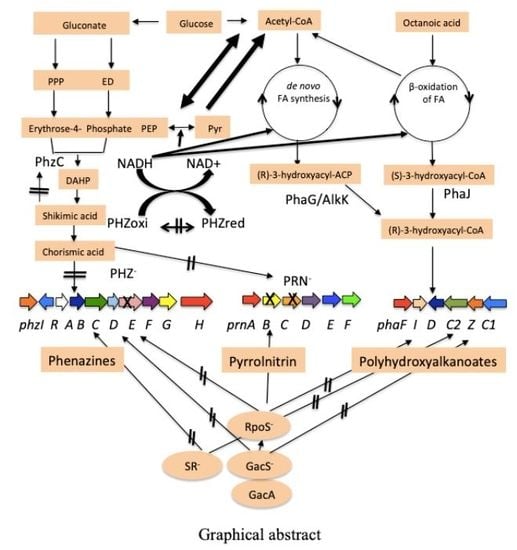
Polyhydroxyalkanoate (PHA) Polymer Accumulation and pha Gene Expression in Phenazine (phz-) and Pyrrolnitrin (prn-) Defective Mutants of Pseudomonas chlororaphis PA23
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain/Mutants and Primers
2.2. Culture Media and Growth Conditions
2.3. Growth of P. chlororaphis Strains and Analyses of PHA Synthesis
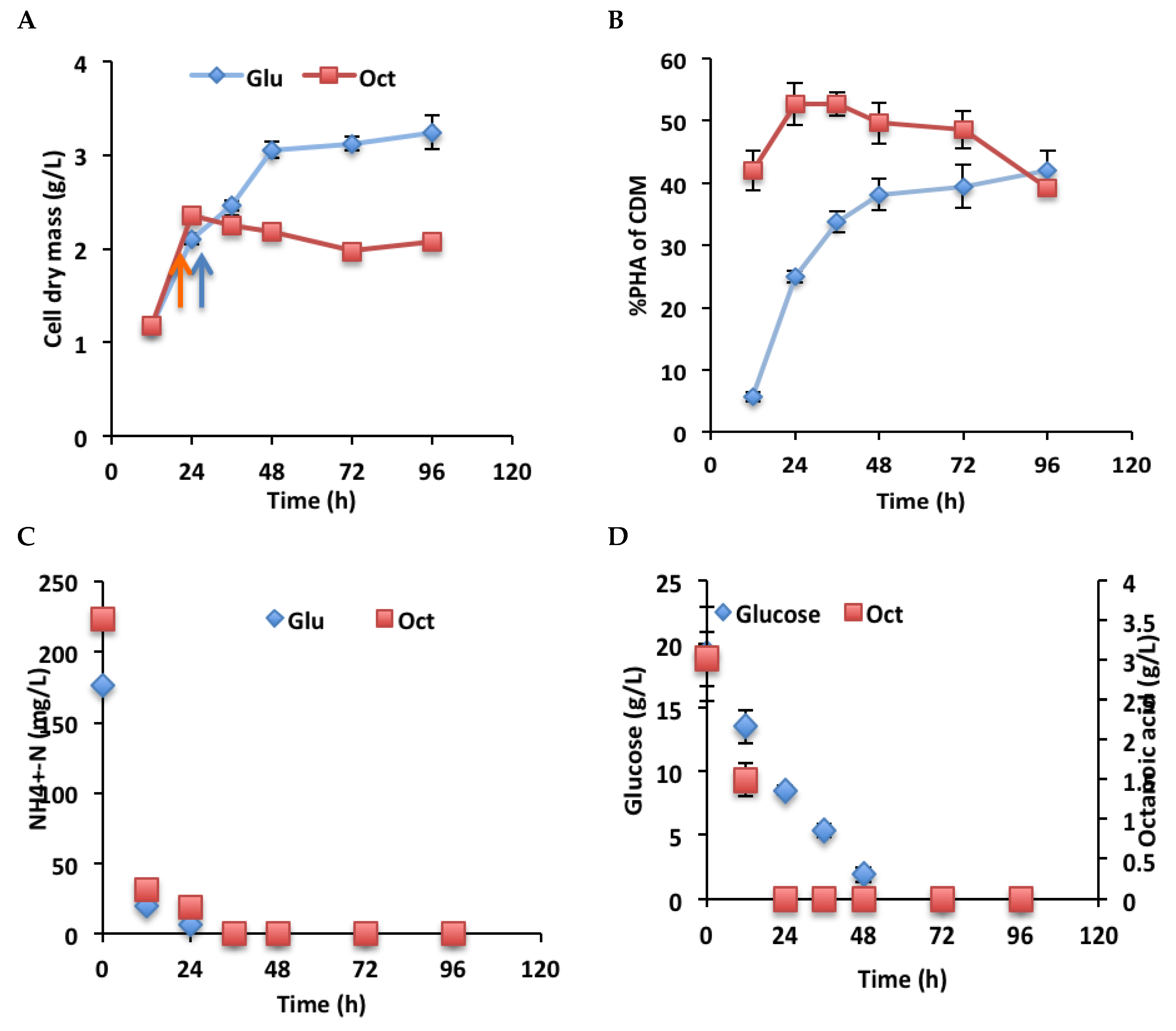
2.4. PHA Production by P. chlororaphis PA23 at Different Time Points
2.5. RNA Isolation, cDNA Synthesis and Gene Expression
2.6. Statistical Analysis
3. Results
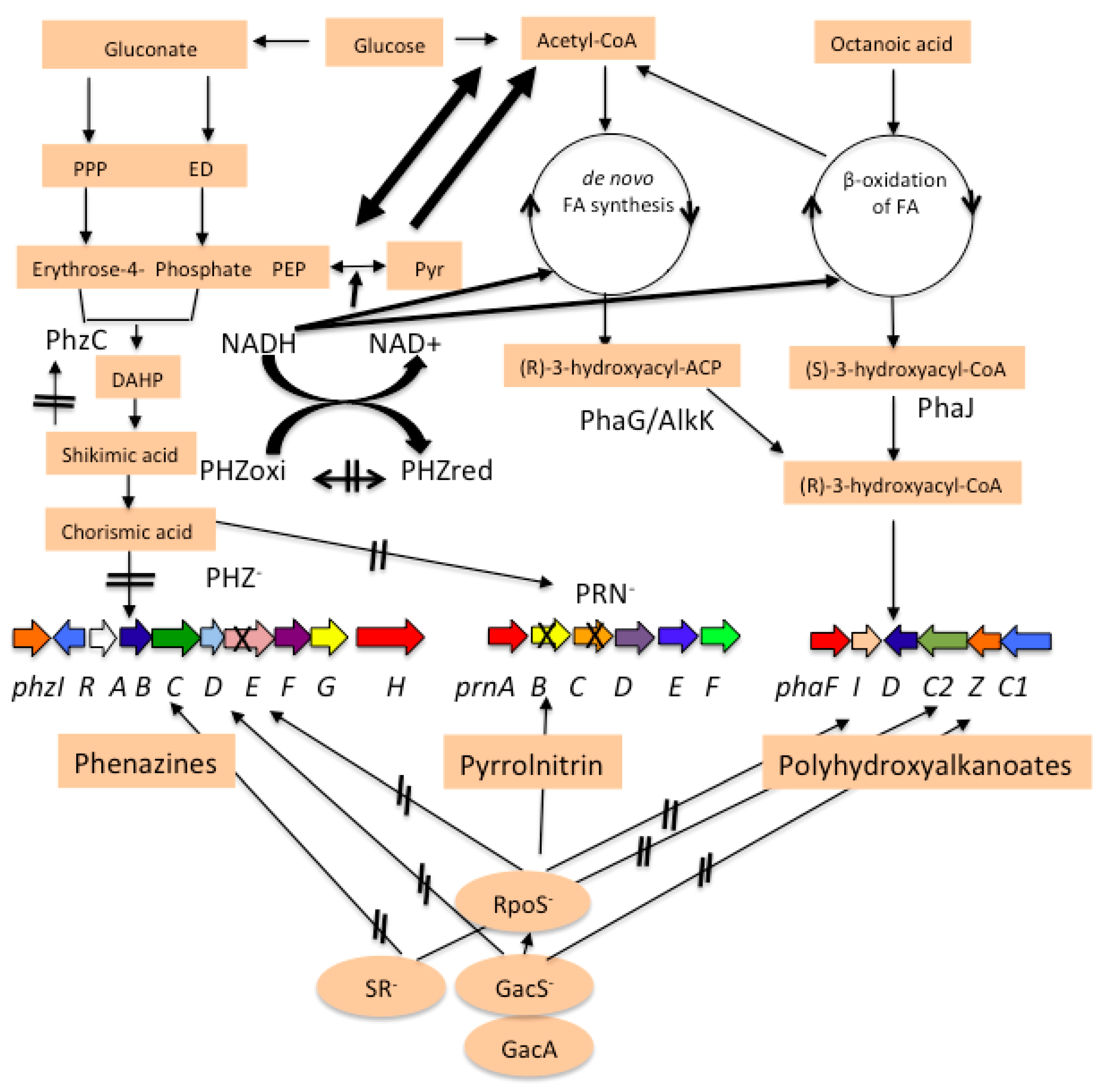
3.1. pha Cluster in P. chlororaphis PA23
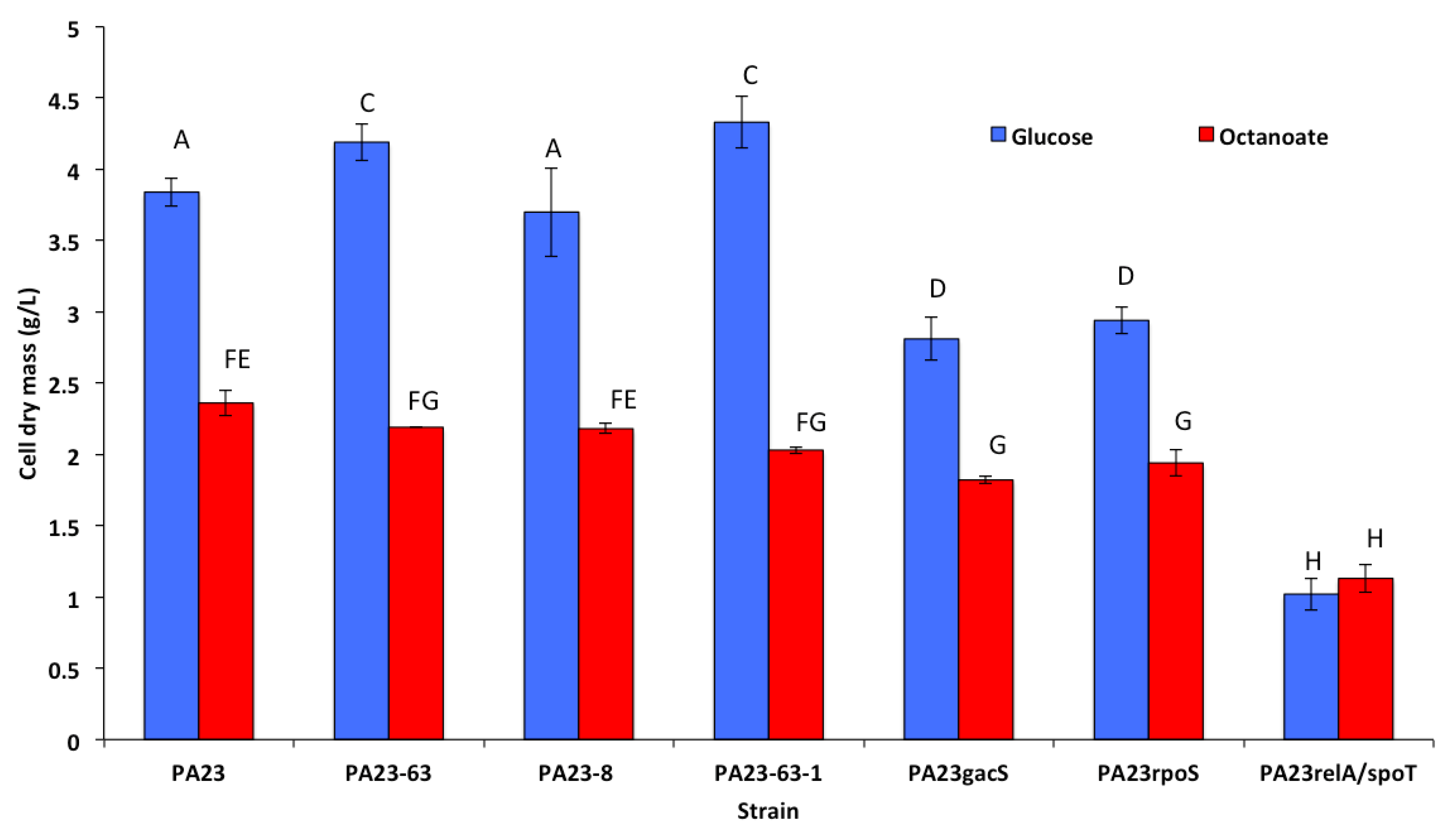
3.2. Growth and Cell Dry Mass of P. chlororaphis 23 and It Mutant Derivatives
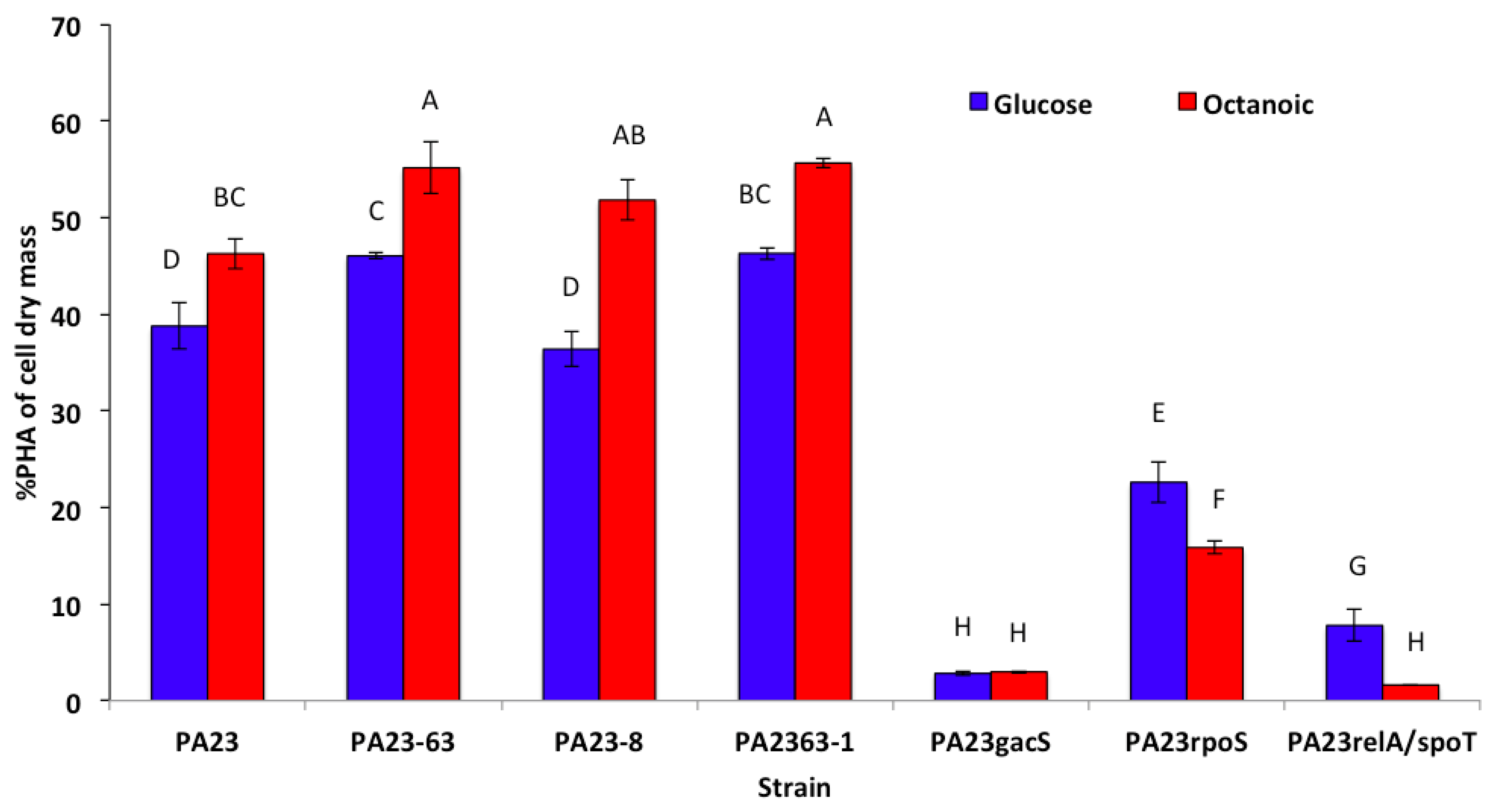
3.3. PHA Production by P. chlororaphis PA23 and It Mutant Derivatives
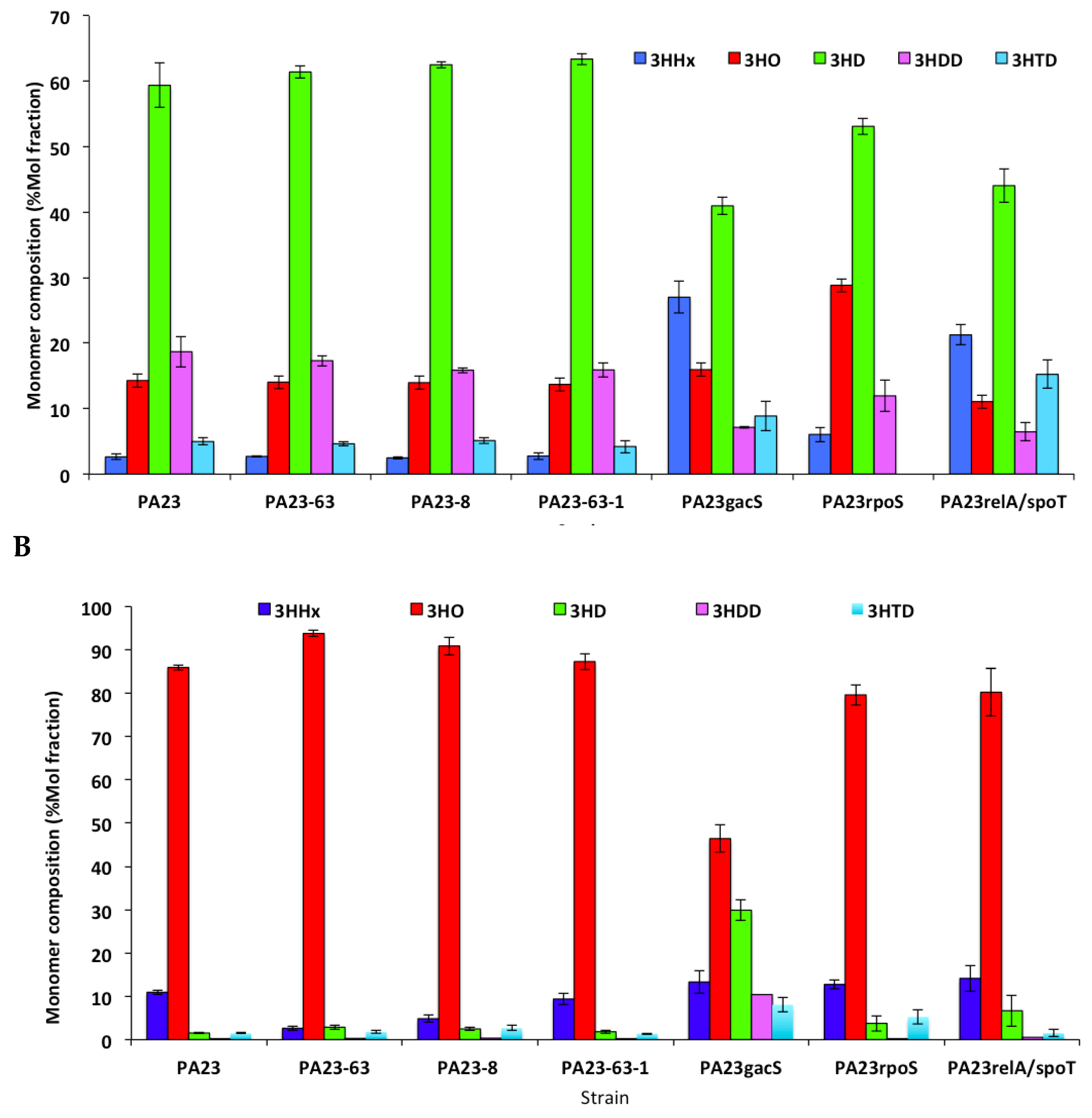
3.4. Monomer Composition of PHAs Produced by P. chlororaphis PA23 and It Derivative Mutants
3.5. Growth and PHA Production by P. chlororaphis PA23 in Glucose and Octanoic Acid Medium at Different Time Points
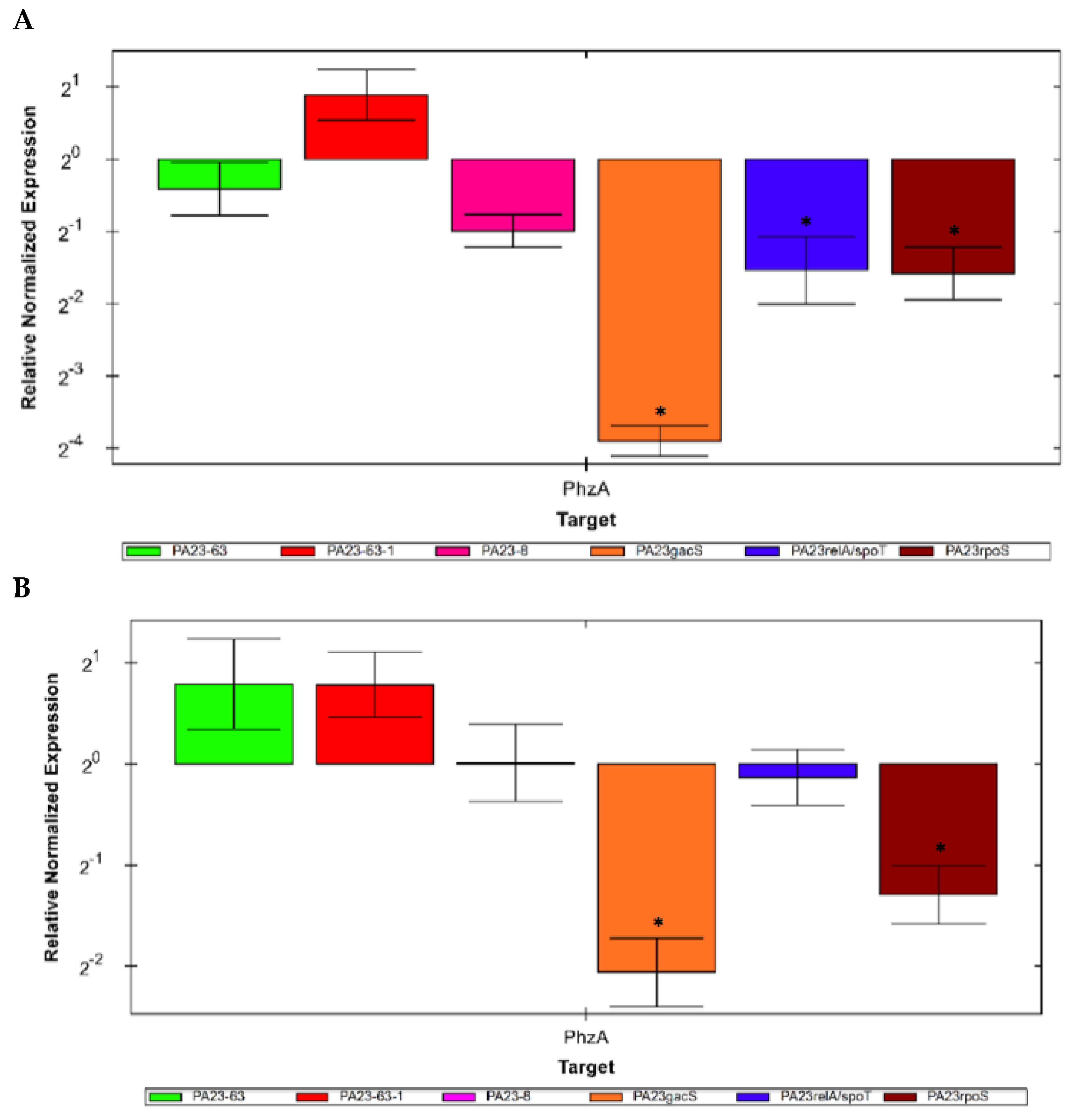
3.6. Expression of the phzA Gene in Mutants Defective in PHZ and/or PRN Production
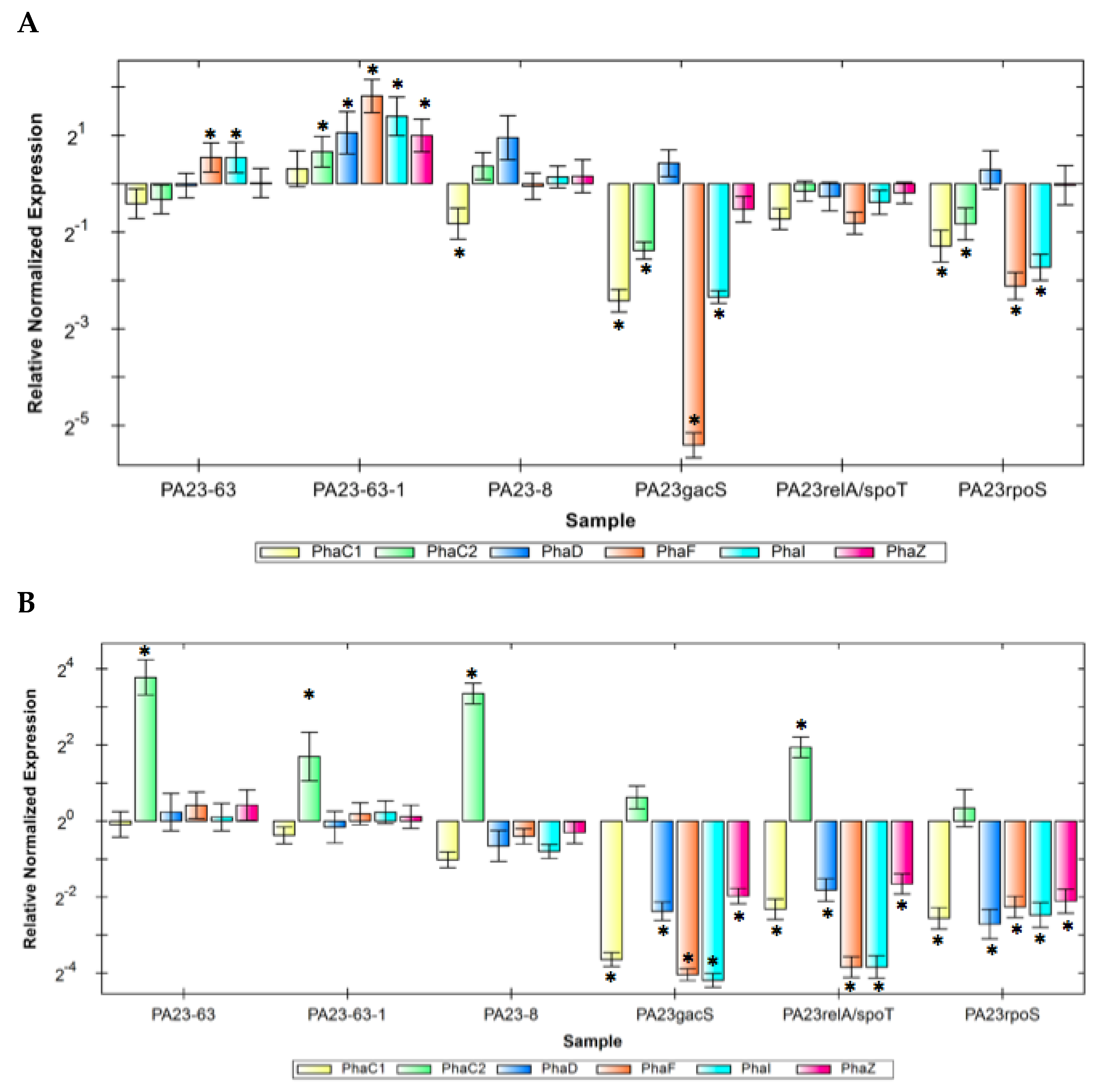
3.7. Expression of pha Genes in the PA23 PHZ and/or PRN Mutants
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Fernando, W.G.D.; Nakkeeran, S.; Zhang, Y.; Savchuk, S. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary by Pseudomonas and Bacillus species on canola petals. Crop Prot. 2007, 26, 100–107. [Google Scholar] [CrossRef]
- Savchuk, S.C.; Dilantha Fernando, W.G. Effect of timing of application and population dynamics on the degree of biological control of Sclerotinia sclerotiorum by bacterial antagonists. FEMS Microbiol. Ecol. 2004, 49, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Poritsanos, N.; Selin, C.; Fernando, W.G.D.; Nakkeeran, S.; de Kievit, T.R. A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can. J. Microbiol. 2006, 52, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fernando, W.G.D.; de Kievit, T.R.; Berry, C.; Daayf, F.; Paulitz, T.C. Detection of antibiotic-related genes from bacterial biocontrol agents with polymerase chain reaction. Can. J. Microbiol. 