Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle
Abstract
1. Introduction
2. Materials and Methods
2.1. Crystallization
2.2. Sample Preparation for the Fixed-Target Scan
2.3. XFEL Data Collection
2.4. Data Processing
2.5. Initial Structure Determination
3. Results
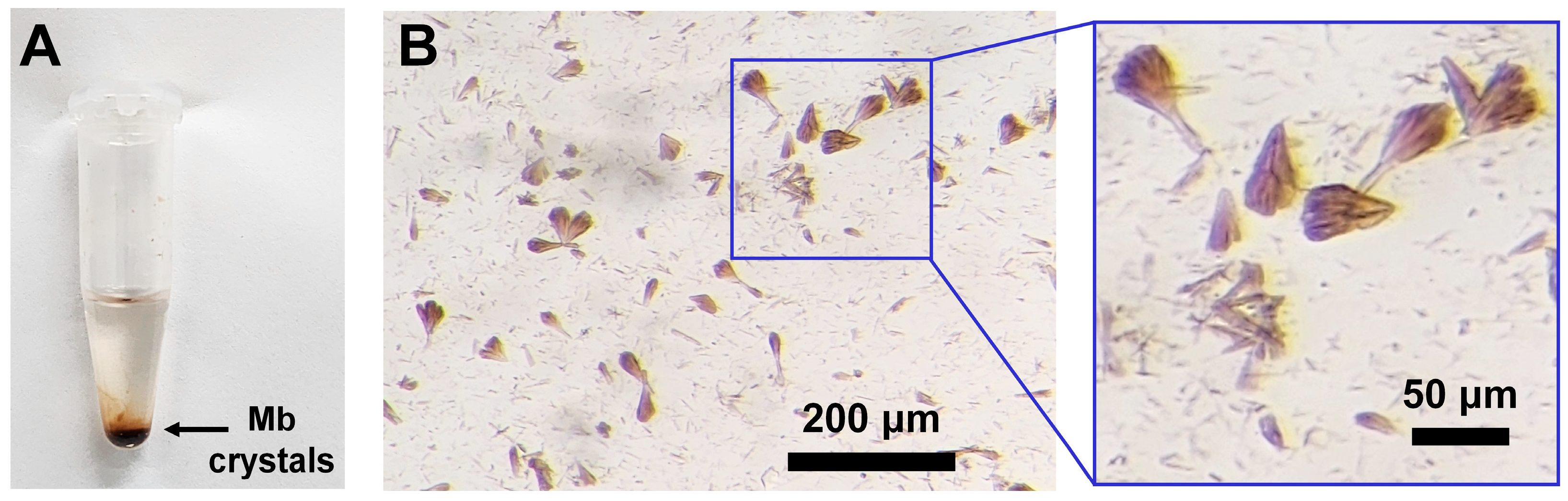
3.1. Mb Crystallization
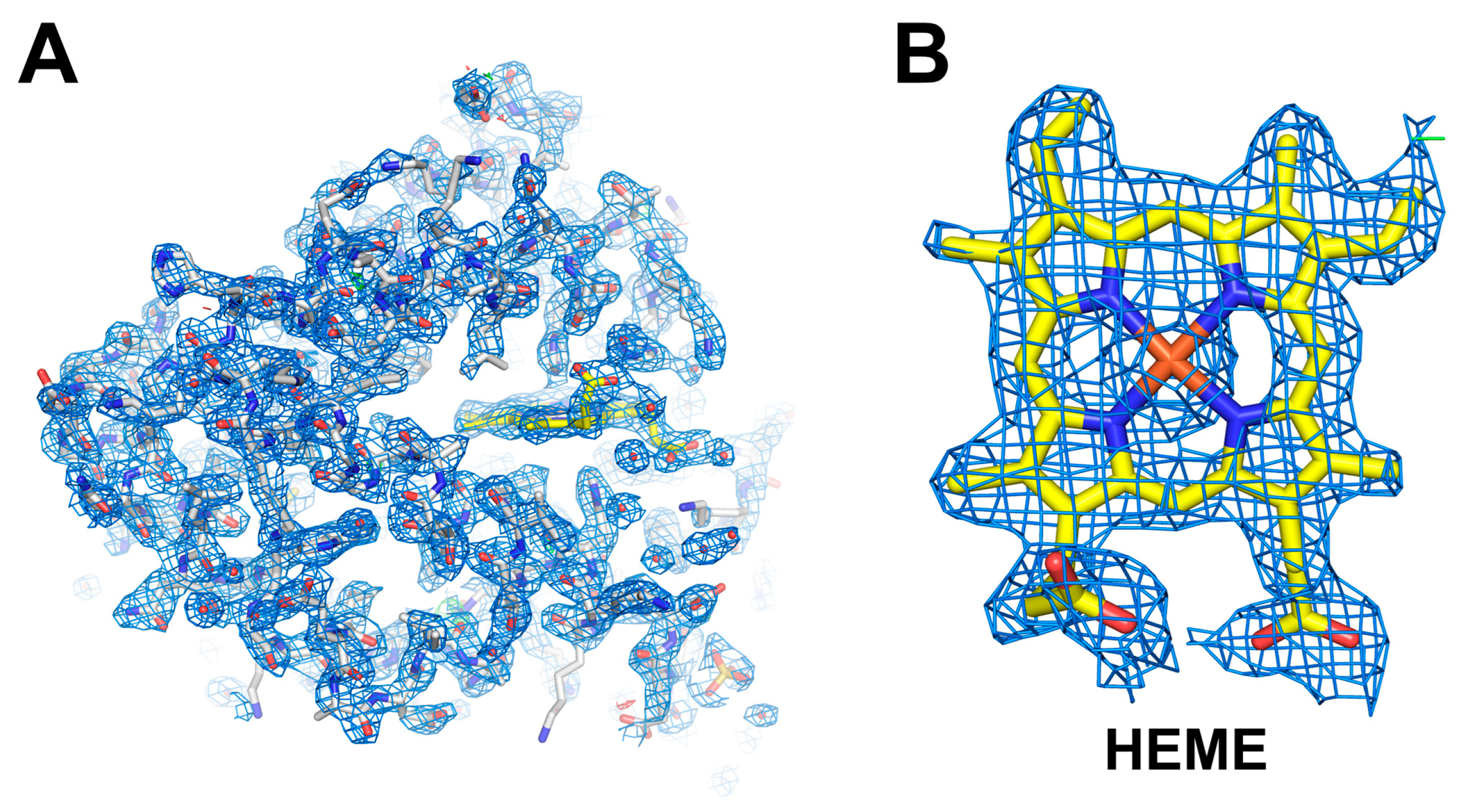
3.2. XFEL Data Processing for Mb
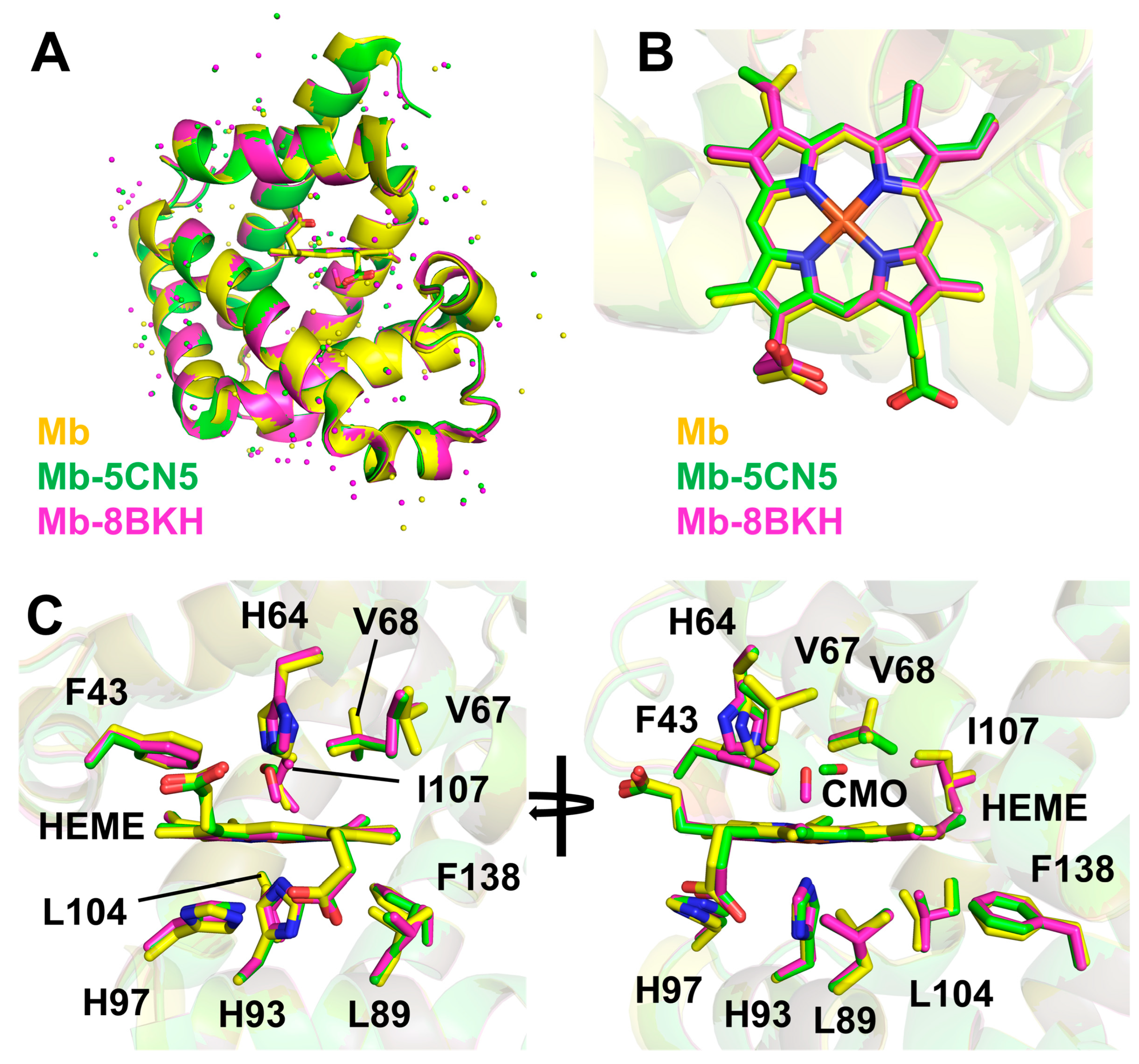
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Mb | Myoglobin |
| SFX | Serial Femtosecond Crystallography |
| XFEL | X-ray Free Electron Laser |
References
- Wittenberg, B.A.; Wittenberg, J.B. Myoglobin-mediated oxygen delivery to mitochondria of isolated cardiac myocytes. Proc. Natl. Acad. Sci. USA 1987, 84, 7503–7507. [Google Scholar] [CrossRef]
- Gros, G.; Wittenberg, B.A.; Jue, T. Myoglobin’s old and new clothes: From molecular structure to function in living cells. J. Exp. Biol. 2010, 213, 2713–2725. [Google Scholar] [CrossRef]
- Wittenberg, J.B.; Wittenberg, B.A. Myoglobin function reassessed. J. Exp. Biol. 2003, 206, 2011–2020. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of Myoglobin: A Three-Dimensional Fourier Synthesis at 2 Å. Resolution. Nature 1960, 185, 422–427. [Google Scholar] [CrossRef]
- Carlsson, M.L.R.; Kanagarajan, S.; Bülow, L.; Zhu, L.-H. Plant based production of myoglobin—A novel source of the muscle heme-protein. Sci. Rep. 2020, 10, 920. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Liang, G.; Dong, J.; Zhou, J.; Li, J.; Chen, J.; Du, G.; Chen, J.; Wang, Z.; Zhao, X. Effect of myoglobin on the flavor, color and texture of high-moisture soy protein concentrate -wheat gluten extrudates. Food Chem. 2025, 473, 143102. [Google Scholar] [CrossRef] [PubMed]
- Plebani, M.; Zaninotto, M. Diagnostic strategies using myoglobin measurement in myocardial infarction. Clin. Chim. Acta 1998, 272, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Vojtěchovský, J.; Chu, K.; Berendzen, J.; Sweet, R.M.; Schlichting, I. Crystal Structures of Myoglobin-Ligand Complexes at Near-Atomic Resolution. Biophys. J. 1999, 77, 2153–2174. [Google Scholar] [CrossRef]
