Clog and Release, and Reverse Motions of DNA in a Nanopore
Abstract
1. Introduction
2. Materials and Methods
3. Results
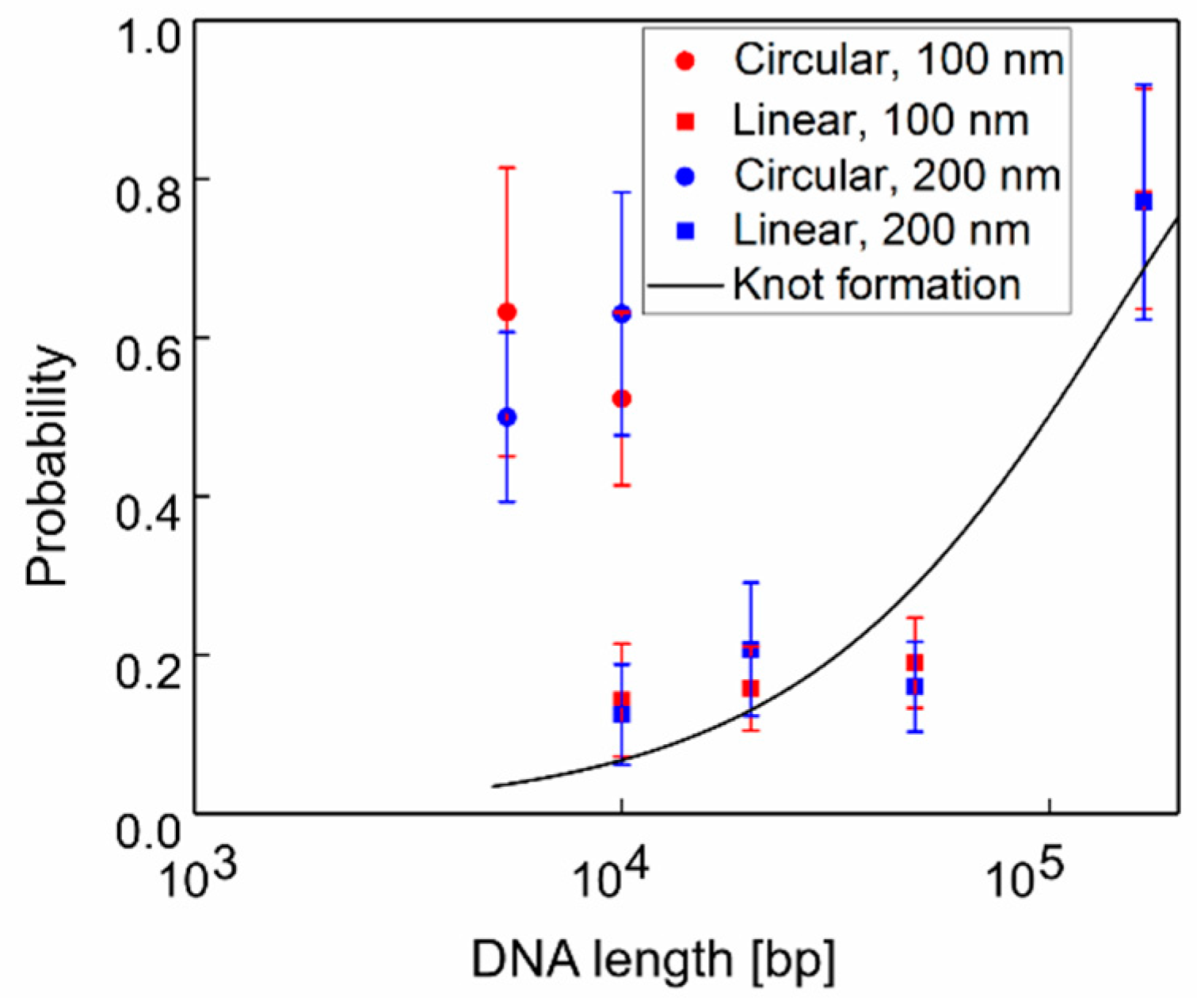
3.1. Clog Probabilities of Various DNA Length and Configurations
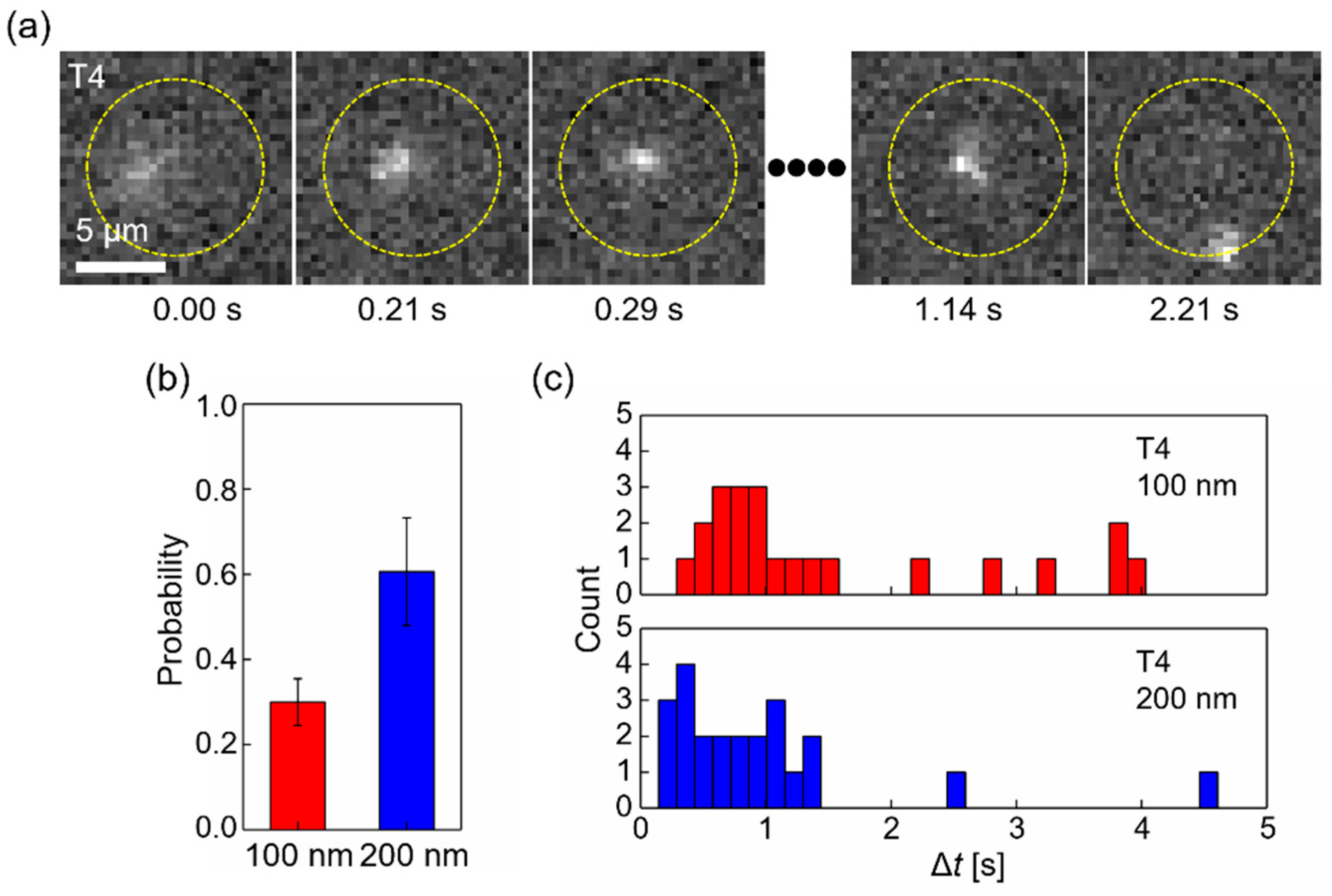
3.2. Release of T4 DNA to the cis Side after Clogging
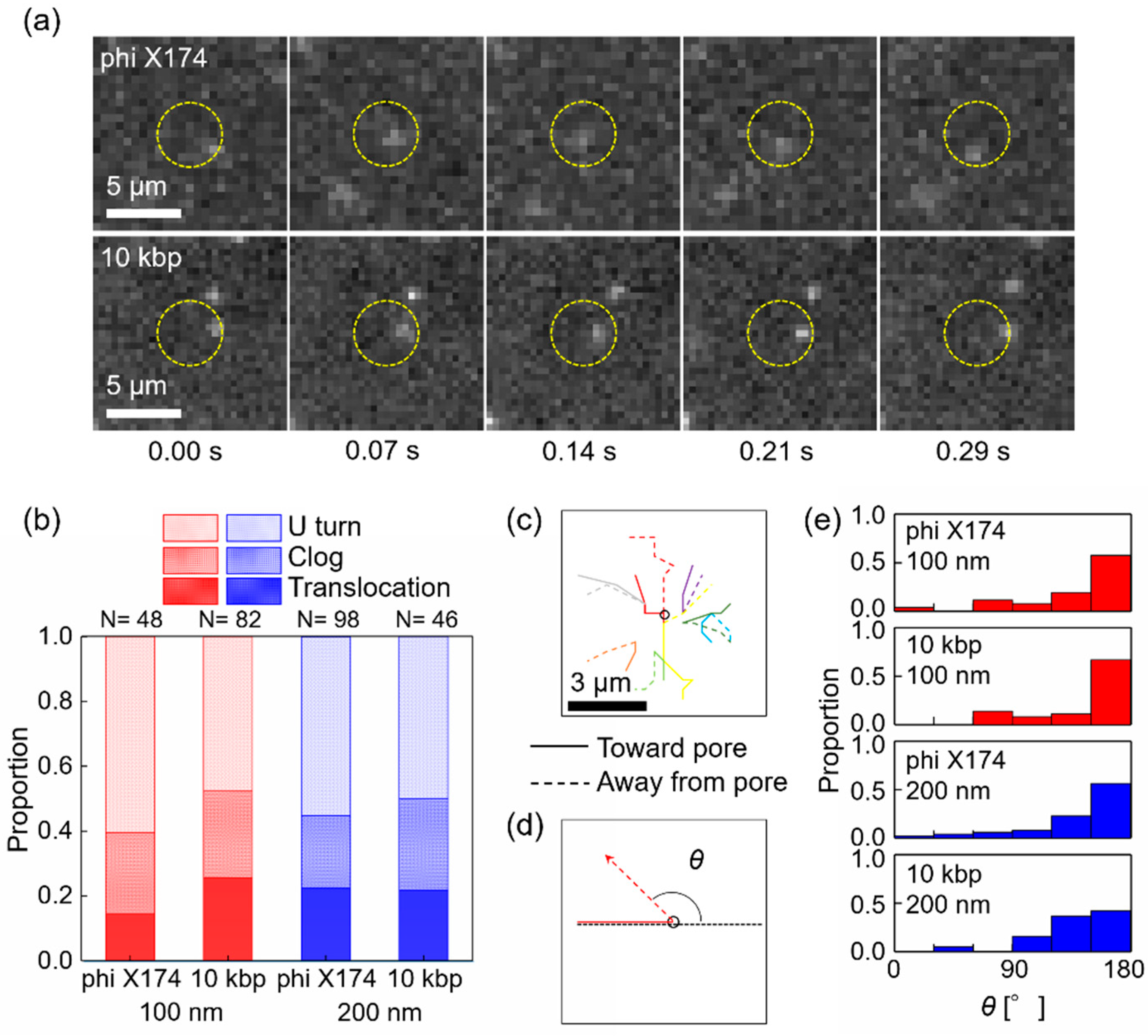
3.3. U Turn of Circular DNA at Nanopore
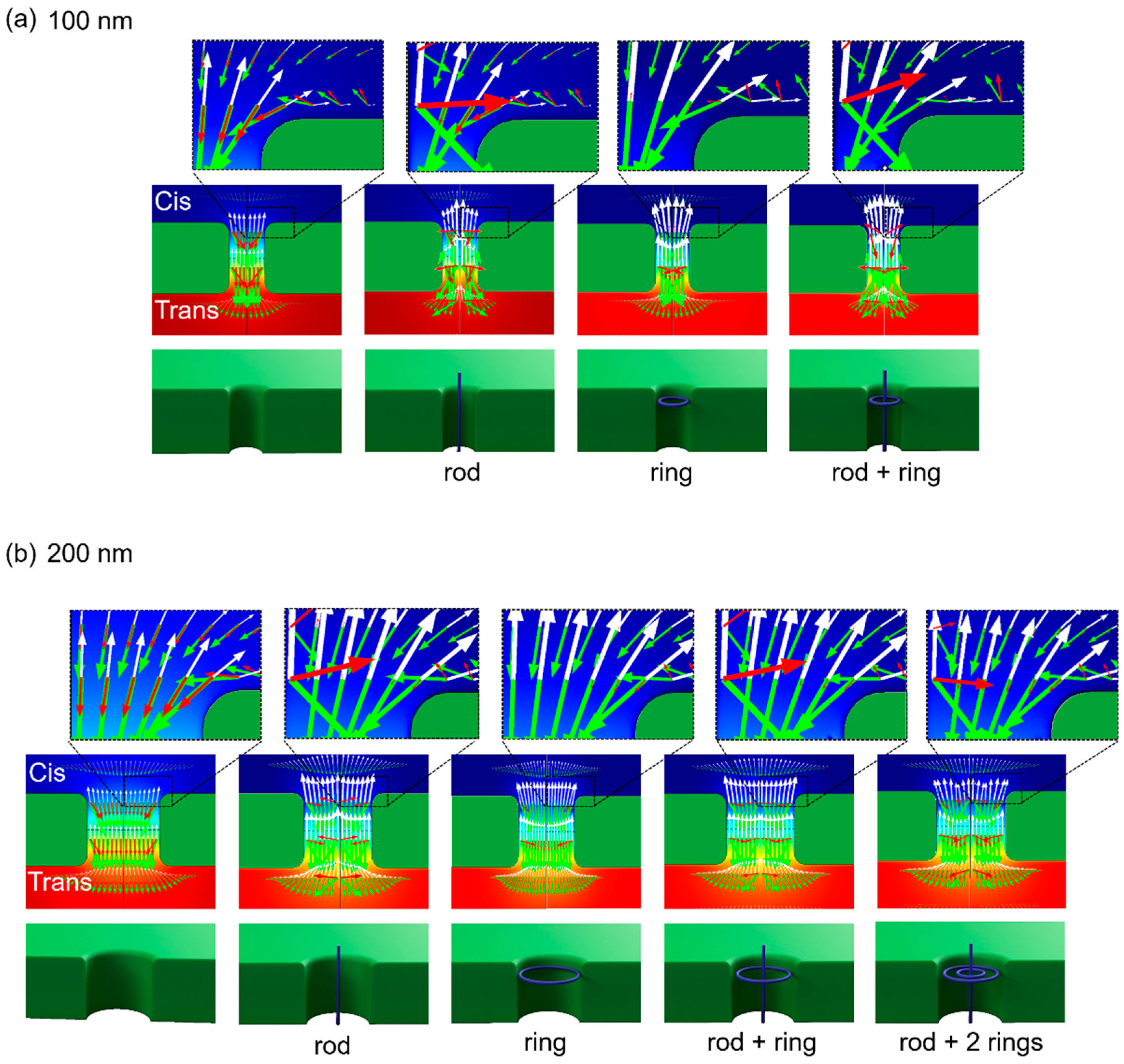
3.4. FEM-Based Numerical Estimation of DNA Motions Near Nanopore
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2007, 2, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Branton, D.; Deamer, D.W.; Marziali, A.; Bayley, H.; Benner, S.A.; Butler, T.; Di Ventra, M.; Garaj, S.; Hibbs, A.; Huang, X.; et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 2008, 26, 1146–1153. [Google Scholar] [CrossRef] [PubMed]
- Deamer, D.; Akeson, M.; Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 2016, 34, 518–524. [Google Scholar] [CrossRef]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- Keyser, U.F. Controlling molecular transport through nanopores. J. R. Soc. Interface R. Soc. 2011, 8, 1369–1378. [Google Scholar] [CrossRef]
- Lee, K.; Park, K.B.; Kim, H.J.; Yu, J.S.; Chae, H.; Kim, H.M.; Kim, K.B. Recent Progress in Solid-State Nanopores. Adv. Mater 2018, 30, e1704680. [Google Scholar] [CrossRef] [PubMed]
- Ando, G.; Hyun, C.; Li, J.; Mitsui, T. Directly observing the motion of DNA molecules near solid-state nanopores. ACS Nano 2012, 6, 10090–10097. [Google Scholar] [CrossRef]