2006, 52, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S., III; Pierson, E.A. Metabolism and function of phenazines in bacteria: Impacts on the behavior of bacteria in the environment and biotechnological processes. Appl. Microbiol. Biotechnol. 2010, 86, 1659–1670. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Parejko, J.A.; Mavrodi, O.V.; Kwak, Y.S.; Weller, D.M.; Blankenfeldt, W.; Thomashow, L.S. Recent insights into the diversity, frequency and ecological roles of phenazines in fluorescent Pseudomonas spp. Environ. Microbiol. 2013, 15, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Thomashow, L. Phenazines in the environment: Microbes, habitats, and ecological relevance. In Microbial Phenazines; Chincholkar, S., Thomashow, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 199–216. ISBN 978-3-64-240573-0. [Google Scholar]
- Xu, Y. Genomic features and regulation of phenazine biosynthesis in the rhizosphere strain Pseudomonas aeruginosa M18. In Microbial Phenazines; Chincholkar, S., Thomashow, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 177–198. ISBN 978-3-64-240573-0. [Google Scholar]
- Van Rij, E.T.; Wesselink, M.; Chin-A-Woeng, T.F.; Bloemberg, G.V.; Lugtenberg, B.J. Influence of environmental conditions on the production of phenazine-1-carboxamide by Pseudomonas chlororaphis PCL1391. Mol. Plant-Microbe Interact. 2004, 17, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Duffy, B.K.; Defago, G. Controlling instability in gacS-gacA regulatory genes during inoculant production of Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 2000, 66, 3142–3150. [Google Scholar] [CrossRef] [PubMed]
- Blankenfeldt, W. The Biosynthesis of phenazines. In Microbial phenazines: Biosynthesis, Agriculture and Health; Chincholkar, S., Thomashow, L., Eds.; Springer: Heidelberg, Germany, 2013; pp. 1–18. [Google Scholar]
- Selin, C.; Habibian, R.; Poritsanos, N.; Sarangi, N.P.; Dilantha Fernando, W.G.; de Kievit, T.R. Phenazines are not essential for Pseudomonas chlororaphis PA23 biocontrol of Sclerotinia sclerotiorum, but do play a role in biofilm formation. FEMS Microbiol. Ecol. 2010, 71, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; Dilantha Fernando, W.G.; de Kievit, T.R. The PhzI/PhzR quorum-sensing system is required for pyrrolnitrin and phenazine production, and exhibits cross-regulation with RpoS in Pseudomonas chlororaphis PA23. Microbiology 2012, 158, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Nandi, M.; Selin, C.; Brawerman, G.; Dilantha Fernando, W.G.; de Kievit, T.R. The global regulator ANR is essential for Pseudomonas chlororaphis strain PA23 biocontrol. Microbiology 2016, 162, 2159–2169. