenol in water. Anal. Chem. 2009, 81, 1615–1621. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Berti, L.; Medintz, I.L. Materials for fluorescence resonanceenergy transfer analysis: Beyond traditional donor-acceptor combinations. Angew. Chem. Int. Ed. 2006, 45, 4562–4589. [Google Scholar] [CrossRef]
- Abu-Alsoud, G.F.; Hawboldt, K.A.; Bottaro, C.S. Comparison of four adsorption isotherm models for characterizing molecular recognition of individual phenolic compounds in porous tailor-made molecularly imprinted polymer films. ACS Appl. Mater. Interfaces 2020, 12, 11998–12009. [Google Scholar] [CrossRef]


| Samples | Added (μM) | Measured (μM) | Recovery (%) | RSD (n = 3, %) |
|---|---|---|---|---|
| Lake water | 0.1000 | 0.09682 | 96.8 | 3.4 |
| 1.000 | 0.9733 | 97.3 | 4.2 | |
| River water | 0.1000 | 0.1019 | 101.9 | 3.8 |
| 1.000 | 1.0433 | 104.3 | 4.7 | |
| Tap water | 0.1000 | 0.1008 | 100.8 | 2.8 |
| 1.000 | 0.9887 | 98.9 | 3.2 |
| Recognition Element | Detection Technique | Linear Range (mol L−1) | LOD (mol L−1) | References |
|---|---|---|---|---|
| / | μ-SPE HPLC | 5.81 × 10−9–1.16 × 10−6 | 1.16 × 10−9 | [7] |
| / | SPE-HPLC-MS | 2.91 × 10−6–1.16 × 10−4 | 1.16 × 10−9 | [8] |
| MnO2 NR-IL | DPV | 7.0 × 10−8–1.0 × 10−4 | 1.0 × 10−8 | [11] |
| GCE−ND | Voltammetry | 5.81 × 10−6–4.65 × 10−4 | 9.3 × 10−7 | [12] |
| / | LDS-SD DLLME SDME-HPLC | 5.81 × 10−7–2.90 × 10−5 | 1.12 × 10−8 | [19] |
| N-Cu-MOFs | Electrochemical | 1.0 × 10−8–5.83 × 10−7 | 3.0 × 10−9 | [23] |
| rGO/GCE | Amperometric | 1.0 × 10−5–5.0 × 10−5 | 2.3 × 10−6 | [26] |
| MIPs/QDs@SiO2 | FL sensing | 2.0 × 10−6–3.0 × 10−5 | 1.7 × 10−7 | [42] |
| MIP@QDs | FL sensing | 2.9 × 10−8–2.90 × 10−6 | 3.2 × 10−9 | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Jiao, P.; Ma, Y.; Wei, Y. Molecular Imprinted ZnS Quantum Dots-Based Sensor for Selective Sulfanilamide Detection. Polymers 2022, 14, 3540. https://doi.org/10.3390/polym14173540
Zhang X, Jiao P, Ma Y, Wei Y. Molecular Imprinted ZnS Quantum Dots-Based Sensor for Selective Sulfanilamide Detection. Polymers. 2022; 14(17):3540. https://doi.org/10.3390/polym14173540
Chicago/Turabian StyleZhang, Xin, Pengfei Jiao, Yihan Ma, and Yuping Wei. 2022. "Molecular Imprinted ZnS Quantum Dots-Based Sensor for Selective Sulfanilamide Detection" Polymers 14, no. 17: 3540. https://doi.org/10.3390/polym14173540
APA StyleZhang, X., Jiao, P., Ma, Y., & Wei, Y. (2022). Molecular Imprinted ZnS Quantum Dots-Based Sensor for Selective Sulfanilamide Detection. Polymers, 14(17), 3540. https://doi.org/10.3390/polym14173540






