Structures, Characterization and DFT Studies of Four Novel Nickel Phenanthroline Complexes
Abstract
1. Introduction
2. Experimental
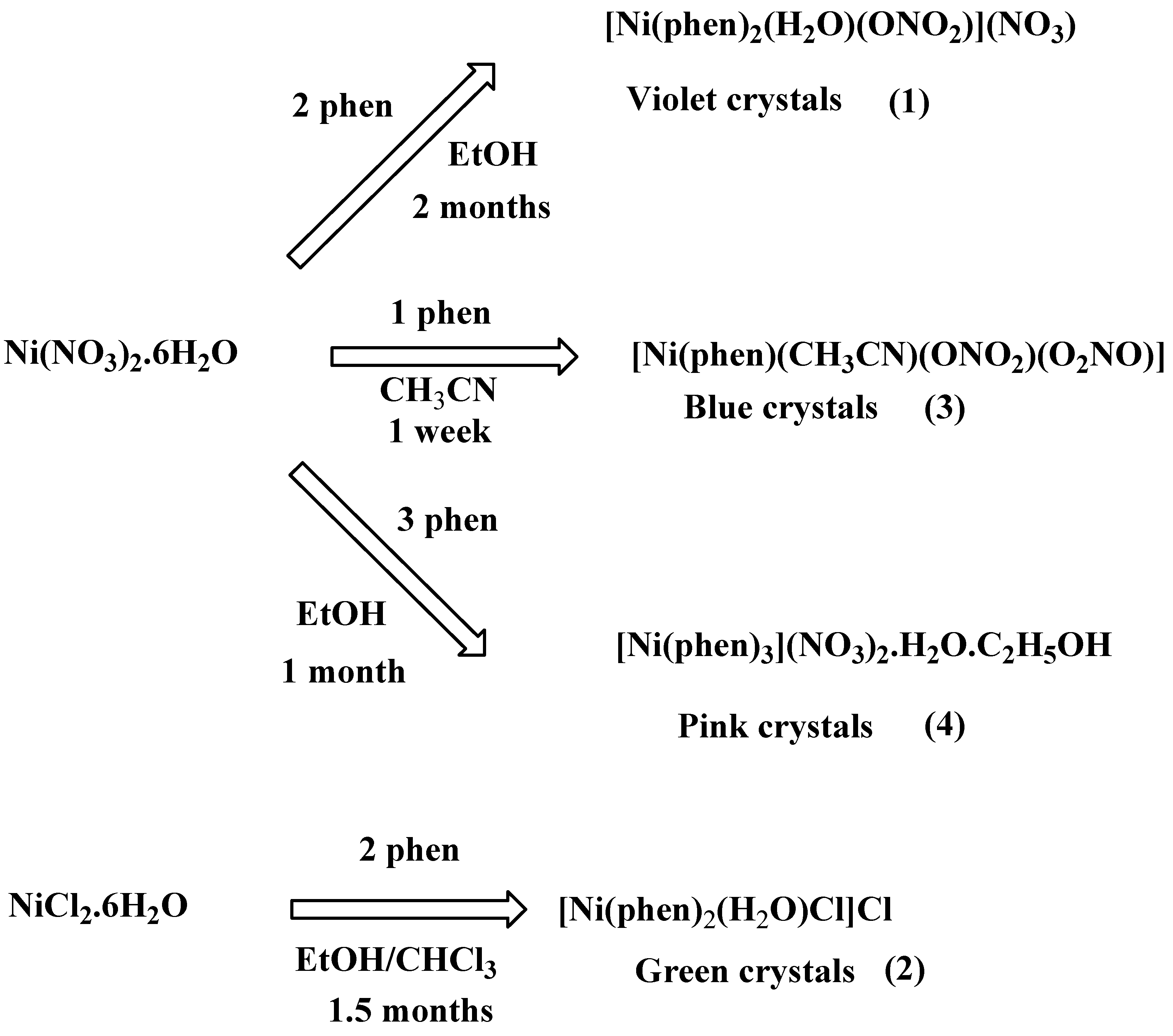
2.1. Synthesis
2.2. FTIR Vibration cm−1
2.3. X-ray Structure Determination
3. Results and Discussion
3.1. Synthesis of Crystals
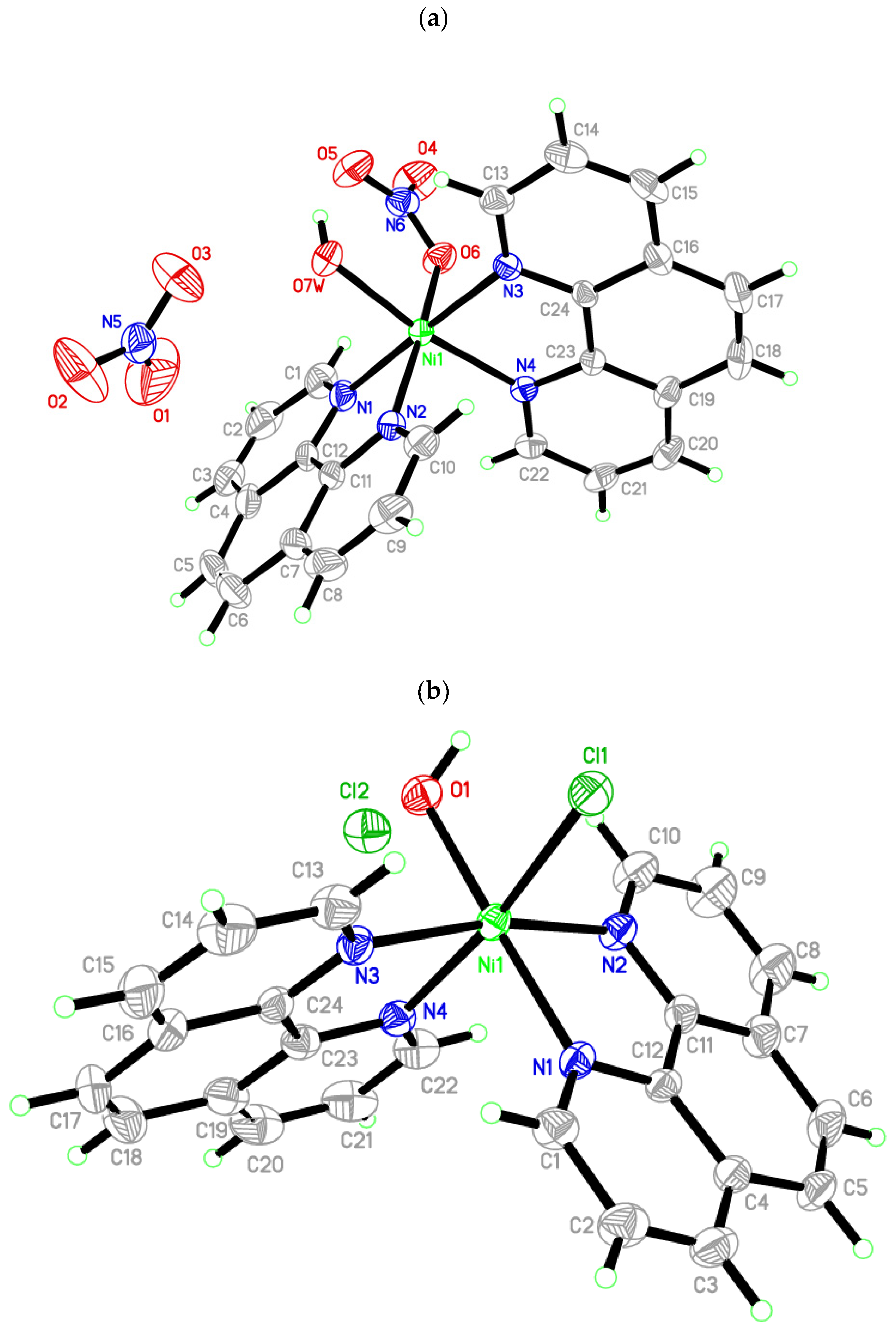
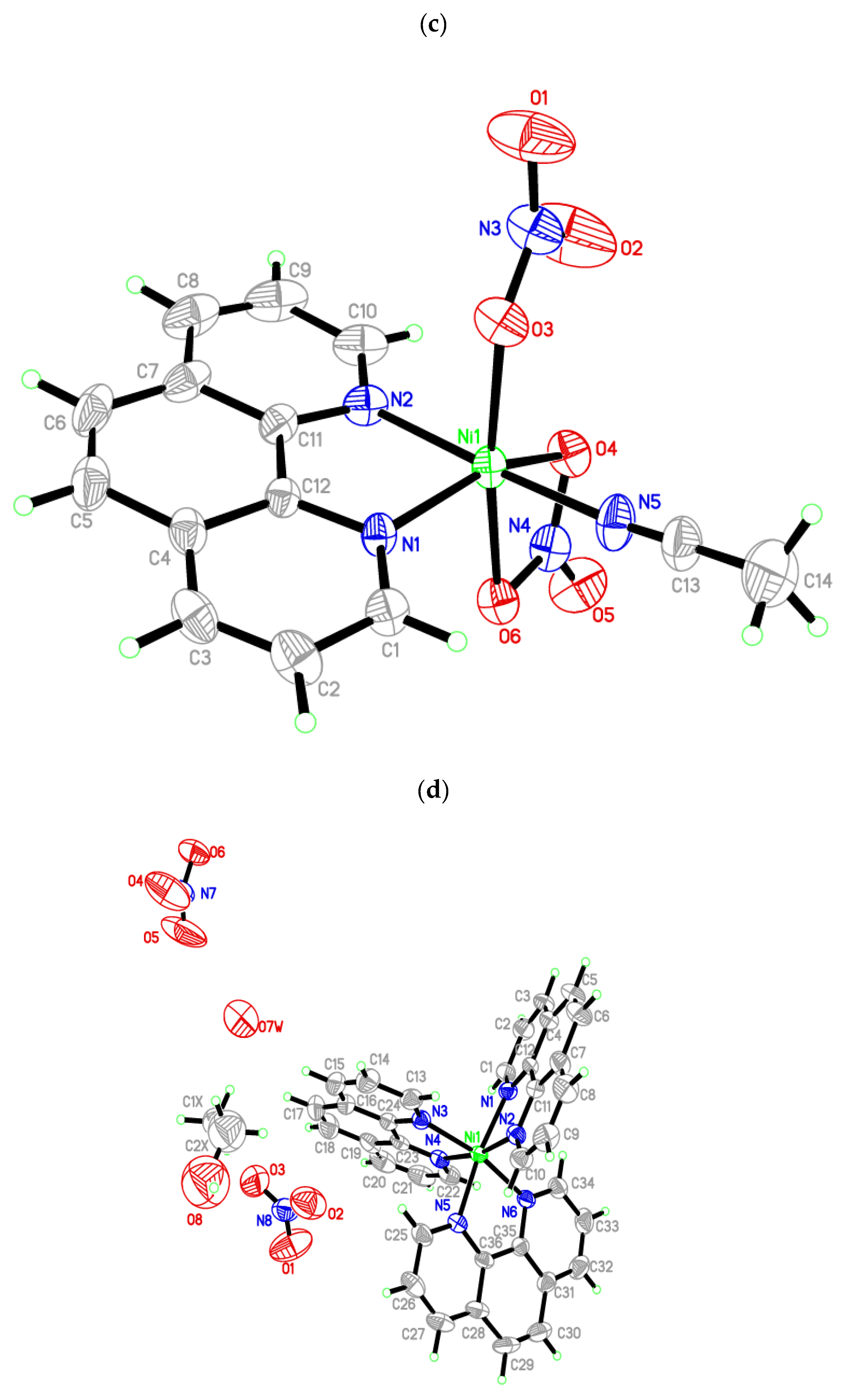
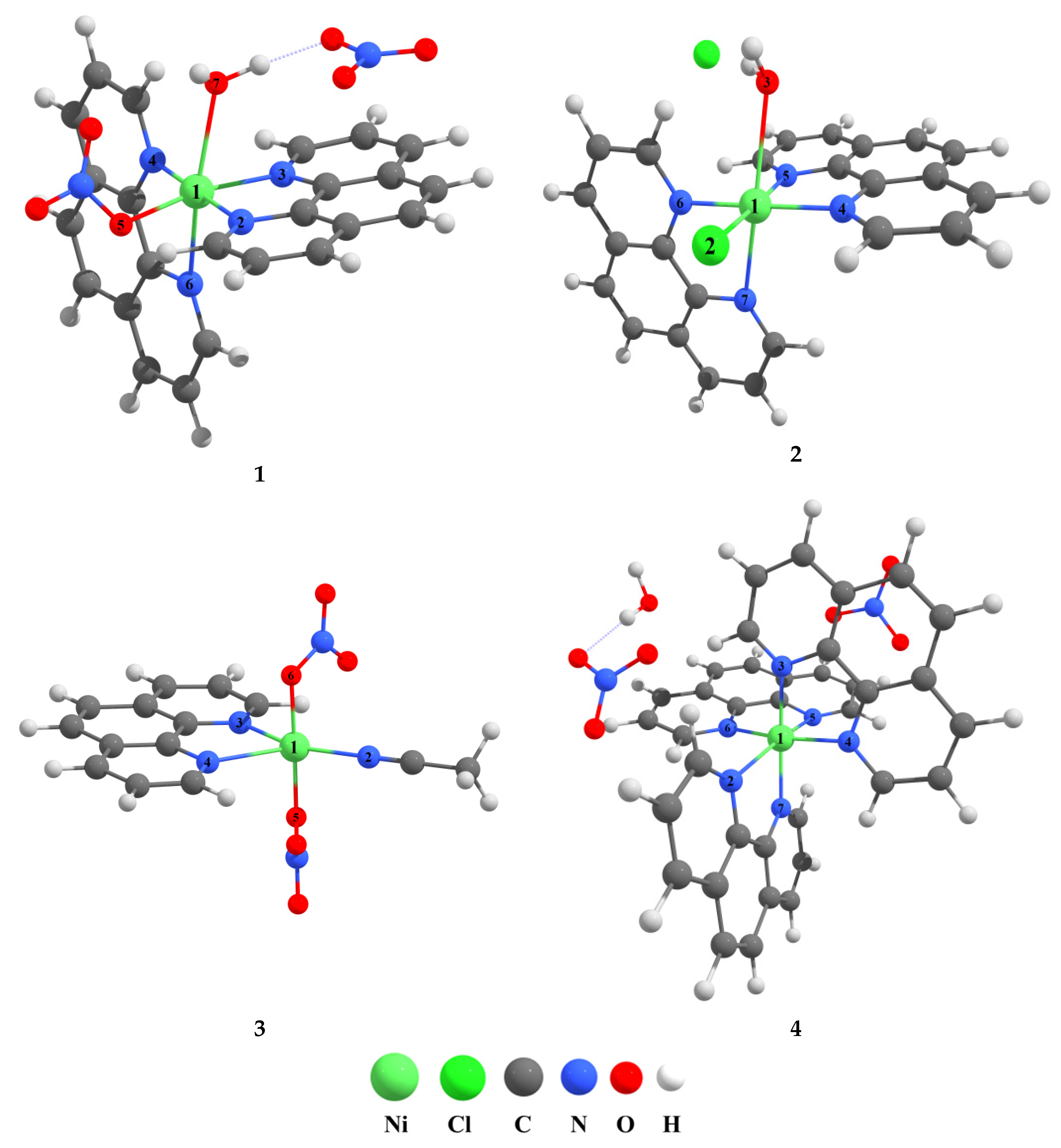
3.2. Molecular Structure of Crystals
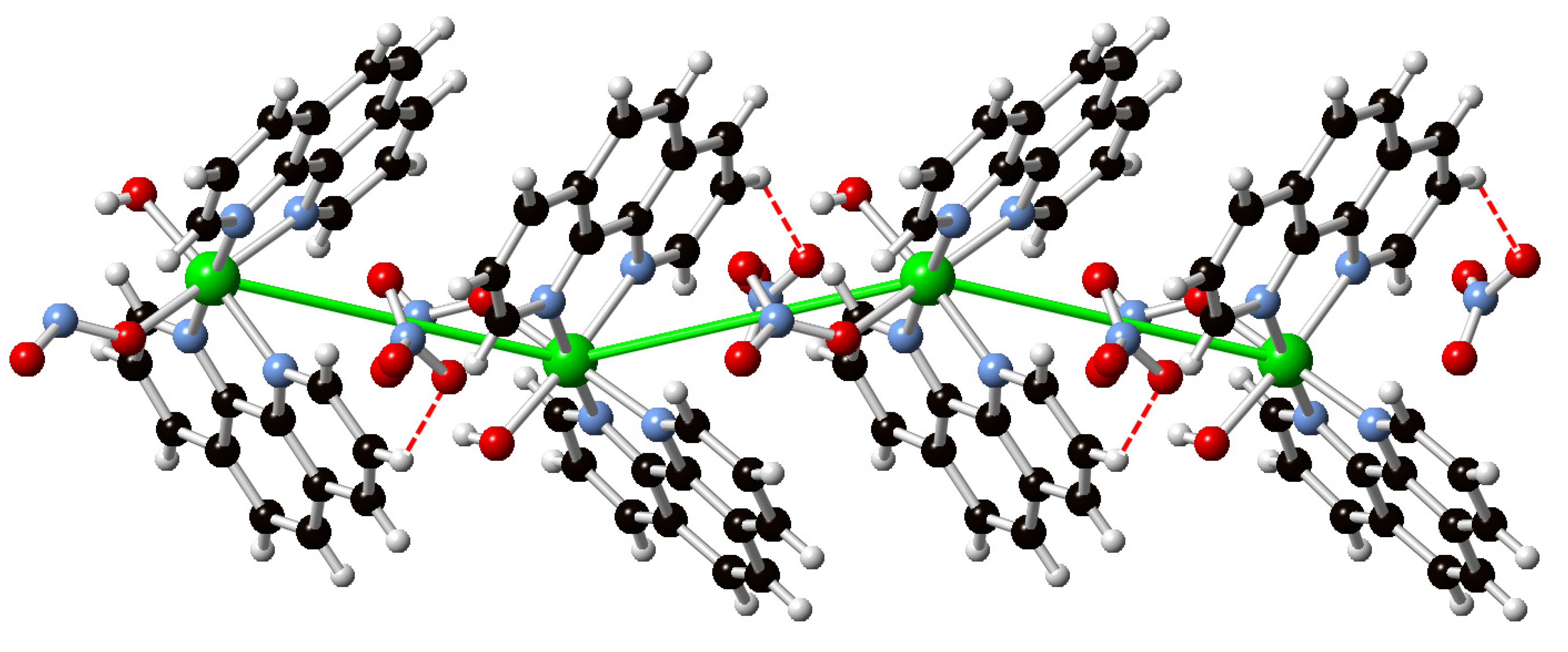
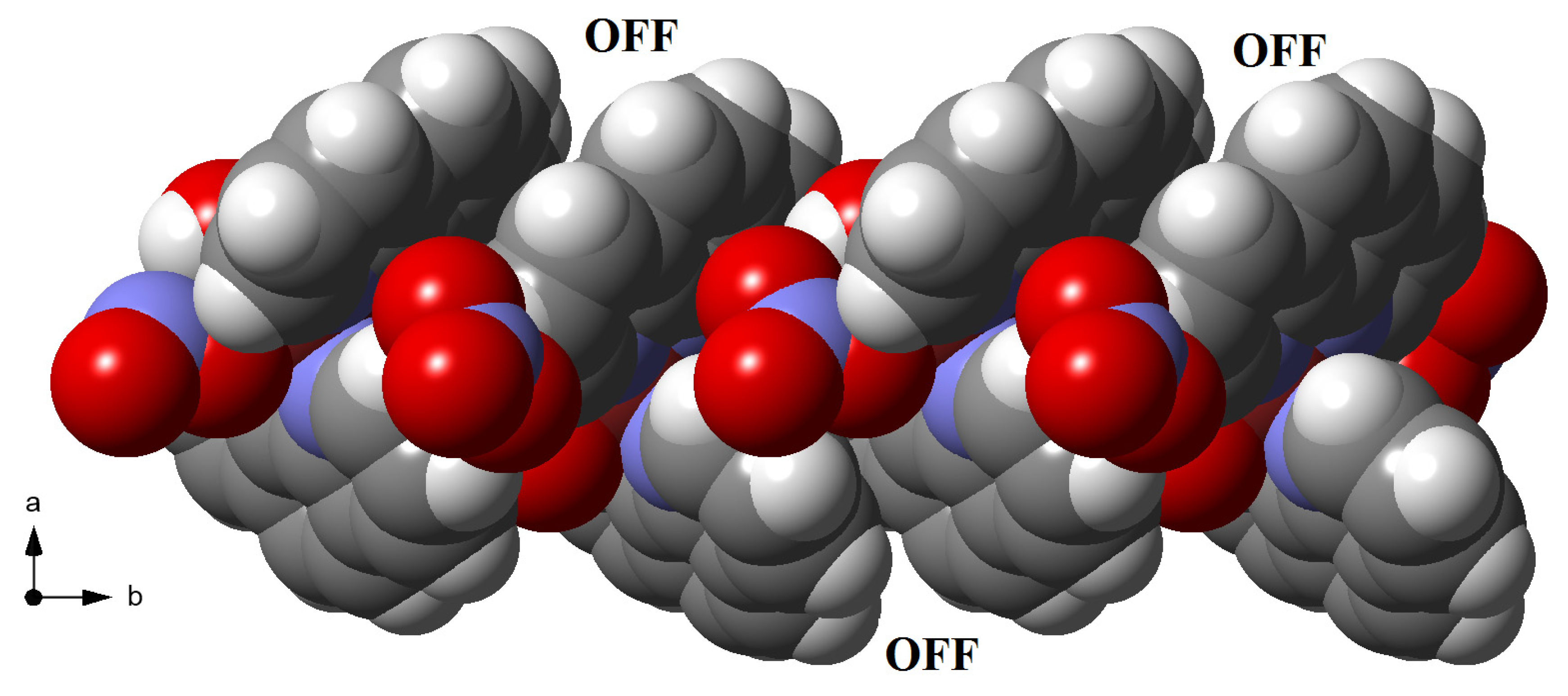
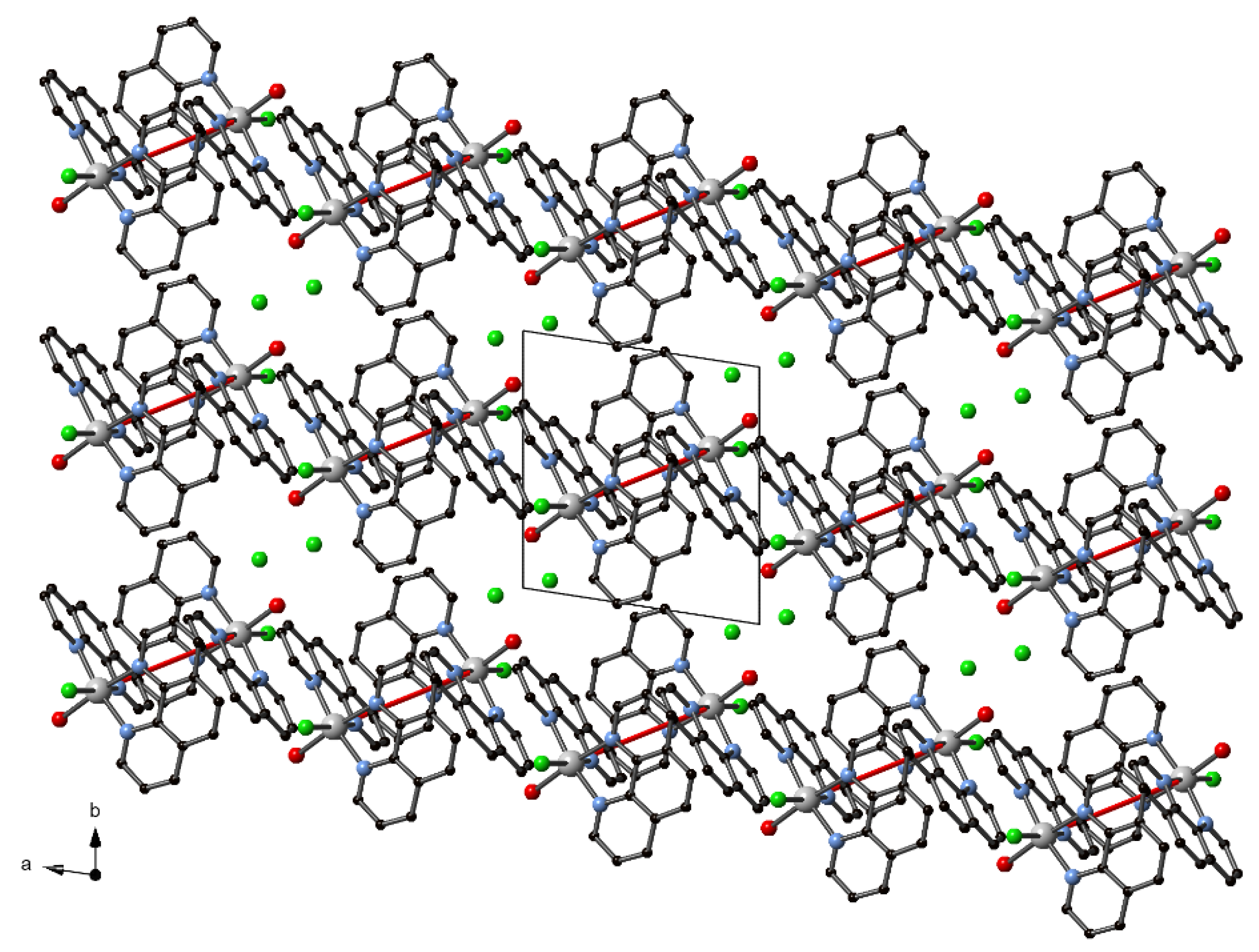
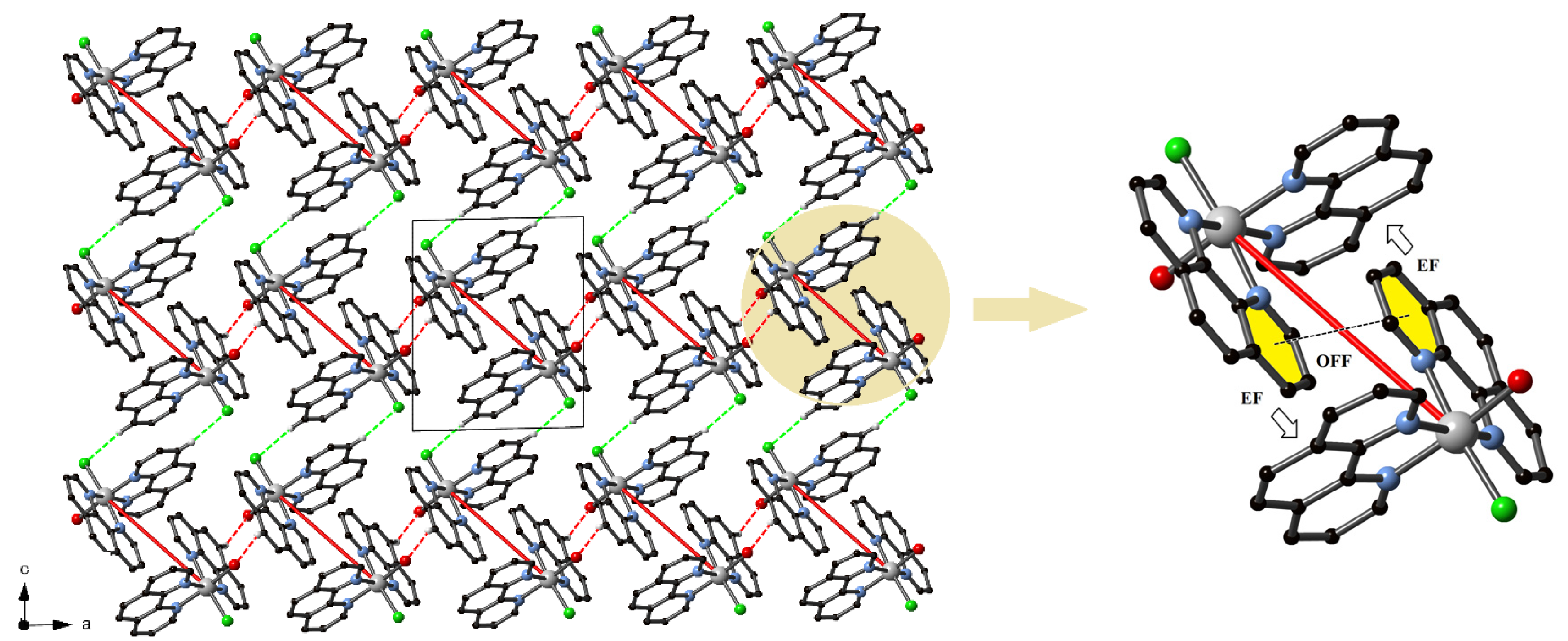
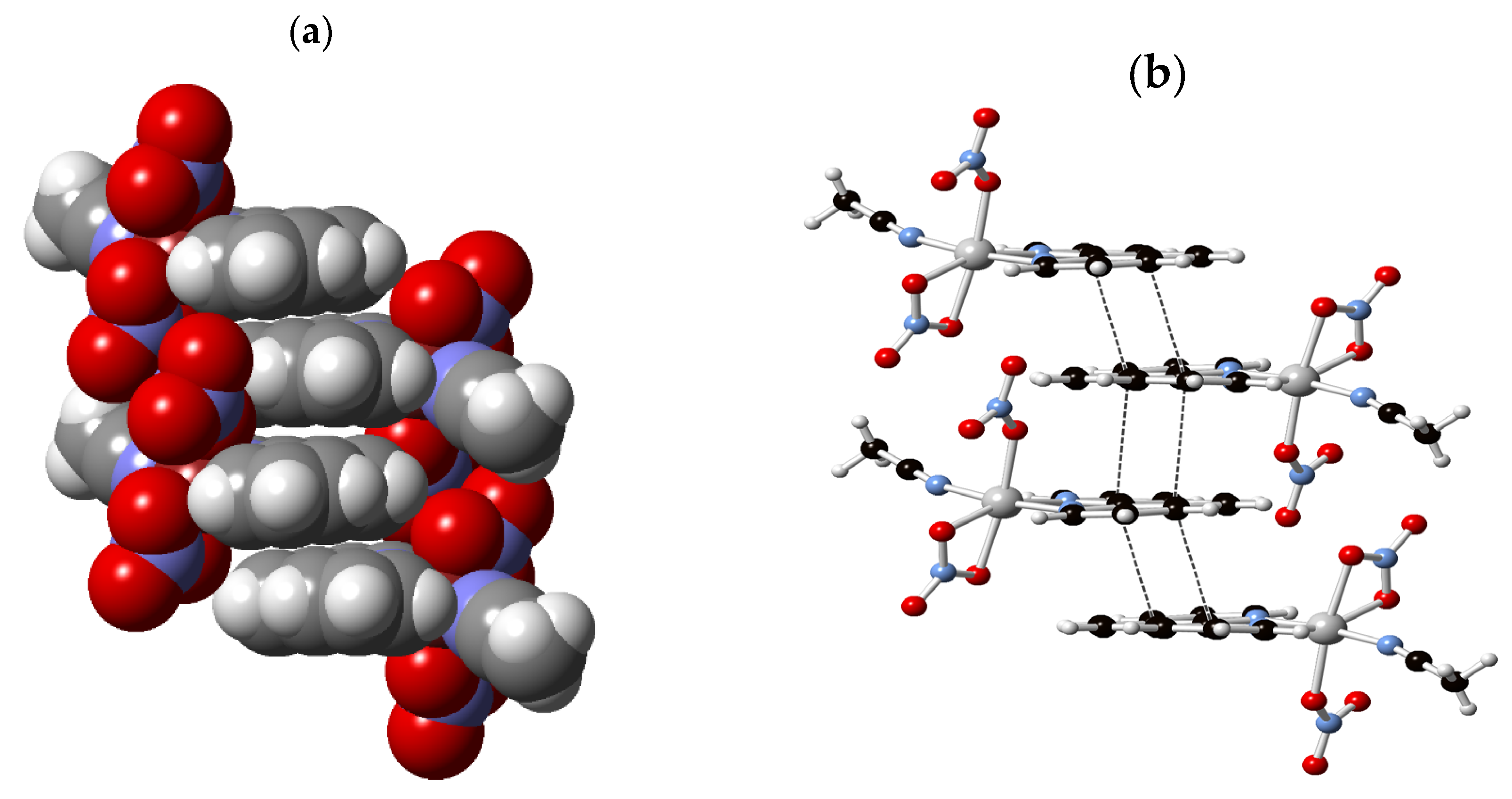
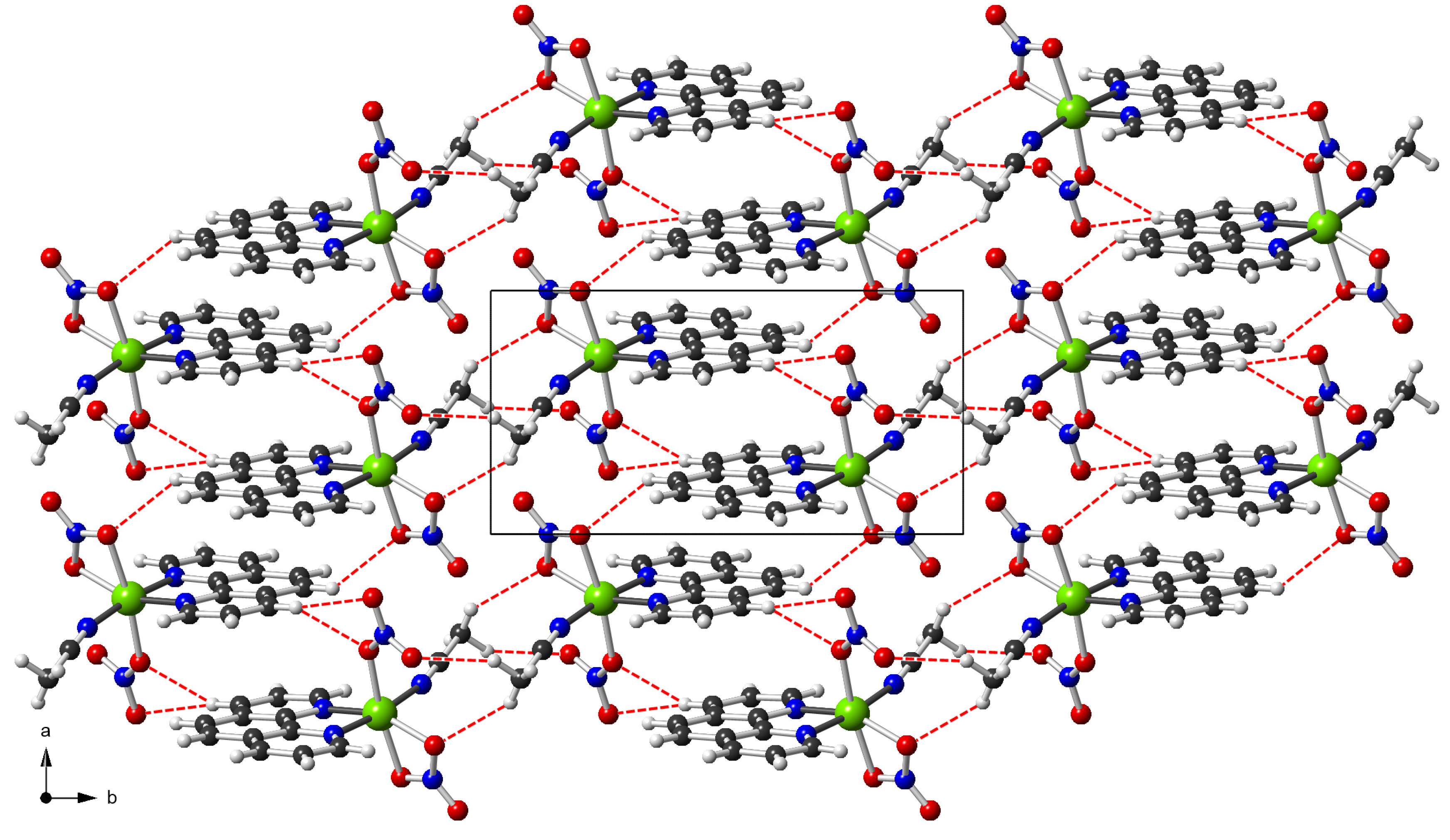
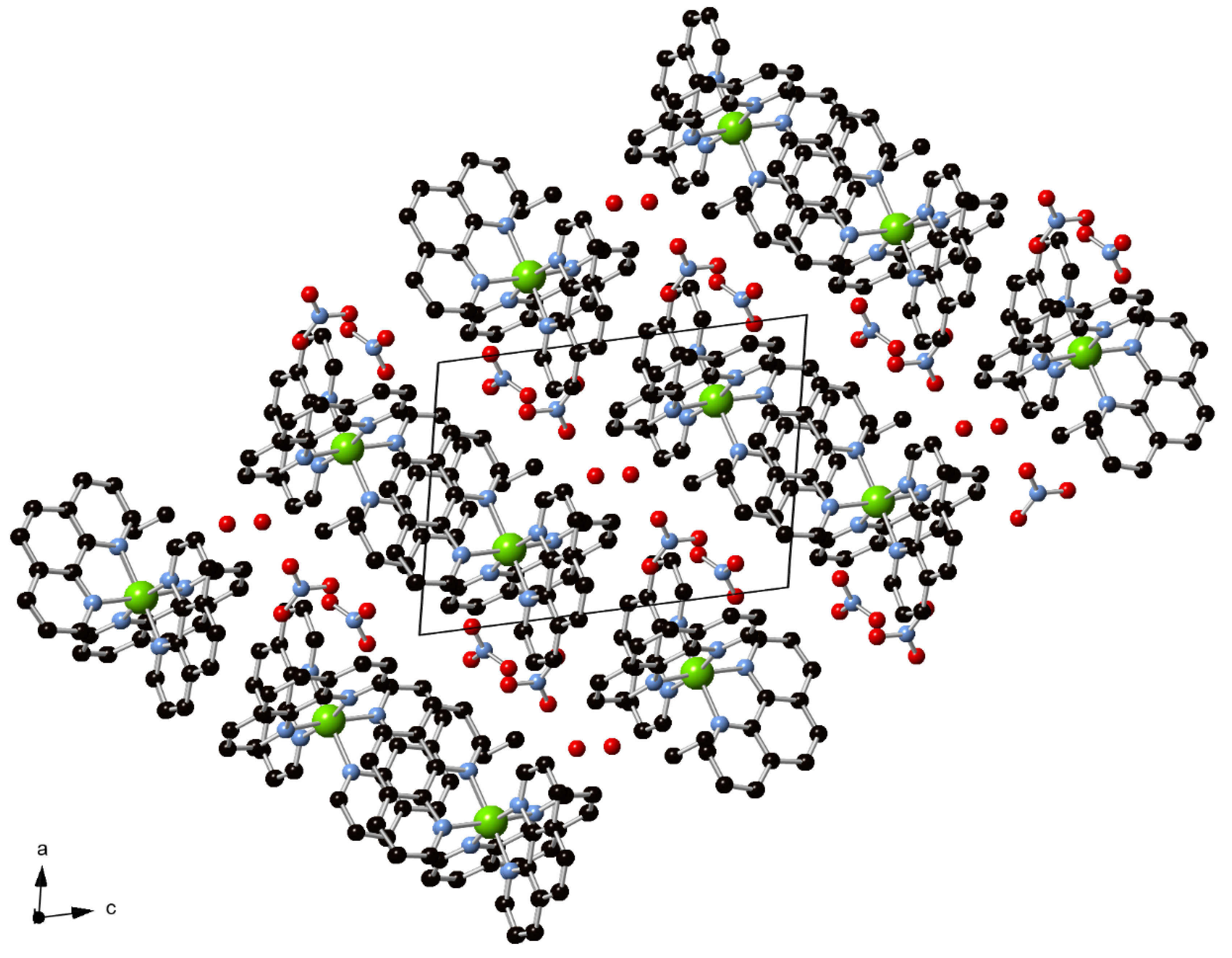
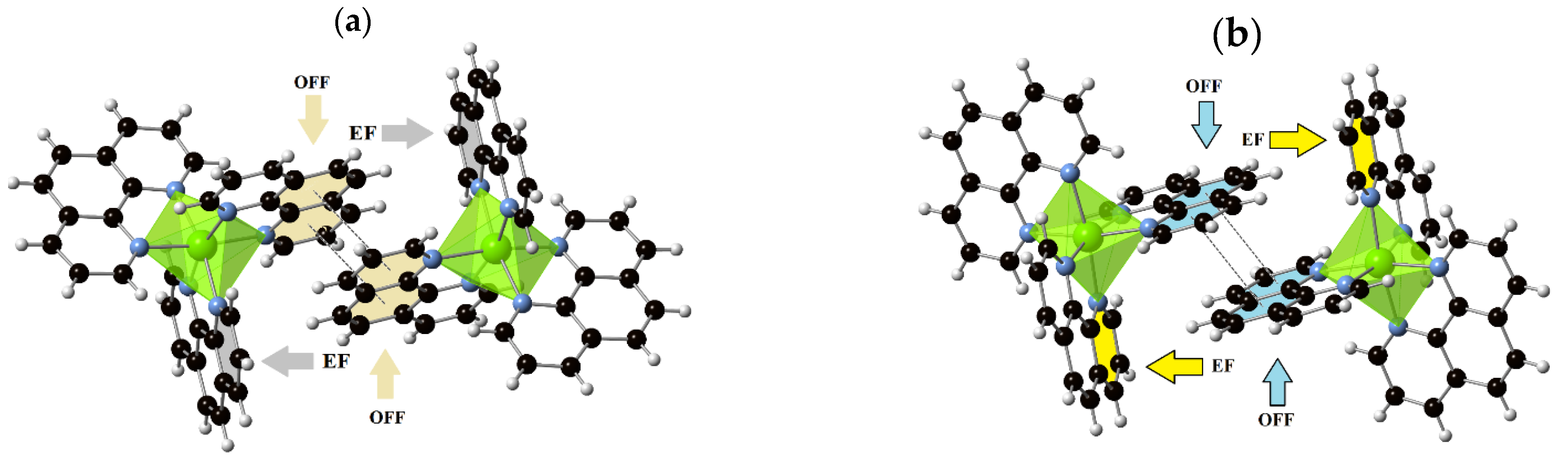
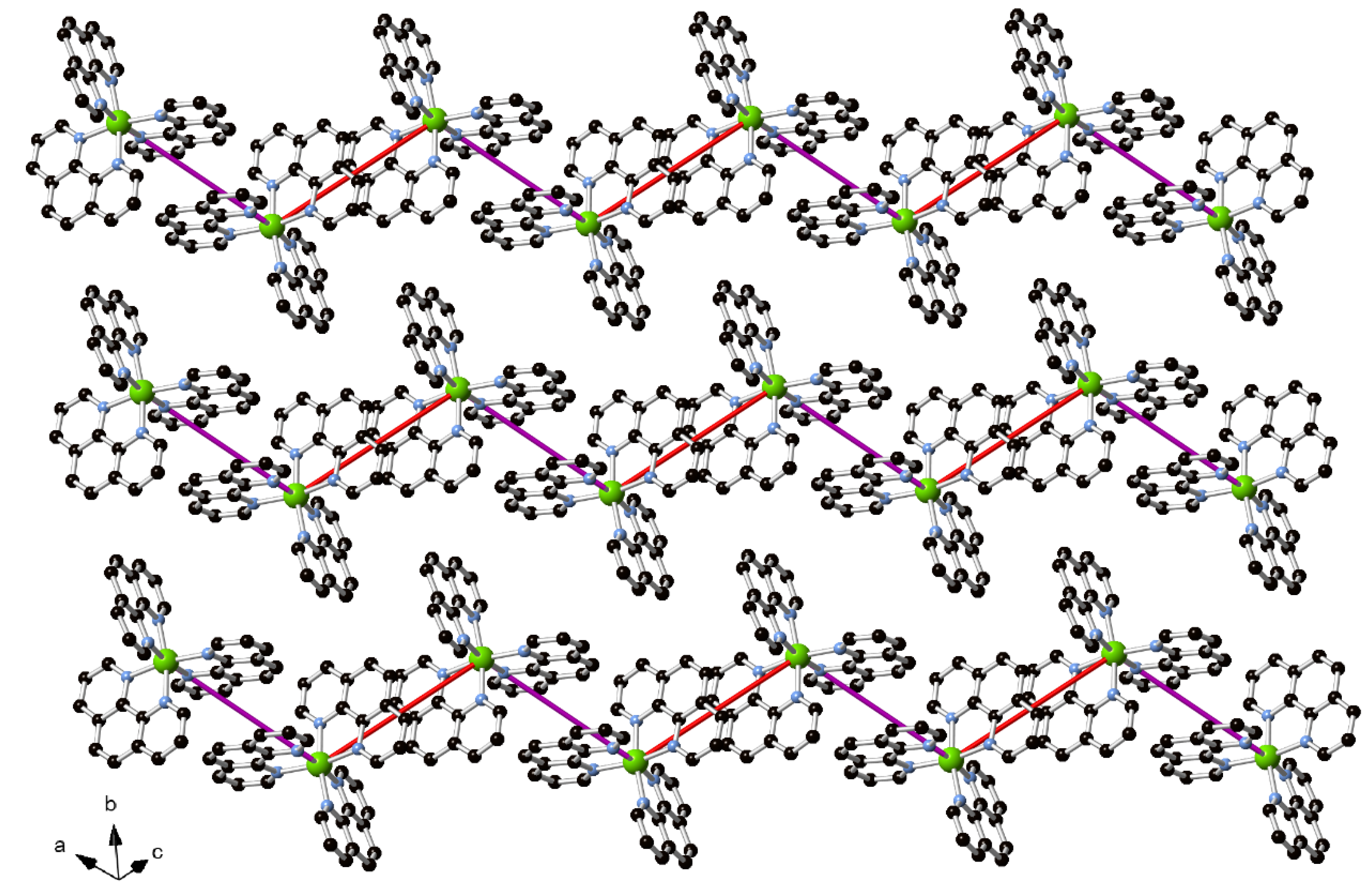
3.3. Supramolecular Features
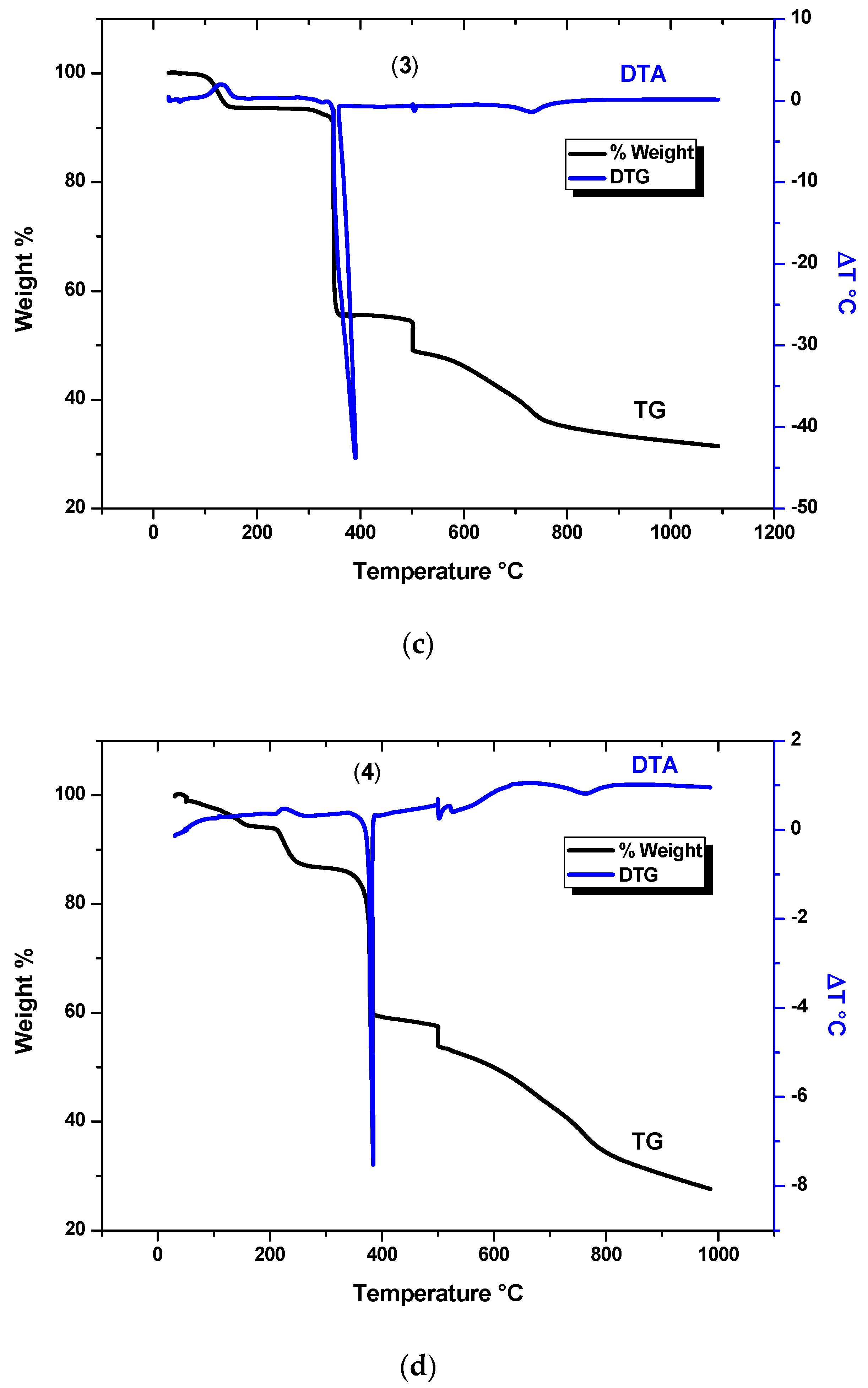
3.4. Thermal Analysis
3.5. DFT Studies
3.5.1. Computational Details
3.5.2. Geometry Optimization
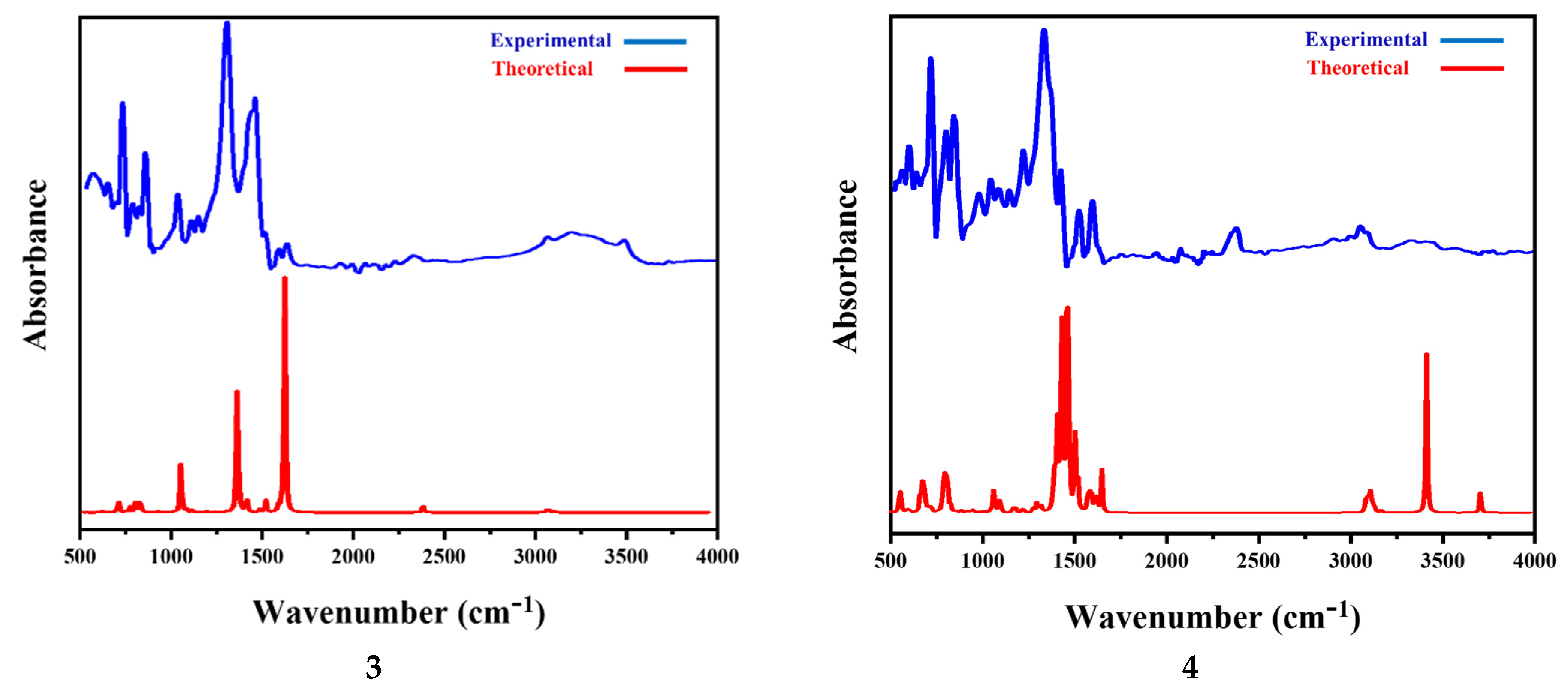
3.5.3. Vibrational Spectral Analysis
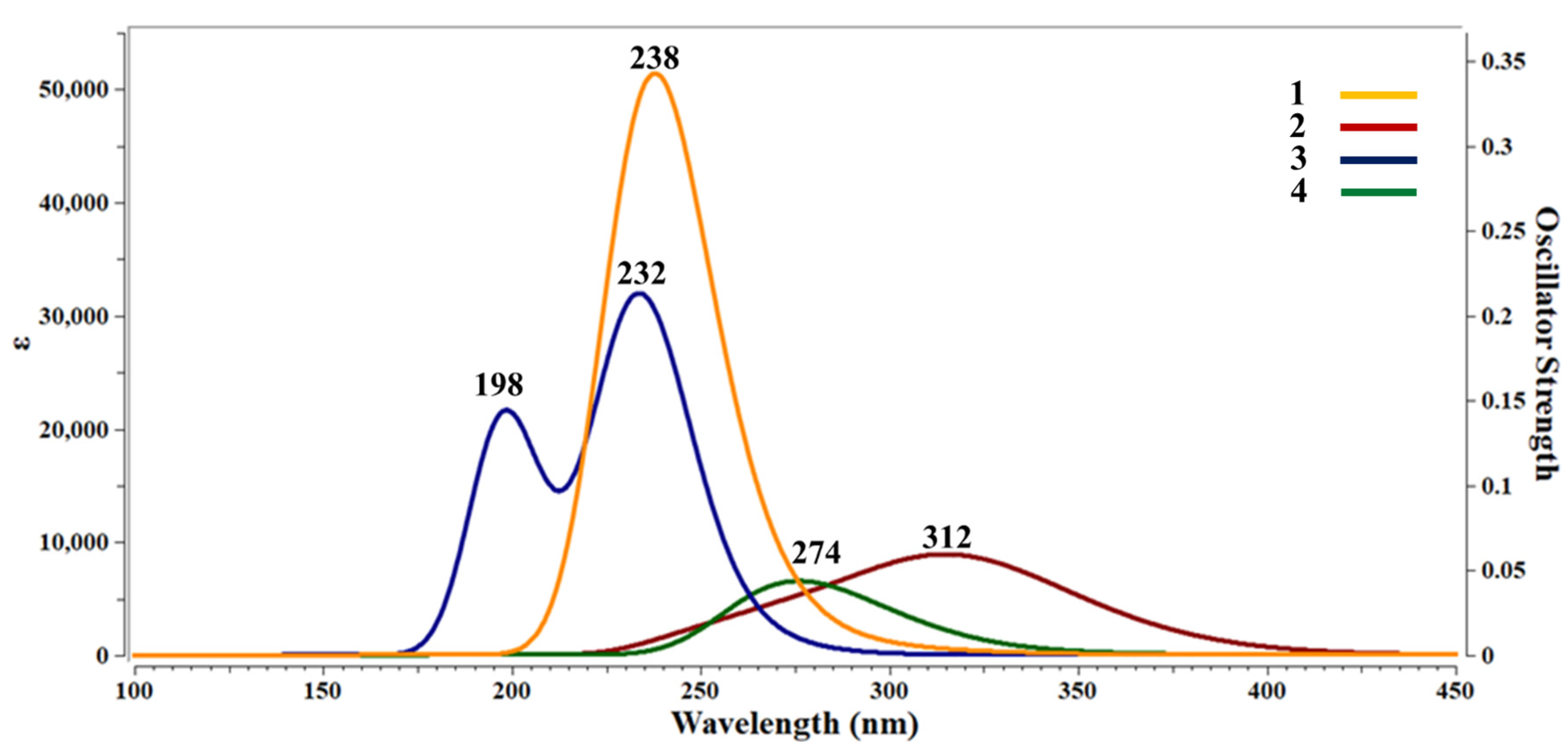
3.5.4. UV-Visible Spectroscopic Study
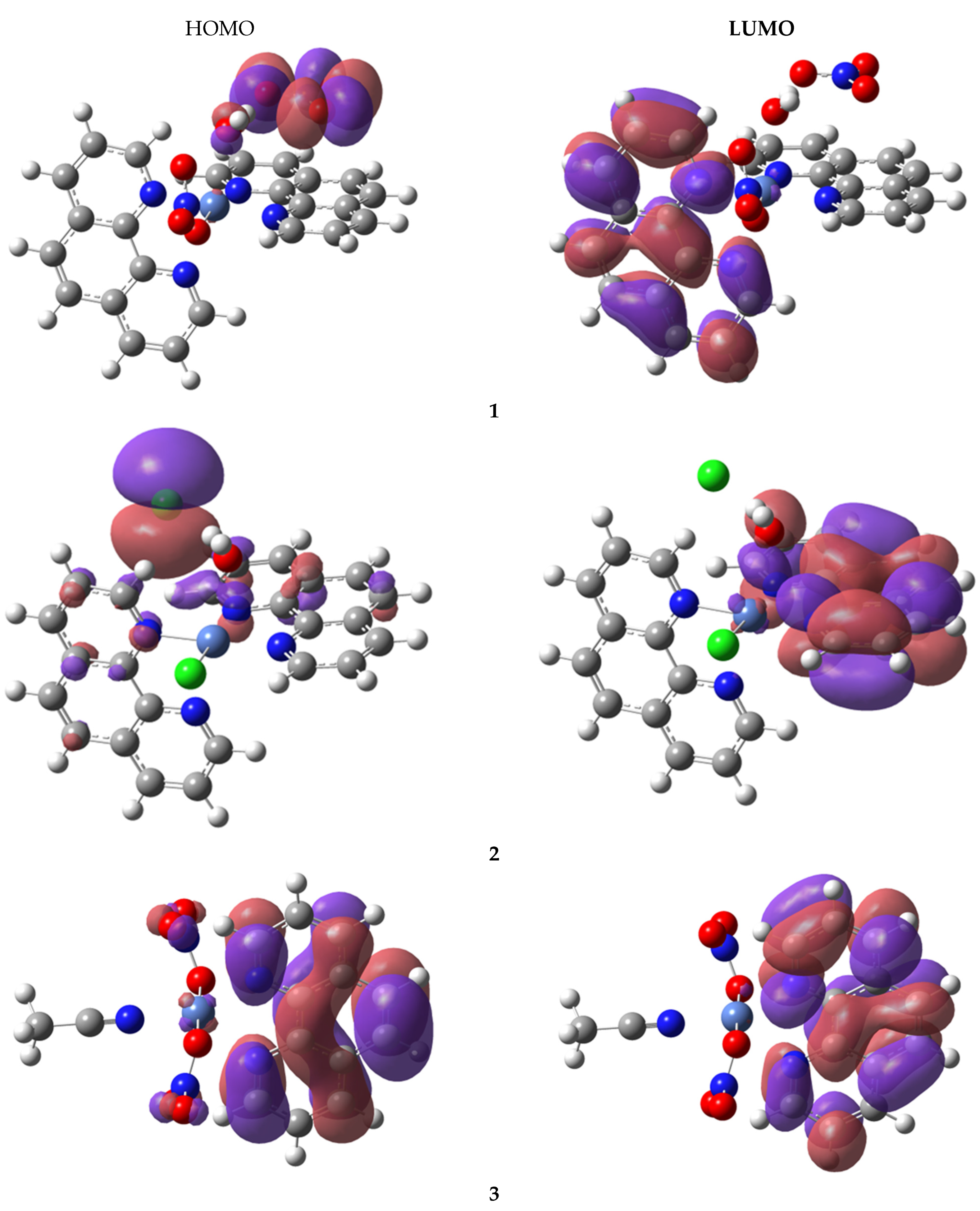
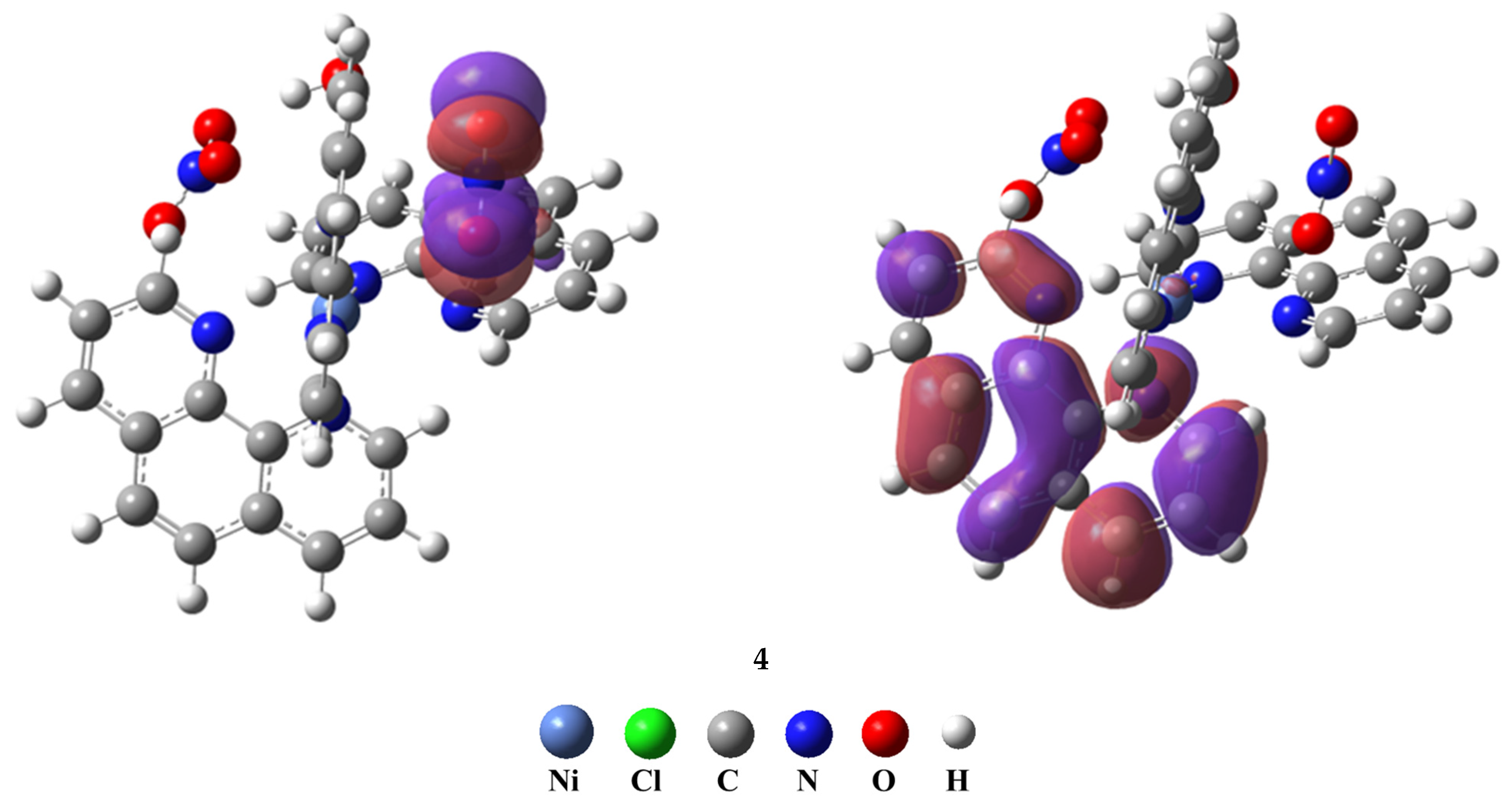
3.5.5. Frontier Molecular Orbitals (FMO) Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Preston, H.; Kennard, C. Stereochemistry of rigid chelate–metal complexes. Part II. Crystal structure of di-µ-chloro-sym-trans-dichloro-bis-(2,9-dimethyl-1,10-phenanthroline) dinickel (II)–2-chloroform. J. Chem. Soc. A Inorg. Phys. Theor. 1969, 2682–2685. [Google Scholar] [CrossRef]