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Kim, B.S. Valorization of polyhydroxyalkanoates production process by co-synthesis of value-added products. Bioresour. Technol. 2018, 269, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Pierson, L.S., III; Pierson, E.A. Phenazine antibiotic production in Pseudomonas aureofaciens: Role in rhizosphere ecology and pathogen suppression. FEMS Microbiol. Lett. 1996, 136, 101–108. [Google Scholar] [CrossRef]
- Yun, H.S.; Kim, D.Y.; Chung, C.W.; Kim, H.W.; Yang, Y.K.; Rhee, Y.H. Characterization of a tacky poly[3-hydroxyalkanoate] produced by Pseudomonas chlororaphis HS21 from palm kernel oil. J. Microbiol. Biotechnol. 2003, 13, 64–69. [Google Scholar] [CrossRef]
- Muhr, A.; Rechberger, E.M.; Salerno, A.; Reiterer, A.; Malli, K.; Strohmeier, K.; Shober, S.; Mittlebach, M.; Koller, M. Novel description of mcl-PHA biosynthesis by Pseudomonas chlororaphis from animal-derived waste. J. Biotechnol. 2013, 165, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Solaiman, D.K.Y.; Ashby, R.D.; Crocker, N.; Lai, B.H.; Zerkowski, J.A. Rhamnolipid and poly[hydroxyalkanoate] biosynthesis in 3-hydroxyacyl-ACP: CoA transacylase [phaG]-knockouts of Pseudomonas chlororaphis. Biocatal. Agric. Biotechnol. 2014, 3, 159–166. [Google Scholar] [CrossRef]
- Sharma, P.K.; Fu, J.; Cicek, N.; Sparling, R.; Levin, D.B. Kinetics of medium-chain-length polyhydroxyalkanoate production by a novel isolate of Pseudomonas putida LS46. Can. J. Microbiol. 2012, 58, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Walsh, M.; O’Connor, K.; Babu, R.; Woods, T.; Kenny, S. Plant oils and products of their hydrolysis as substrates for polyhydroxyalkanoate synthesis. Chem. Biochem. Eng. Q. 2015, 29, 123–133. [Google Scholar] [CrossRef]
- Sharma, P.K.; Fu, J.; Zhang, X.; Fristensky, B.; Sparling, R.; Levin, D.B. Genome features of Pseudomonas putida LS46, a novel polyhydroxyalkanoate producer and its comparison with other P. putida strains. AMB Express 2014, 4, 37. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.K.; Munir, R.I.; de Kievit, T.; Levin, D.B. Synthesis of polyhydroxyalkanoates [PHAs] from vegetable oils and free fatty acids by wild-type and mutant strains of Pseudomonas chlororaphis. Can. J. Microbiol. 2017, 63, 1009–1024. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Xu, J.; Gutierrez, M.; Yoo, T.; Cho, Y.H.; Yoon, S.C. Metabolic relationship between polyhydroxyalkanoic acid and rhamnolipid synthesis in Pseudomonas aeruginosa: Comparative 13C NMR analysis of the products in wild-type and mutants. J. Biotechnol. 2011, 151, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Manuel, J.; Selin, C.; Fernando, W.G.; de Kievit, T. Stringent response mutants of Pseudomonas chlororaphis PA23 exhibit enhanced antifungal activity against Sclerotinia sclerotiorum in vitro. Microbiology 2012, 158, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Girard, G.; van Rij, E.T.; Lugtenberg, B.J.J.; Bloemberg, G.V. Regulatory roles of psrA and rpoS in phenazine-1-carboxamide synthesis by Pseudomonas chlororaphis PCL1391. Microbiology 2006, 152, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Selin, C.; Manuel, J.; Fernando, W.G.D.; de Kievit, T. Expression of the Pseudomonas chlororaphis strain PA23 Rsm system is under control of GacA, RpoS, PsrA, quorum sensing and the stringent response. Biol. Control 2014, 69, 24–33. [Google Scholar] [CrossRef]
- Wang, D.; Lee, S.H.; Seeve, C.; Yu, J.M.; Pierson, L.S., III; Pierson, E.A. Roles of the Gac-Rsm pathway in the regulation of phenazine biosynthesis in Pseudomonas chlororaphis 30–84. MicrobiologyOpen 2013, 2, 505–524. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Yang, S.; Fang, Y.; Yang, R.; Mou, D.; Cui, J.; Wen, L. RpoS as an intermediate in RsmA-dependent regulation of secondary antifungal metabolites biosynthesis in Pseudomonas sp. M18. FEMS Microbiol. Lett. 2006, 268, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Ryan, W.J.; O’Leary, N.D.; O’Mahony, M.; Dobson, A.D.W. GacS-dependent regulation of polyhydroxyalkanoate synthesis in Pseudomonas putida CA-3. Appl. Environ. Microbiol. 2013, 79, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Raiger-Lustman, L.J.; Ruiz, J.A. The alternative sigma factor, rpoS affects polyhydroxyalkanoate metabolism in Pseudomonas putida. FEMS Microbiol. Lett. 2008, 284, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.A.; López, N.I.; Méndez, B.S. rpoS gene expression in carbon-starved cultures of the polyhydroxyalkanoate-accumulating species Pseudomonas oleovorans. Curr. Microbiol. 2004, 48, 396–400. [Google Scholar] [CrossRef] [PubMed]
