Crystal Structure of the Biocide Methylisothiazolinone
Abstract
1. Introduction
2. Materials and Methods
2.1. In Situ Cryocrystallization
2.2. X-Ray Intensity Data Collection and Processing
2.3. Structure Solution and Refinement
2.4. Computational Methods
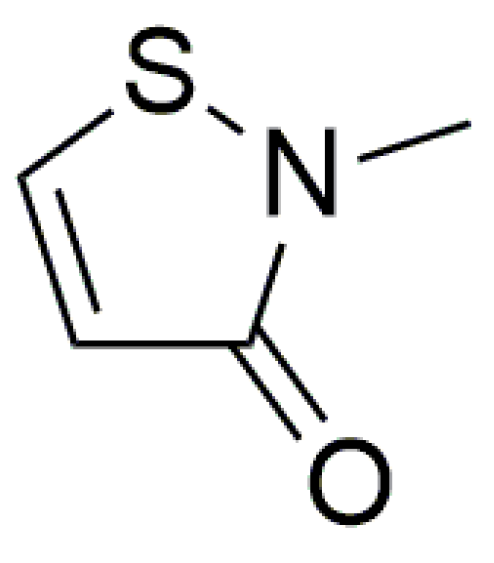
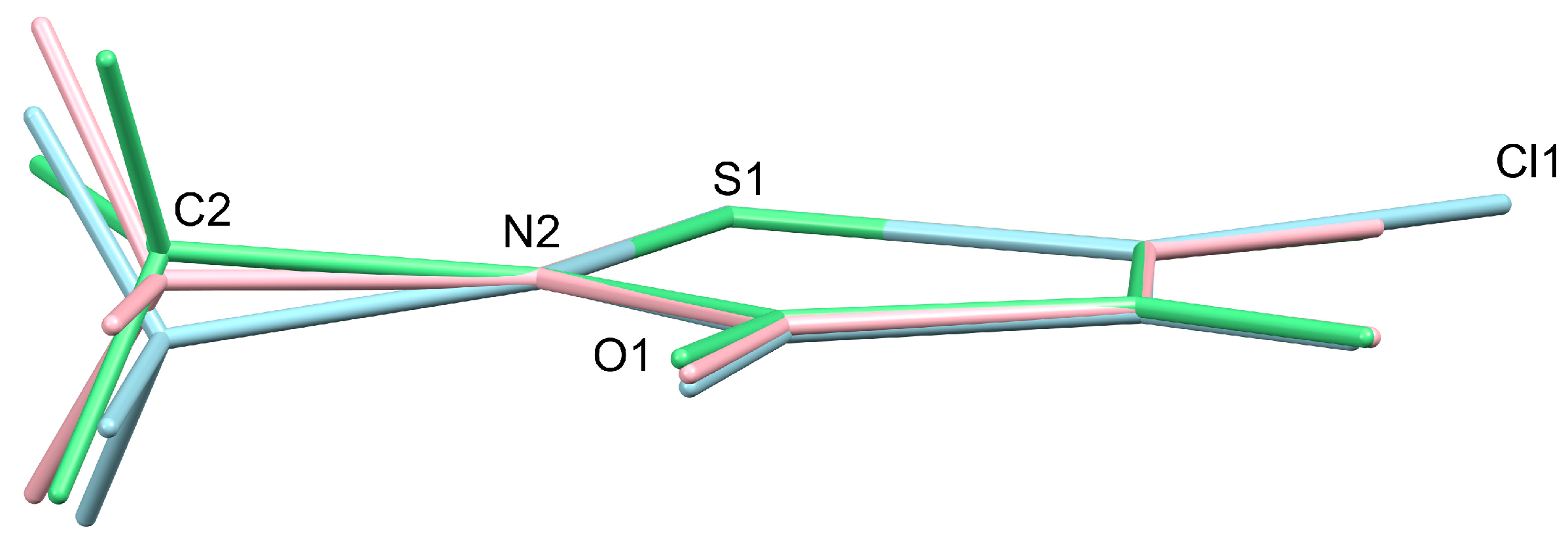
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morley, J.O.; Oliver Kapur, A.J.; Charlton, M.H. Structure–activity relationships in 3-isothiazolones. Org. Biomol. Chem. 2005, 3, 3713–3719. [Google Scholar] [CrossRef] [PubMed]
- Lundov, M.D.; Johansen, J.D.; Zachariae, C.; Moesby, L. Low-level efficacy of cosmetic preservatives. Int. J. Cosmet. Sci. 2011, 33, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Collier, P.J.; Ramsey, A.J.; Austin, P.; Gilbert, P. Growth inhibitory and biocidal activity of some isothiazolone biocides. J. Appl. Bacteriol. 1990, 69, 569–577. [Google Scholar] [CrossRef]
- Wisastra, R.; Ghizzoni, M.; Maarsingh, H.; Minnaard, A.J.; Haisma, H.J.; Dekker, F.J. Isothiazolones; thiol-reactive inhibitors of cysteine protease cathepsin B and histone acetyltransferase PCAF. Org. Biomol. Chem. 2011, 9, 1817–1822. [Google Scholar] [CrossRef] [PubMed]
- Crow, W.D.; Leonard, N.J. 3-Isothiazolone-cis-3-Thiocyanoacrylamide Equilibria1,2. J. Org. Chem. 1965, 30, 2660–2665. [Google Scholar] [CrossRef]
- Silva, V.; Silva, C.; Soares, P.; Garrido, E.M.; Borges, F.; Garrido, J. Isothiazolinone Biocides: Chemistry, Biological, and Toxicity Profiles. Molecules 2020, 25, 991. [Google Scholar] [CrossRef] [PubMed]
- Song, M.-K.; Baek, Y.-W.; Kim, D.I.; Yoon, S.-H.; Lee, K. Effects of stabilizer magnesium nirate on CMIT/MIT-induced respiratory toxicity. Toxicol. Res. 2023, 39, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, E.; Waeber, R.; Niederer, M.; Maurer, T.; Maly, P.; Scherer, S. Preservation of products with MCI/MI in Switzerland. Contact Dermat. 2001, 45, 257–264. [Google Scholar] [CrossRef]
- Ghose, A.K.; Viswanadhan, V.N.; Wendoloski, J.J. Prediction of Hydrophobic (Lipophilic) Properties of Small Organic Molecules Using Fragmental Methods: An Analysis of ALOGP and CLOGP Methods. J. Phys. Chem. A 1998, 102, 3762–3772. [Google Scholar] [CrossRef]
- Alvarez-Sánchez, R.; Basketter, D.; Pease, C.; Lepoittevin, J.-P. Studies of Chemical Selectivity of Hapten, Reactivity, and Skin Sensitization Potency. 3. Synthesis and Studies on the Reactivity toward Model Nucleophiles of the 13C-Labeled Skin Sensitizers, 5-Chloro-2-methylisothiazol-3-one (MCI) and 2-Methylisothiazol-3-one (MI). Chem. Res. Toxicol. 2003, 16, 627–636. [Google Scholar]