- Bencini, A.; Lippolis, V. 1, 10-Phenanthroline: A versatile building block for the construction of ligands for various purposes. Coord. Chem. Rev. 2010, 254, 2096–2180. [Google Scholar] [CrossRef]
- Teng, Q.; Huynh, H.V. A unified ligand electronic parameter based on 13CNMR spectroscopy of N-heterocyclic carbene complexes. Dalton Trans. 2017, 46, 614–627. [Google Scholar] [CrossRef]
- Brewer, B.; Brooks, N.R.; Abdul-Halim, S.; Sykes, A.G. Differential metathesis reactions of 2,2′-bipyridine and 1,10-phenanthroline complexes of cobalt (II) and nickel (II): Cocrystallization of ionization isomers {[cis-Ni (phen)2(H2O)2][cis-Ni(phen)2 (H2O)Cl]}(PF6)3·4.5 H2O, and a synthetic route to asymmetric tris-substituted complexes. J. Chem. Crystallogr. 2003, 33, 651–662. [Google Scholar]
- Chesnut, D.J.; Haushalter, R.C.; Zubieta, J. The hydrothermal synthesis and structural characterization of a new class of compounds: Nickel organoamine-halocadmates. Inorg. Chim. Acta 1999, 292, 41–51. [Google Scholar] [CrossRef]
- Norman, R.E.; Xie, M. Nickel(II) 1,10-Phenanthroline complexes: Cis-[Aqua(Bromo) bis (1,10-Phenanthroline)Nickel(II)] Bromide Trihydrate and (tris (1,10-Phenanthroline)Nickel(II)] Bromide Octahydrate. J. Coord. Chem. 2004, 57, 425–434. [Google Scholar] [CrossRef]