- Fologea, D.; Ledden, B.; McNabb, D.S.; Li, J. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007, 91, 539011–539013. [Google Scholar] [CrossRef]
- Li, J.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J.A. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003, 2, 611–615. [Google Scholar] [CrossRef]
- Storm, A.J.; Storm, C.; Chen, J.H.; Zandbergen, H.; Joanny, J.F.; Dekker, C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005, 5, 1193–1197. [Google Scholar] [CrossRef]
- Larkin, J.; Henley, R.Y.; Muthukumar, M.; Rosenstein, J.K.; Wanunu, M. High-bandwidth protein analysis using solid-state nanopores. Biophys. J. 2014, 106, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Talaga, D.S.; Li, J. Single-molecule protein unfolding in solid state nanopores. J. Am. Chem. Soc. 2009, 131, 9287–9297. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, M.; Rahong, S.; Iizumi, Y.; Okazaki, T.; Taniguchi, M.; Kawai, T. Single-molecule sensing electrode embedded in-plane nanopore. Sci. Rep. 2011, 1, 46. [Google Scholar] [CrossRef] [PubMed]
- Garaj, S.; Hubbard, W.; Reina, A.; Kong, J.; Branton, D.; Golovchenko, J.A. Graphene as a subnanometre trans-electrode membrane. Nature 2010, 467, 190–193. [Google Scholar] [CrossRef] [PubMed]