- Juengert, J.R.; Christoph, M.; Borisava, M.; Mayer, C.; Wolz, C.; Brigham, C.J.; Sinskey, A.J.; Jendrossek, D. Absence of ppGpp leads to increased mobilization of intermediately accumulated poly [3-hydroxybutyrate] in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2017, 83, 13–16. [Google Scholar] [CrossRef] [PubMed]
- Brigham, C.J.; Speth, D.R.; Rha, C.; Sinskey, A.J. Whole-genome microarray and gene deletion studies reveal regulation of the polyhydroxyalkanoate production cycle by the stringent response in Ralstonia eutropha H16. Appl. Environ. Microbiol. 2012, 78, 8033–8044. [Google Scholar] [CrossRef] [PubMed]
- Mozejko-Ciesielska, J.; Dabrowska, D.; Szalewska-Palasz, A.; Ciesielski, S. Medium-chain-length polyhydroxyalkanoates synthesis by Pseudomonas putida KT2440 relA/spoT mutant: Bioprocess characterization and transcriptome analysis. AMB Express 2017, 7, 92. [Google Scholar] [CrossRef] [PubMed]
- Loewen, P.C.; Villenueva, J.; Dilantha Fernando, W.G.; de Kievit, T. Genome Sequence of Pseudomonas chlororaphis strain PA23. Genome Annoc. 2014, 2, e00689-14. [Google Scholar] [CrossRef] [PubMed]
- Braunegg, G.; Sonnleitner, B.; Lafferty, R.M. A rapid gas chromatographic method for the determination of polyhydroxybutyric acid in microbial biomass. Eur. J. Appl. Microbiol. Biotechnol. 1978, 6, 29–37. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative gene expression data using real time quantitative PCR and the 2−δδCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Prieto, A.; Escapa, I.F.; Martínez, V.; Dinjaski, N.; Herencias, C.; Peña, F.; Natalia Tarazona, N.; Revelles, O. A holistic view of polyhydroxyalkanoate metabolism in Pseudomonas putida. Environ. Microbiol. 2016, 18, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Liu, T.; Zheng, Z.; Chen, J.C.; Chen, G.Q. Polyhydroxyalkanoate synthases PhaC1 and PhaC2 from Pseudomonas stutzeri 1317 had different substrate specificities. FEMS Microbiol. Lett. 2004, 234, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Mavrodi, D.V.; Blankenfeldt, W.; Thomashow, L.S.; Mentel, M. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Ann. Rev. Phytopathol. 2006, 44, 417–445. [Google Scholar] [CrossRef] [PubMed]