- Prior, T.J.; Rujiwatra, A.; Chimupala, Y. [Ni(1,10-phenanthroline)2(H2O)2](NO3)2: A Simple Coordination Complex with a Remarkably Complicated Structure that Simplifies on Heating. Crystals 2011, 1, 178–194. [Google Scholar] [CrossRef]
- Tang, X.Y.; Qiu, Y.C.; Sun, F.; Yue, S.T. Diaquabis(1,10-phenanthroline-k2N,N′)nickel(II) diperchlorate 0.4-hydrate. Acta Crystallogr. 2007, E63, m2515. [Google Scholar]
- Zhu, L.; Zhang, L.H.; Liu, B.Y.; An, Z. Crystal structure of diaquabis(1,10-phenanthroline)nickel(II) sulfate hydrate (1:5.6), [Ni(H2O)2(C12H8N2)2][SO4]·5.6H2O. Z. Kristallogr.-New Cryst. Struct. 2006, 221, 451–452. [Google Scholar]
- Ferbinteanu, M.; Cimpoesu, F.; Mariusandruh, A.; Rochon, F.D. Solid-state chemistry of [Ni(AA)3][PdCl4]·nH2O complexes (AA = bipy, phen) and crystal structures of cis-diaquabis(phenanthroline)nickel(II) tetrachlorozincate and cis-dichloro-bis(bipyridine)nickel(II). Polyhedron 1998, 17, 3671–3679. [Google Scholar] [CrossRef]
- Oresmaa, L.; Haukka, M.; Vainiotalo, P.; Pakkanen, T.A. Ab Initio Calculations and Mass Spectrometric Determination of the Gas-Phase Proton Affinities of 4,4′-Disubstituted 2,2′-Bipyridines. J. Org. Chem. 2002, 67, 8216–8219. [Google Scholar] [CrossRef]
- Reyzer, M.; Brodbelt, J. Ligand displacement reactions of dimer and trimer pyridyl ligand/transition metal complexes1. Int. J. Mass Spectrom. 1999, 182, 311–322. [Google Scholar] [CrossRef]
- Ha, K. Crystal structure of cis-bis (2,2′-bipyridine-κ2N,N′) bis (thiocyanato-κN) nickel (II), C22H16N6NiS2. Z. Für Krist.-New Cryst. Struct. 2017, 232, 151–152. [Google Scholar]