- Yusko, E.C.; Johnson, J.M.; Majd, S.; Prangkio, P.; Rollings, R.C.; Li, J.; Yang, J.; Mayer, M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nanotechnol. 2011, 6, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Han, A.P.; Schurmann, G.; Mondin, G.; Bitterli, R.A.; Hegelbach, N.G.; de Rooij, N.F.; Staufer, U. Sensing protein molecules using nanofabricated pores. Appl. Phys. Lett. 2006, 88, 093901. [Google Scholar] [CrossRef]
- McMullen, A.; de Haan, H.W.; Tang, J.X.; Stein, D. Stiff filamentous virus translocations through solid-state nanopores. Nat. Commun. 2014, 5, 4171. [Google Scholar] [CrossRef]
- Arima, A.; Tsutsui, M.; Harlisa, I.H.; Yoshida, T.; Tanaka, M.; Yokota, K.; Tonomura, W.; Taniguchi, M.; Okochi, M.; Washio, T.; et al. Selective detections of single-viruses using solid-state nanopores. Sci. Rep. 2018, 8, 16305. [Google Scholar] [CrossRef]
- Ryuzaki, S.; Tsutsui, M.; He, Y.; Yokota, K.; Arima, A.; Morikawa, T.; Taniguchi, M.; Kawai, T. Rapid structural analysis of nanomaterials in aqueous solutions. Nanotechnology 2017, 28, 155501. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Chen, K.; Ghosal, S.; Ricci, M.; Keyser, U.F. Asymmetric dynamics of DNA entering and exiting a strongly confining nanopore. Nat. Commun. 2017, 8, 380. [Google Scholar] [CrossRef]
- Squires, T.M.; Quake, S.R. Microfluidics: Fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005, 77, 977–1026. [Google Scholar] [CrossRef]
- Wong, C.T.; Muthukumar, M. Polymer capture by electro-osmotic flow of oppositely charged nanopores. J. Chem. Phys. 2007, 126, 164903. [Google Scholar] [CrossRef] [PubMed]