- De Eugenio, L.I.; Galán, B.; Escapa, I.F.; Maestro, B.; Sanz, J.M.; García, J.L.; Prieto, M.A. The PhaD regulator controls the simultaneous expression of the pha genes involved in polyhydroxyalkanoate metabolism and turnover in Pseudomonas putida KT2442. Environ. Microbiol. 2010, 12, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Conte, E.; Catara, V.; Greco, S.; Russo, M.; Alicata, R.; Strano, L.; Lombardo, A.; Di Silvestro, S.; Catara, A. Regulation of polyhydroxyalkanoate synthases (phaC1 and phaC2) gene expression in Pseudomonas corrugata. Appl. Microbiol. Biotechnol. 2006, 72, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Newman, D.K. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nat. Chem. Biol. 2006, 2, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.F.; Ai, G.M.; Zheng, Q.X.; Liu, C.; Jiang, C.Y.; Liu, L.X.; Liu, Y.M.; Yang, C.; Liu, S.J. Metabolic flux responses to genetic modification for shikimic acid production by Bacillus subtilis strains. Microb. Cell Fact. 2014, 13, 40. [Google Scholar] [CrossRef] [PubMed]
- Krämer, M.; Bongaerts, J.; Bovenberg, R.; Kremer, S.; Müller, U.; Orf, S.; Wubbolts, M.; Raeven, L. Metabolic engineering for microbial production of shikimic acid. Metab. Eng. 2003, 5, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Jin, K.; Zhou, L.; Jiang, H.; Sun, S.; Fang, Y.; Liu, J.; Zhang, X.; He, Y.W. Engineering the central biosynthetic and secondary metabolic pathways of Pseudomonas aeruginosa strain PA1201 to improve phenazine-1-carboxylicacid production. Metab. Eng. 2015, 32, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Blankenfeldt, W.; Parsons, J.F. The structural biology of phenazine biosynthesis. Curr. Opin. Struct. Biol. 2014, 29, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Guo, S.; Iqbal, H.M.N.; Hu, H.; Wang, W.; Zhang, X. Engineering Pseudomonas for phenazine biosynthesis, regulation, and biotechnological applications: A review. World J. Microbiol. Biotechnol. 2017, 33, 191. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wang, Z.; Huang, X.; Hu, H.; Wang, W.; Zhang, X. Developing genome-reduced Pseudomonas chlororaphis strains for the production of secondary metabolites. BMC Genom. 2017, 18, 715. [Google Scholar] [CrossRef] [PubMed]
- Price-Whelan, A.; Dietrich, L.E.P.; Newman, D.K. Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 2007, 189, 6372–6381. [Google Scholar] [CrossRef] [PubMed]