- Arning, J.; Matzke, M.; Stolte, S.; Nehen, F.; Bottin-Weber, U.; Böschen, A.; Abdulkarim, S.; Jastorff, B.; Ranke, J. Analyzing Cytotoxic Effects of Selected Isothiazol-3-one Biocides Using the Toxic Ratio Concept and Structure−Activity Relationship Considerations. Chem. Res. Toxicol. 2009, 22, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Hunziker, N. The ‘Isothiazolinone Story’. Dermatology 2009, 184, 85–86. [Google Scholar] [CrossRef]
- Isaksson, M. Successful inhibition of allergic contact dermatitis caused by methylchloroisothiazolinone/methylisothiazolinone with topical glutathione. Contact Dermat. 2015, 73, 126–128. [Google Scholar] [CrossRef] [PubMed]
- Gruvberger, B.; Bruze, M. Can Glutathione-Containing Emollients Inactivate methylchloroisothiazolinone/methylisothiazolinone? Contact Dermat. 1998, 38, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Crow, W.D.; Leonard, N.J. A synthesis of 3-isothiazolones. Tetrahedron Lett. 1964, 5, 1477–1480. [Google Scholar] [CrossRef]
- Leonard, N.J.; Edwin Wilson, G. Rearrangement of a 1,4-thiazepine ring and formation of 3-isothiazolones. Tetrahedron Lett. 1964, 5, 1471–1475. [Google Scholar] [CrossRef]
- Rothe, H.; Ryan, C.A.; Page, L.; Vinall, J.; Goebel, C.; Scheffler, H.; Toner, F.; Roper, C.; Kern, P.S. Application of in vitro skin penetration measurements to confirm and refine the quantitative skin sensitization risk assessment of methylisothiazolinone. Regul. Toxicol. Pharmacol. 2017, 91, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Virgilio, J.A.; Manowitz, M.; Heilweil, E. 2-Alkyl-3-Haloisothiazolium Salts and Their Derivatives 1979. U.S. Patent 4,281,136, 27 July 1981. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Kleemiss, F.; Dolomanov, O.V.; Bodensteiner, M.; Peyerimhoff, N.; Midgley, L.; Bourhis, L.J.; Genoni, A.; Malaspina, L.A.; Jayatilaka, D.; Spencer, J.L.; et al. Accurate crystal structures and chemical properties from NoSpherA2. Chem. Sci. 2021, 12, 1675–1692. [Google Scholar] [CrossRef]
- Midgley, L.; Bourhis, L.J.; Dolomanov, O.V.; Grabowsky, S.; Kleemiss, F.; Puschmann, H.; Peyerimhoff, N. Vanishing of the atomic form factor derivatives in non-spherical structural refinement—A key approximation scrutinized in the case of Hirshfeld atom refinement. Acta Crystallogr. A 2021, 77, 519–533. [Google Scholar] [CrossRef] [PubMed]
- Bourhis, L.J.; Dolomanov, O.V.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. The anatomy of a comprehensive constrained, restrained refinement program for the modern computing environment—Olex2 dissected. Acta Crystallogr. A 2015, 71, 59–75. [Google Scholar] [CrossRef]
- Neese, F. A perspective on the future of quantum chemical software: The example of the ORCA program package. Faraday Discuss. 2024, 254, 295–314. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F.; Ahlrichs, R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.T.A.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Hertwig, R.H.; Koch, W. On the parameterization of the local correlation functional. What is Becke-3-LYP? Chem. Phys. Lett. 1997, 268, 345–351. [Google Scholar] [CrossRef]