- Postnikova, G.B.; Shekhovtsova, E.A. Myoglobin: Oxygen Depot or Oxygen Transporter to Mitochondria? A Novel Mechanism of Myoglobin Deoxygenation in Cells (review). Biochemistry 2018, 83, 168–183. [Google Scholar] [CrossRef]
- Strandberg, B. Chapter 1: Building the Ground for the First Two Protein Structures: Myoglobin and Haemoglobin. J. Mol. Biol. 2009, 392, 2–10. [Google Scholar] [CrossRef]
- Brunori, M.; Bourgeois, D.; Vallone, B. The structural dynamics of myoglobin. J. Struct. Biol. 2004, 147, 223–234. [Google Scholar] [CrossRef]
- Kaieda, S.; Halle, B. Internal Water and Microsecond Dynamics in Myoglobin. J. Phys. Chem. B 2013, 117, 14676–14687. [Google Scholar] [CrossRef]
- Kern, D.; Zuiderweg, E.R.P. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003, 13, 748–757. [Google Scholar] [CrossRef]
- de Chadarevian, S. John Kendrew and myoglobin: Protein structure determination in the 1950s. Protein Sci. 2018, 27, 1136–1143. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Radiation Damage on Thaumatin: A Case Study of Crystals That Are Larger Than the Microfocusing X-ray Beam. Appl. Sci. 2023, 13, 1876. [Google Scholar] [CrossRef]
- Nam, K.H. Radiation Damage on Selenomethionine-Substituted Single-Domain Substrate-Binding Protein. Crystals 2023, 13, 1620. [Google Scholar] [CrossRef]
- Nam, K.H. Guide to serial synchrotron crystallography. Curr. Res. Struct. Biol. 2024, 7, 100131. [Google Scholar] [CrossRef]
- Nam, K.H. Temperature-Dependent Structural Changes of the Active Site and Substrate-Binding Cleft in Hen Egg White Lysozyme. Crystals 2025, 15, 111. [Google Scholar] [CrossRef]
- Garman, E.F. Radiation damage in macromolecular crystallography: What is it and why should we care? Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Shelley, K.L.; Garman, E.F. Quantifying and comparing radiation damage in the Protein Data Bank. Nat. Commun. 2022, 13, 1314. [Google Scholar] [CrossRef]