- Blunt, W.; Dartiailh, C.; Sparling, R.; Gapes, D.; Levin, D.B.; Cicek, N. Microaerophilic environments improve the productivity of medium chain length polyhydroxyalkanoate biosynthesis from fatty acids in Pseudomonas putida LS46. Process Biochem. 2017, 59, 18–25. [Google Scholar] [CrossRef]
- Poblete-Castro, I.; Escapa, I.; Jager, C.; Puchalka, J.; Lam, C.; Schomburg, D.; Prieto, M.; Santos, M.D. The metabolic response of P. putida KT2442 producing high levels of polyhydroxyalkanoate under single and multiple-nutrient-limited growth: Highlights from a multi-level omics approach. Microb. Cell Fact. 2012, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Acuña, J.M.B.; Bielecka, A.; Häussler, S.; Schobert, M.; Jahn, M.; Wittmann, C.; Jahn, D.; Poblete-Castro, I. Production of medium chain length polyhydroxyalkanoate in metabolic flux optimized Pseudomonas putida. Microb. Cell Fact. 2014, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.Y.; Kim, M.K.; Park, Y.H.; Lee, S.Y. Regulatory effects of cellular nicotinamide nucleotides and enzyme activities on poly(3-hydroxybutyrate) synthesis in recombinant Escherichia coli. Biotechnol. Bioeng. 1996, 52, 707–712. [Google Scholar] [CrossRef]
- Escapa, I.F.; García, J.L.; Bühler, B.; Blank, L.M.; Prieto, M.A. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida. Environ. Microbiol. 2012, 14, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; de Roo, G.; Ruth, K.; Witholt, B.; Zinn, M.; Thöny-Meyer, L. Simultaneous accumulation and degradation of polyhydroxyalkanoates: Futile cycle or clever regulation? Biomacromolecules 2009, 10, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Domröse, A.; Weihmann, R.; Thies, S.; Jaeger, K.E.; Drepper, T.; Loeschck, A. Rapid generation of recombinant Pseudomonas putida secondary metabolite producers using yTREX. J. Synth. Syst. Biotechnol. 2017, 2, 310–319. [Google Scholar] [CrossRef] [PubMed]
| Strain/Mutant | Relevant Genotype, Phenotype or Sequence | Reference |
|---|---|---|
| P. chlororaphis PA23 | RifR; WT [soybean root tip isolate] PHZ+PRN+ | [2] |
| PA23-63 | PHZ- RifR phzE::Tn5-OT182 genomic fusion | [12] |
| PA23-8 | PRN- RifR prnBC deletion mutant | [12] |
| PA23-63-1 | PHZ- PRN- RifR phzE::Tn5-OT182 genomic fusion; prnBC deletion mutant | [12] |
| PA23rpoS | PA23 with pKNOCK inserted into rpoS | [25] |
| PA23gacS | PHZ- RifR gacS::Tn5-OT182 genomic fusion | [3] |