- Tsierkezos, N.G.; Diefenbach, M.; Roithová, J.; Schröder, D.; Schwarz, H. Competitive Complexation of Gaseous MnII by 1, 10-Phenanthroline, 2,2′-Bipyridine, and 4, 5-Diazafluorene. Inorg. Chem. 2005, 44, 4969–4978. [Google Scholar] [CrossRef]
- Woodward, J.D.; Backov, R.; Abboud, K.A.; Ohnuki, H.; Meisel, M.W.; Talham, D.R. Structural, thermal, and magnetic properties of three transition metal-4,4′-bipyridine coordination polymers:[Ni(4,4′-bipy) 3(H2O)2](ClO4)2 1.4(4,4′-bipy)·3(H2O);[Co(4,4′-bipy)3(H2O)2](ClO4)2·1.4(4,4′-bipy)· 3(H2O);[Cu(4,4′-bipy)3(DMSO)2](ClO4)2 2(4,4′-bipy). Polyhedron 2003, 22, 2821–2830. [Google Scholar]
- Alramadhan, S.A.; Hammud, H.H. Graphene Nickel Silica Supported Nanocomposites, Application in Water Treatment. Appl. Nanosci. 2020, 11, 273–291. [Google Scholar] [CrossRef]
- Hammud, H.H.; Karnati, R.K.; Alotaibi, N.; Hussain, S.G. Thirumurugan Prakasam, Cobalt–carbon silica nanoparticles for uptake of cationic and anionic dye from polluted water. Molecules 2021, 26, 7489. [Google Scholar] [CrossRef]
- Alotaibi, N.; Hammud, H.H.; Al Otaibi, N. Electrocatalytic Properties of 3D Hierarchical Graphitic Carbon Cobalt Nanoparticles for Urea Oxidation. ACS Omega 2020, 5, 26038–26048. [Google Scholar] [CrossRef]
- Hammud, H.H.; Alotaibi, N.; Al Otaibi, N.; Aljaafari, A.; Ahmed, F.; Azam, A.; Prakasam, T. Hierarchical Porous Carbon Cobalt Nanocomposites based Sensor for Fructose. Chemosensors 2021, 9, 6. [Google Scholar] [CrossRef]