- Pfanzagl, V.; Beale, J.H.; Michlits, H.; Schmidt, D.; Gabler, T.; Obinger, C.; Djinović-Carugo, K.; Hofbauer, S. X-ray–induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner. J. Biol. Chem. 2020, 295, 13488–13501. [Google Scholar] [CrossRef]
- Della Longa, S.; Arcovito, A.; Benfatto, M.; Congiu-Castellano, A.; Girasole, M.; Hazemann, J.L.; Lo Bosco, A. Redox-Induced Structural Dynamics of Fe-Heme Ligand in Myoglobin by X-Ray Absorption Spectroscopy. Biophys. J. 2003, 85, 549–558. [Google Scholar] [CrossRef]
- Boutet, S.; Lomb, L.; Williams, G.J.; Barends, T.R.M.; Aquila, A.; Doak, R.B.; Weierstall, U.; DePonte, D.P.; Steinbrener, J.; Shoeman, R.L.; et al. High-Resolution Protein Structure Determination by Serial Femtosecond Crystallography. Science 2012, 337, 362–364. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Stauch, B.; Cherezov, V.; Schlichting, I. Serial femtosecond crystallography. Nat. Rev. Methods Primers 2022, 2, 59. [Google Scholar] [CrossRef] [PubMed]
- Chapman, H.N.; Caleman, C.; Timneanu, N. Diffraction before destruction. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130313. [Google Scholar] [CrossRef] [PubMed]
- Neutze, R.; Wouts, R.; van der Spoel, D.; Weckert, E.; Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 2000, 406, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Orville, A.M. Recent results in time resolved serial femtosecond crystallography at XFELs. Curr. Opin. Struct. Biol. 2020, 65, 193–208. [Google Scholar] [CrossRef]
- Park, J.; Nam, K.H. Recent chemical mixing devices for time-resolved serial femtosecond crystallography. TrAC Trends Anal. Chem. 2024, 172, 117554. [Google Scholar] [CrossRef]
- Park, J.; Nam, K.H. Experimental approaches for time-resolved serial femtosecond crystallography at PAL-XFEL. In Time-Resolved Methods in Structural Biology; Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2024; pp. 131–160. [Google Scholar]