- van der Heyden, F.H.; Bonthuis, D.J.; Stein, D.; Meyer, C.; Dekker, C. Electrokinetic energy conversion efficiency in nanofluidic channels. Nano Lett. 2006, 6, 2232–2237. [Google Scholar] [CrossRef] [PubMed]
- Laohakunakorn, N.; Ghosal, S.; Otto, O.; Misiunas, K.; Keyser, U.F. DNA interactions in crowded nanopores. Nano Lett. 2013, 13, 2798–2802. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Gu, J.; Brandin, E.; Kim, Y.R.; Wang, Q.; Branton, D. Probing Single DNA Molecule Transport Using Fabricated Nanopores. Nano Lett. 2004, 4, 2293–2298. [Google Scholar] [CrossRef] [PubMed]
- Mihovilovic, M.; Hagerty, N.; Stein, D. Statistics of DNA capture by a solid-state nanopore. Phys. Rev. Lett. 2013, 110, 028102. [Google Scholar] [CrossRef] [PubMed]
- Plesa, C.; Verschueren, D.; Pud, S.; van der Torre, J.; Ruitenberg, J.W.; Witteveen, M.J.; Jonsson, M.P.; Grosberg, A.Y.; Rabin, Y.; Dekker, C. Direct observation of DNA knots using a solid-state nanopore. Nat. Nanotechnol. 2016, 11, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Rosa, A.; Di Ventra, M.; Micheletti, C. Topological jamming of spontaneously knotted polyelectrolyte chains driven through a nanopore. Phys. Rev. Lett. 2012, 109, 118301. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Kato, Y.; Ishida, K.; Hyun, C.; Li, J.; Mitsui, T. DNA motion induced by electrokinetic flow near an Au coated nanopore surface as voltage controlled gate. Nanotechnology 2015, 26, 065502. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Sakashita, N.; Ishida, K.; Mitsui, T. Gate-Voltage-Controlled Threading DNA into Transistor Nanopores. J. Phys. Chem. B 2018, 122, 827–833. [Google Scholar] [CrossRef]