- Hammud, H.H.; Traboulsi, H.; Karnati, R.K.; Hussain, S.G.; Bakir, E. Hierarchical Graphitic Carbon-Encapsulating Cobalt Nanoparticles for Catalytic Hydrogenation of 2,4-Dinitrophenol. Catalysts 2022, 12, 39. [Google Scholar] [CrossRef]
- Krebsz, M.; Kótai, L.; Sajó, I.E.; Váczi, T.; Pasinszki, T. Carbon Microsphere-Supported Metallic Nickel Nanoparticles as Novel Heterogeneous Catalysts and Their Application for the Reduction of Nitrophenol. Molecules 2021, 26, 5680. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G. SHELXTL-PC (Version 5.1); Siemens analytical instruments, Inc.: Madison, WI, USA, 1997. [Google Scholar]
- Li, S.-D.; Su, F.; Zhou, C.-Y.; Hu, Q.-L.; Li, Y.-Q.; Wang, Z.-J. Syntheses, structures and magnetic properties of two CoII/NiII isostructural coordination polymers based on an asymmetric semirigid tricarboxylate ligand. Acta Cryst. 2022, C78, 23–29. [Google Scholar] [CrossRef]
- Nath, H.; Sharma, P.; Frontera, A.; Barcelo-Oliver, M.; Verma, A.K.; Das, J.; Bhattacharyya, M.K. Phenanthroline-based Ni(II) coordination compounds involving unconventional discrete fumarate-water-nitrate clusters and energetically significant cooperative ternary π-stacked assemblies: Antiproliferative evaluation and theoretical studies. J. Mol. Struct. 2022, 1248, 131424. [Google Scholar] [CrossRef]
- Ren, X.-C.; Li, J.; Wang, W.-H.; Chen, X.; Zhang, B.; Li, L.-Z. [NH4][Ni(phen)3]BiI6: Synthesis, structure, photocatalytic property and theoretical study of a discrete iodobismuthate. Inorg. Chem. Commun 2021, 130, 108714. [Google Scholar] [CrossRef]