- Barends, T.R.M.; Foucar, L.; Ardevol, A.; Nass, K.; Aquila, A.; Botha, S.; Doak, R.B.; Falahati, K.; Hartmann, E.; Hilpert, M.; et al. Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation. Science 2015, 350, 445–450. [Google Scholar] [CrossRef]
- Barends, T.R.M.; Gorel, A.; Bhattacharyya, S.; Schirò, G.; Bacellar, C.; Cirelli, C.; Colletier, J.-P.; Foucar, L.; Grünbein, M.L.; Hartmann, E.; et al. Influence of pump laser fluence on ultrafast myoglobin structural dynamics. Nature 2024, 626, 905–911. [Google Scholar] [CrossRef]
- Oghbaey, S.; Sarracini, A.; Ginn, H.M.; Pare-Labrosse, O.; Kuo, A.; Marx, A.; Epp, S.W.; Sherrell, D.A.; Eger, B.T.; Zhong, Y.; et al. Fixed target combined with spectral mapping: Approaching 100% hit rates for serial crystallography. Acta Crystallogr. D Struct. Biol. 2016, 72, 944–955. [Google Scholar] [CrossRef]
- Owen, R.L.; Axford, D.; Sherrell, D.A.; Kuo, A.; Ernst, O.P.; Schulz, E.C.; Miller, R.J.; Mueller-Werkmeister, H.M. Low-dose fixed-target serial synchrotron crystallography. Acta Crystallogr. D Struct. Biol. 2017, 73, 373–378. [Google Scholar] [CrossRef]
- Calvey, G.D.; Katz, A.M.; Zielinski, K.A.; Dzikovski, B.; Pollack, L. Characterizing Enzyme Reactions in Microcrystals for Effective Mix-and-Inject Experiments using X-ray Free-Electron Lasers. Anal. Chem. 2020, 92, 13864–13870. [Google Scholar] [CrossRef]
- Mehrabi, P.; Bücker, R.; Bourenkov, G.; Ginn, H.M.; von Stetten, D.; Müller-Werkmeister, H.M.; Kuo, A.; Morizumi, T.; Eger, B.T.; Ou, W.L.; et al. Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison. Sci. Adv. 2021, 7, eabf1380. [Google Scholar] [CrossRef]
- Brucker, E.A.; Olson, J.S.; Phillips, G.N.; Dou, Y.; Ikeda-Saito, M. High Resolution Crystal Structures of the Deoxy, Oxy, and Aquomet Forms of Cobalt Myoglobin. J. Biol. Chem. 1996, 271, 25419–25422. [Google Scholar] [CrossRef]