- Gunther, K.; Mertig, M.; Seidel, R. Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res. 2010, 38, 6526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Zhang, F.; van Kan, J.A.; van der Maarel, J.R. Effects of electrostatic screening on the conformation of single DNA molecules confined in a nanochannel. J. Chem. Phys. 2008, 128, 225109. [Google Scholar] [CrossRef] [PubMed]
- Kundukad, B.; Yan, J.; Doyle, P.S. Effect of YOYO-1 on the mechanical properties of DNA. Soft Matter 2014, 10, 9721–9728. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Ghosal, S.; Hu, G. Hydrodynamic flow in the vicinity of a nanopore induced by an applied voltage. Nanotechnology 2013, 24, 245202. [Google Scholar] [CrossRef]
- Constantin, D.; Siwy, Z.S. Poisson-Nernst-Planck model of ion current rectification through a nanofluidic diode. Phys. Rev. E Stat. Nonline Soft Matter Phys. 2007, 76, 041202. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Tsutsui, M.; Fan, C.; Taniguchi, M.; Kawai, T. Controlling DNA translocation through gate modulation of nanopore wall surface charges. ACS Nano 2011, 5, 5509–5518. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.C.; Balducci, A.; Doyle, P.S. Ionic effects on the equilibrium dynamics of DNA confined in nanoslits. Nano Lett. 2008, 8, 1683–1688. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, K.D. DNA electrophoresis in microfabricated devices. Rev. Mod. Phys. 2010, 82, 2903–2947. [Google Scholar] [CrossRef]