- Weigend, F. Accurate Coulomb-fitting basis sets for H to Rn. Phys. Chem. Chem. Phys. 2006, 8, 1057–1065. [Google Scholar] [CrossRef]
- Fletcher, R. Practical Methods of Optimization, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2000. [Google Scholar]
- Dovesi, R.; Erba, A.; Orlando, R.; Zicovich-Wilson, C.M.; Civalleri, B.; Maschio, L.; Rérat, M.; Casassa, S.; Baima, J.; Salustro, S.; et al. Quantum-mechanical condensed matter simulations with CRYSTAL. WIREs Comput. Mol. Sci. 2018, 8, e1360. [Google Scholar] [CrossRef]
- Dovesi, R.; Pascale, F.; Civalleri, B.; Doll, K.; Harrison, N.M.; Bush, I.; D’Arco, P.; Noël, Y.; Rérat, M.; Carbonnière, P.; et al. The CRYSTAL code, 1976–2020 and beyond, a long story. J. Chem. Phys. 2020, 152, 204111. [Google Scholar] [CrossRef] [PubMed]
- Bursch, M.; Mewes, J.-M.; Hansen, A.; Grimme, S. Best-Practice DFT Protocols for Basic Molecular Computational Chemistry. Angew. Chem. Int. Ed. 2022, 61, e202205735. [Google Scholar] [CrossRef] [PubMed]
- Perdew, J.P.; Burke, K.; Ernzerhof, M. Generalized Gradient Approximation Made Simple. Phys. Rev. Lett. 1996, 77, 3865–3868. [Google Scholar] [CrossRef]
- Grimme, S.; Ehrlich, S.; Goerigk, L. Effect of the damping function in dispersion corrected density functional theory. J. Comput. Chem. 2011, 32, 1456–1465. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Hansen, A.; Brandenburg, J.G.; Bannwarth, C. Dispersion-Corrected Mean-Field Electronic Structure Methods. Chem. Rev. 2016, 116, 5105–5154. [Google Scholar] [CrossRef] [PubMed]
- Grimme, S.; Antony, J.; Ehrlich, S.; Krieg, H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010, 132, 154104. [Google Scholar] [CrossRef]
- Groom, C.R.; Bruno, I.J.; Lightfoot, M.P.; Ward, S.C. The Cambridge Structural Database. Acta Crystallogr. Sect. B 2016, 72, 171–179. [Google Scholar] [CrossRef]
- Kato, M.; Fujihara, T.; Yano, D.; Nagasawa, A. 5-Chloro-2-methylisothiazolin-3-one: Intermolecular two-dimensional networks via unusual C-Cl...O&z-dbnd;C interactions. Acta Crystallogr. Sect. E 2007, 63, o3097. [Google Scholar]
- Sekine, A.; Mitsumori, T.; Uekusa, H.; Ohashi, Y.; Yagi, M. Crystal Structure of a Host-guest Complex of Gallic Acid Methyl Ester and 5-Chloro-2-methyl-4-isothiazoline-3-one. Anal. Sci. X-Ray Struct. Anal. Online 2003, 19, x47–x48. [Google Scholar] [CrossRef][Green Version]
- Sekine, A.; Jomoto, K.; Uekusa, H.; Ohashi, Y.; Yagi, M. Crystal Structure of a Host-guest Complex of 4,4′-Ethylidenebisphenol and 5-Chloro-2-methyl-4-isothiazoline-3-one. Anal. Sci. X-Ray Struct. Anal. Online 2003, 19, x45–x46. [Google Scholar] [CrossRef][Green Version]