- Deng, Y.; Albert, T.; Van Stappen, C.; Palacios, P.M.; Amador, M.L.; Dwaraknath, S.; Guo, Y.; Moënne-Loccoz, P.; Lu, Y. Unveiling an Ultrareduced State of Heme in Myoglobin. J. Am. Chem. Soc. 2025, 147, 29983–29993. [Google Scholar] [CrossRef]
- Nam, K.H. Real-time monitoring of large-scale crystal growth using batch crystallization for serial crystallography. J. Cryst. Growth 2023, 614, 127219. [Google Scholar] [CrossRef]
- Lee, D.; Baek, S.; Park, J.; Lee, K.; Kim, J.; Lee, S.J.; Chung, W.K.; Lee, J.L.; Cho, Y.; Nam, K.H. Nylon mesh-based sample holder for fixed-target serial femtosecond crystallography. Sci. Rep. 2019, 9, 6971. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, S.; Nam, K.H.; Kim, B.; Ko, I.S. Current status of the CXI beamline at the PAL-XFEL. J. Korean Phys. Soc. 2016, 69, 1089–1093. [Google Scholar] [CrossRef]
- Kang, H.S.; Min, C.K.; Heo, H.; Kim, C.; Yang, H.; Kim, G.; Nam, I.; Baek, S.Y.; Choi, H.J.; Mun, G.; et al. Hard X-ray free-electron laser with femtosecond-scale timing jitter. Nat. Photonics 2017, 11, 708–713. [Google Scholar] [CrossRef]
- Kim, J.; Kim, H.Y.; Park, J.; Kim, S.; Kim, S.; Rah, S.; Lim, J.; Nam, K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick-Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018, 25, 289–292. [Google Scholar] [CrossRef]
- Mariani, V.; Morgan, A.; Yoon, C.H.; Lane, T.J.; White, T.A.; O’Grady, C.; Kuhn, M.; Aplin, S.; Koglin, J.; Barty, A.; et al. OnDA: Online data analysis and feedback for serial X-ray imaging. J. Appl. Crystallogr. 2016, 49, 1073–1080. [Google Scholar] [CrossRef]
- Barty, A.; Kirian, R.A.; Maia, F.R.; Hantke, M.; Yoon, C.H.; White, T.A.; Chapman, H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014, 47, 1118–1131. [Google Scholar] [CrossRef]
- White, T.A. Processing serial crystallography data with CrystFEL: A step-by-step guide. Acta Crystallogr. D Struct. Biol. 2019, 75, 219–233. [Google Scholar] [CrossRef]