- Gershow, M.; Golovchenko, J.A. Recapturing and trapping single molecules with a solid-state nanopore. Nat. Nanotechnol. 2007, 2, 775–779. [Google Scholar] [CrossRef]
- Nakane, J.; Akeson, M.; Marziali, A. Evaluation of nanopores as candidates for electronic analyte detection. Electrophoresis 2002, 23, 2592–2601. [Google Scholar] [CrossRef]
- Dai, L.; Renner, C.B.; Doyle, P.S. The polymer physics of single DNA confined in nanochannels. Adv. Colloid Interface Sci. 2016, 232, 80–100. [Google Scholar] [CrossRef] [PubMed]
- Vlassarev, D.M.; Golovchenko, J.A. Trapping DNA near a solid-state nanopore. Biophys. J. 2012, 103, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Conlisk, A.T. Forces affecting double-stranded DNA translocation through synthetic nanopores. Biomed. Microdevices 2011, 13, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Stein, D.; Deurvorst, Z.; van der Heyden, F.H.; Koopmans, W.J.; Gabel, A.; Dekker, C. Electrokinetic concentration of DNA polymers in nanofluidic channels. Nano Lett. 2010, 10, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Mitscha-Baude, G.; Buttinger-Kreuzhuber, A.; Tulzer, G.; Heitzinger, C. Adaptive and iterative methods for simulations of nanopores with the PNP–Stokes equations. J. Comput. Phys. 2017, 338, 452–476. [Google Scholar] [CrossRef]