- Wang, F.; Chen, C.; Deng, G.; Xi, C. Concise Approach to Benzisothiazol-3(2H)-one via Copper-Catalyzed Tandem Reaction of o-Bromobenzamide and Potassium Thiocyanate in Water. J. Org. Chem. 2012, 77, 4148–4151. [Google Scholar] [CrossRef] [PubMed]
- Kitaigorodsky, A.I. Molecular Crystals and Molecules; Academic Press: New York, NY, USA, 1973. [Google Scholar]
- Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer model energies and energy frameworks: Extension to metal coordination compounds, organic salts, solvates and open-shell systems IUCrJ 2017, 4, 575–587. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]



| empirical formula | C4H5NOS |
| Mr | 115.157 |
| T (K) | 100(2) |
| λ (Å) | 0.71073 |
| crystal system | triclinic |
| space group | P-1 |
| a (Å) | 5.6037(6) |
| b (Å) | 6.4473(7) |
| c (Å) | 7.6391(9) |
| α (°) | 67.857(2) |
| β (°) | 77.541(3) |
| γ (°) | 84.523(2) |
| V (Å3) | 249.59(5) |
| Z | 2 |
| pcalc (mg m−3) | 1.532 |
| μ (mm−1) | 0.507 |
| F(000) | 120.313 |
| crystal size (mm) | 0.51 × 0.44 × 0.30 |
| θ range for data collection (°) | 2.94–41.67 |
| reflections collected/unique | 17,950/3288 |
| Rint | 0.0391 |
| observed reflections [I > 2σ(I)] | 2786 |
| data/restraints/parameters | 3288/0/134 |
| goodness-of-fit on F2 | 1.1698 |
| R1 [I > 2σ(I)] | 0.0244 |
| wR2 (all data) | 0.0598 |
| Δpmax, Δpmin (e Å−3) | 0.383, −0.247 |
| Distance | Angle | ||
|---|---|---|---|
| S1-N2 | 1.6851(5) | N2-S1-C5 | 90.81(2) |
| S1-C5 | 1.7096(6) | C3-C4-C5 | 112.68(4) |
| C4-C5 | 1.3509(7) | C4-C3-N2 | 108.79(4) |
| C3-C4 | 1.4533(7) | S1-N2-C3 | 114.93(4) |
| C3-O1 | 1.2373(6) | S1-N2-C4 | 122.41(4) |
| C3-N2 | 1.3734(6) | C2-N2-C4 | 122.34(4) |
| C2-N2 | 1.4508(7) | N2-C3-O1 | 122.59(4) |
| C-Haverage | 1.06(1) | C4-C3-O1 | 128.61(4) |
| Interaction | C-H | H···O | C···O | Angle |
|---|---|---|---|---|
| C5-H5···O1 1 | 1.056(10) | 2.163(11) | 2.9635(7) | 130.9(8) |
| C4-H4···O1 2 | 1.077(9) | 2.437(9) | 3.4733(7) | 160.9(9) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goddard, R.; Seidel, R.W.; Patzer, M.; Nöthling, N. Crystal Structure of the Biocide Methylisothiazolinone. Crystals 2024, 14, 1100. https://doi.org/10.3390/cryst14121100
Goddard R, Seidel RW, Patzer M, Nöthling N. Crystal Structure of the Biocide Methylisothiazolinone. Crystals. 2024; 14(12):1100. https://doi.org/10.3390/cryst14121100
Chicago/Turabian StyleGoddard, Richard, Rüdiger W. Seidel, Michael Patzer, and Nils Nöthling. 2024. "Crystal Structure of the Biocide Methylisothiazolinone" Crystals 14, no. 12: 1100. https://doi.org/10.3390/cryst14121100
APA StyleGoddard, R., Seidel, R. W., Patzer, M., & Nöthling, N. (2024). Crystal Structure of the Biocide Methylisothiazolinone. Crystals, 14(12), 1100. https://doi.org/10.3390/cryst14121100