- Gevorkov, Y.; Yefanov, O.; Barty, A.; White, T.A.; Mariani, V.; Brehm, W.; Tolstikova, A.; Grigat, R.R.; Chapman, H.N. XGANDALF—Extended gradient descent algorithm for lattice finding. Acta Crystallogr. A Found. Adv. 2019, 75, 694–704. [Google Scholar] [CrossRef]
- Yefanov, O.; Mariani, V.; Gati, C.; White, T.A.; Chapman, H.N.; Barty, A. Accurate determination of segmented X-ray detector geometry. Opt. Express 2015, 23, 28459–28470. [Google Scholar] [CrossRef]
- Liebschner, D.; Afonine, P.V.; Baker, M.L.; Bunkoczi, G.; Chen, V.B.; Croll, T.I.; Hintze, B.; Hung, L.W.; Jain, S.; McCoy, A.J.; et al. Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 2019, 75, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Maurus, R.; Bogumil, R.; Nguyen, N.T.; Mauk, A.G.; Brayer, G. Structural and spectroscopic studies of azide complexes of horse heart myoglobin and the His-64→Thr variant. Biochem. J. 1998, 332, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Gevorkov, Y.; Barty, A.; Brehm, W.; White, T.A.; Tolstikova, A.; Wiedorn, M.O.; Meents, A.; Grigat, R.-R.; Chapman, H.N.; Yefanov, O. pinkIndexer—A universal indexer for pink-beam X-ray and electron diffraction snapshots. Acta Crystallogr. A Found. Adv. 2020, 76, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Beyerlein, K.R.; White, T.A.; Yefanov, O.; Gati, C.; Kazantsev, I.G.; Nielsen, N.F.-G.; Larsen, P.M.; Chapman, H.N.; Schmidt, S. FELIX: An algorithm for indexing multiple crystallites in X-ray free-electron laser snapshot diffraction images. J. Appl. Crystallogr. 2017, 50, 1075–1083. [Google Scholar] [CrossRef]
- Nam, K.H. Impact of Diffraction Data Volume on Data Quality in Serial Crystallography. Crystals 2025, 15, 104. [Google Scholar] [CrossRef]
- Park, J.; Park, S.; Kim, J.; Park, G.; Cho, Y.; Nam, K.H. Polyacrylamide injection matrix for serial femtosecond crystallography. Sci. Rep. 2019, 9, 2525. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Park, S.; Park, J. Preliminary XFEL data from spontaneously grown endo-1,4-β-xylanase crystals from Hypocrea virens. Acta Crystallogr. F Struct. Biol. Commun. 2022, 78, 226–231. [Google Scholar] [CrossRef] [PubMed]