- Buyukdagli, S.; Ala-Nissila, T. Controlling Polymer Translocation and Ion Transport via Charge Correlations. Langmuir 2014, 30, 12907–12915. [Google Scholar] [CrossRef] [PubMed]
- Suma, A.; Micheletti, C. Pore translocation of knotted DNA rings. Proc. Natl. Acad. Sci. USA 2017, 114, E2991–E2997. [Google Scholar] [CrossRef] [PubMed]
- Krueger, E.; Shim, J.; Fathizadeh, A.; Chang, A.N.; Subei, B.; Yocham, K.M.; Davis, P.H.; Graugnard, E.; Khalili-Araghi, F.; Bashir, R.; et al. Modeling and Analysis of Intercalant Effects on Circular DNA Conformation. ACS Nano 2016, 10, 8910–8917. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubota, T.; Lloyd, K.; Sakashita, N.; Minato, S.; Ishida, K.; Mitsui, T. Clog and Release, and Reverse Motions of DNA in a Nanopore. Polymers 2019, 11, 84. https://doi.org/10.3390/polym11010084
Kubota T, Lloyd K, Sakashita N, Minato S, Ishida K, Mitsui T. Clog and Release, and Reverse Motions of DNA in a Nanopore. Polymers. 2019; 11(1):84. https://doi.org/10.3390/polym11010084
Chicago/Turabian StyleKubota, Tomoya, Kento Lloyd, Naoto Sakashita, Seiya Minato, Kentaro Ishida, and Toshiyuki Mitsui. 2019. "Clog and Release, and Reverse Motions of DNA in a Nanopore" Polymers 11, no. 1: 84. https://doi.org/10.3390/polym11010084
APA StyleKubota, T., Lloyd, K., Sakashita, N., Minato, S., Ishida, K., & Mitsui, T. (2019). Clog and Release, and Reverse Motions of DNA in a Nanopore. Polymers, 11(1), 84. https://doi.org/10.3390/polym11010084