| PA23relA/spoT | PA23 with pKNOCK-Gm inserted into relA; TetR cassette inserted into spoT | [25] |
| PA23rpoS | PA23 with pKNOCK-Tc inserted into rpoS | [13] |
| Primers | ||
| phzA-FOR | 5′-GACTGGCAATGGCACAAC-3′ | [14] |
| phzA-REV | 5′-GCAATAACCTTCGGGATAACC-3′ | [14] |
| phzI-FOR | 5′-CGATGCCGTTGTTCTGG-3′ | [14] |
| phzI-REV | 5′-AGCCGTTCGTAGTGGACTC-3′ | [14] |
| phaF-FOR | 5′-GAAAAAGAAGGCAGCTCGTG-3′ | This study |
| phaF-REV | 5′-ATCGACTTTCTTGCCGACAG-3′ | This study |
| phaI-FOR | 5′-CTACACCAAGGTCGGTCAGG-3′ | This study |
| phaI-REV | 5′-ATCCAGCTGCACTTCGACTT-3′ | This study |
| phaD-FOR | 5′-CTGGGTATCGCTGACCAGTT-3′ | This study |
| phaD-REV | 5′-ACTACCGCTTCCTGTTCCAG-3′ | This study |
| phaC2-FOR | 5′-ATTCCAGATCAGGTCGTTGG-3′ | This study |
| phaC2-REV | 5′-GGTCAGCCTGCTGGATAGTC-3′ | This study |
| phaZ-FOR | 5′-CCTGCCCATAGTCGAGGTAA-3′ | This study |
| phaZ-REV | 5′-CTGGAGCTGGTGTTTCCATT-3′ | This study |
| phaC1-FOR | 5′-GCCAGGTAGGTTTGCAGGTA-3′ | This study |
| phaC1-REV | 5′-TCTGCTCGTATGGTGCTGAC-3′ | This study |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, P.K.; Munir, R.I.; Plouffe, J.; Shah, N.; De Kievit, T.; Levin, D.B. Polyhydroxyalkanoate (PHA) Polymer Accumulation and pha Gene Expression in Phenazine (phz-) and Pyrrolnitrin (prn-) Defective Mutants of Pseudomonas chlororaphis PA23. Polymers 2018, 10, 1203. https://doi.org/10.3390/polym10111203
Sharma PK, Munir RI, Plouffe J, Shah N, De Kievit T, Levin DB. Polyhydroxyalkanoate (PHA) Polymer Accumulation and pha Gene Expression in Phenazine (phz-) and Pyrrolnitrin (prn-) Defective Mutants of Pseudomonas chlororaphis PA23. Polymers. 2018; 10(11):1203. https://doi.org/10.3390/polym10111203
Chicago/Turabian StyleSharma, Parveen K., Riffat I. Munir, Jocelyn Plouffe, Nidhi Shah, Teresa De Kievit, and David B. Levin. 2018. "Polyhydroxyalkanoate (PHA) Polymer Accumulation and pha Gene Expression in Phenazine (phz-) and Pyrrolnitrin (prn-) Defective Mutants of Pseudomonas chlororaphis PA23" Polymers 10, no. 11: 1203. https://doi.org/10.3390/polym10111203
APA StyleSharma, P. K., Munir, R. I., Plouffe, J., Shah, N., De Kievit, T., & Levin, D. B. (2018). Polyhydroxyalkanoate (PHA) Polymer Accumulation and pha Gene Expression in Phenazine (phz-) and Pyrrolnitrin (prn-) Defective Mutants of Pseudomonas chlororaphis PA23. Polymers, 10(11), 1203. https://doi.org/10.3390/polym10111203