- Matsia, S.; Kaoulla, A.; Menelaou, M.; Hatzidimitriou, A.; Papadopoulos, T.; Reimann, M.K.; Pöttgen, R.; Salifoglou, A. Structural speciation in chemical reactivity profiling of binary-ternary systems of Ni(II) with iminodialcohol and aromatic chelators. Polyhedron 2022, 212, 115577. [Google Scholar] [CrossRef]
- Russell, V.; Craig, D.; Scudder, M.; Dance, I. Crystal packing motifs for mixed oxalate/phenanthroline metal complexes. CrystEngComm 2001, 3, 96–106. [Google Scholar] [CrossRef]
- Dance, I.; Scudder, M. Molecules embracing in crystals. CrystEngComm 2009, 11, 2233–2247. [Google Scholar] [CrossRef]
- Shaabani, B.; Mirtamizdoust, B.; Viterbo, D.; Croce, G.; Hammud, H.; Hojati-Lalemi, P.; Khandar, A. Sonochemical synthesis of a novel nano-scale lead(II) coordination polymers: Synthesis, crystal structure, thermal properties and DFT calculations of [Pb(dmp)(μ-N3)(μ-NO3)]n with the novel Pb2-(μ-N3)2-(μ-NO3)2 unit. ZAAC J. Inorg. Gen. Chem. 2011, 637, 713–719. [Google Scholar]
- Mirtamizdoust, B.; Trávníček, Z.; Hanifehpour, Y.; Talemi, P.; Hammud, H.; Joo, S.W. Synthesis and characterization of nano-peanuts of lead(II) coordination polymer [Pb(qcnh)(NO3)2]n with ultrasonic assistance: A new precursor for the preparation of pure-phase nano-sized PbO. Ultrason. Sonochem. 2017, 34, 255–261. [Google Scholar] [CrossRef]