| Data Collection | Mb |
|---|---|
| X-ray Source | NCI, PAL-XFEL |
| Detector | MX225-HS (4 × 4 pixel binning) |
| XFEL energy (ev) | 9500 |
| Pulse width (fs) | ~20 |
| Collected images | 130,665 |
| Hit images | 70,195 |
| Indexed images | 1376 |
| Indexed diffraction pattern | 1389 |
| Space group | P21 |
| Cell dimension | |
| a, b, c (Å) α, β, γ (°) | 63.69, 28.62, 35.59 90.0, 107.16, 90.0 |
| Resolution (Å) | 61.72–2.30 (2.38–2.30) |
| Unique reflections | 5781 (577) |
| Completeness (%) | 99.24 (98.97) |
| Redundancy | 11.0 (7.3) |
| SNR a | 2.07 (1.22) |
| CC1/2 | 0.7261 (0.4101) |
| CC* | 0.917 (0.7627) |
| Rsplit (%) b | 50.27 (82.31) |
| Wilson B-factor (Å2) | 27.66 |
| Molecular Replacement | Mb |
|---|---|
| Top LLG | 960.605 |
| Top TFZ | 24.5 |
| Refinement | |
| Resolution (Å) | 61.42–2.30 (2.53–2.30) |
| Rwork | 0.2468 (0.2402) |
| Rfree | 0.3174 (0.3636) |
| R.m.s.deviation | |
| Bonds (Å) | 0.009 |
| Angles (°) | 1.027 |
| Average B factors (Å2) | |
| Protein | 16.37 |
| Heme | 12.24 |
| Water | 18.12 |
| Ramachandran plot | |
| Favored (%) | 95.30 |
| Allowed (%) | 4.70 |
| PDB code | 9X07 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.; Park, S.; Nam, K.H. Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle. Crystals 2025, 15, 905. https://doi.org/10.3390/cryst15100905
Park J, Park S, Nam KH. Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle. Crystals. 2025; 15(10):905. https://doi.org/10.3390/cryst15100905
Chicago/Turabian StylePark, Jaehyun, Sehan Park, and Ki Hyun Nam. 2025. "Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle" Crystals 15, no. 10: 905. https://doi.org/10.3390/cryst15100905
APA StylePark, J., Park, S., & Nam, K. H. (2025). Preliminary Serial Femtosecond Crystallography Studies of Myoglobin from Equine Skeletal Muscle. Crystals, 15(10), 905. https://doi.org/10.3390/cryst15100905