- Zaworotko, M.J.; Hammud, H.H.; Kabbani, A.; McManus, G.J.; Ghannoum, A.M.; Masoud, M.S. Synthesis and characterization of some transition metal complexes with mixed adenine and acetylacetonate ligands. Crystal structures of solvated complex {[Cu(acac)2(adenine)]•EtOH} and {[Cu(acac)2(adenine)]•DMF•H2O. J. Chem. Crystallogr. 2009, 39, 853–863. [Google Scholar] [CrossRef]
- Hammud, H.H.; Nemer, G.; Sawma, W.; Touma, J.; Barnabe, P.; Bou-Mouglabey, Y.; Ghannoum, A.; El-Hajjar, J.; Usta, J. Copper–adenine complex, a compound, with multi-biochemical targets and potential anti-cancer effect. Chem.-Biol. Interact. 2008, 173, 84–96. [Google Scholar] [CrossRef] [PubMed]
- Zaworotko, M.J.; Hammud, H.H.; Kravtsov, V.C. The co-crystal of iron(II) complex hydrate with hydroxybenzoic acid: [Fe(Phen)3]Cl(p-hydroxybenzoate).2(p-hydroxybenzoic acid).7H2O. J. Chem. Crystallogr. 2007, 37, 219–231. [Google Scholar] [CrossRef]
- Hammud, H.H.; El Hamaoui, B.; Noubani, N.H.; Feng, X.; Wu, Z.-S.; Müllen, K.; Ayub, K. Carbon-Cobalt Nanostructures as an Efficient Adsorbent of Malachite Green. Nanosci. Nanotechnol.-Asia 2018, 8, 263–280. [Google Scholar] [CrossRef]
- Mroueh, M.; Daher, C.; Hariri, E.; Demirdjian, S.; Isber, S.; Choi, E.S.; Mirtamizdoust, B.; Hammud, H.H. Magnetic property, DFT calculation, and biological activity of bis[(μ2-chloro)chloro(1,10-phenanthroline)copper(II)] complex. Chem.-Biol. Interact. 2015, 231, 53–60. [Google Scholar] [CrossRef]
- Hammud, H.H.; Kortz, U.; Bhattacharya, S.; Demirdjian, S.; Hariri, E.; Isber, S.; Choi, E.S.; Mirtamizdoust, B.; Mroueh, M.; Daher, C.F. Structure, DFT studies, Magnetism and Biological activity of Bis[(µ2-azido)-chloro-(1,10-phenanthroline)-copper(II)] complex. Inorg. Chim. Acta 2020, 506, 119533. [Google Scholar] [CrossRef]
- Hammud, H.H.; Zaworotko, M.J.; McManus, G.J.; Tabesh, R.N.; Ibrahim, H.I.M.; Ayub, K.; Ludwig, R. The co-crystal of copper(II) phenanthroline chloride complex hydrate with p-aminobenzoic acid: Structure, cytotoxicity, thermal analysis and DFT calculation. Chem. Mon. 2021, 152, 323–336. [Google Scholar] [CrossRef]
- Frisch, M.; Clemente, F. Gaussian 09, Revision A. 01; Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Eds.; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Khan, S.; Yar, M.; Kosar, N.; Ayub, K.; Arshad, M.; Zahid, M.N.; Mahmood, T. First-principles study for exploring the adsorption behavior of G-series nerve agents on graphdyine surface. Comput. Theor. Chem. 2020, 1191, 113043. [Google Scholar] [CrossRef]
- Roy, T.G.; Hazari, S.K.S.; Dey, B.K.; Miah, H.A.; Olbrich, F.; Rehder, D. Synthesis and antimicrobial activities of isomers of N(4),N(11)-dimethyl-3, 5,7,7,10,12,14,14-octamethyl-1,4,8,11-tetraazacyclotetradecane and their nickel(II) complexes. Inorg. Chem. 2007, 46, 5372–5380. [Google Scholar] [CrossRef]
- Roy, T.G.; Hazari, S.K.S.; Dey, B.K.; Meah, H.A.; Rahman, S.; Kim, D.I.; Park, Y.C. Synthesis, electrolytic behaviour and antimicrobial activities of cadmium complexes of isomers of 3,10-C-meso-3,5,7,7,10,12,14,14-octamethyl-1,4,8,11-tetraazacyclotetradecane. J. Coord. Chem. 2007, 60, 1567–1578. [Google Scholar] [CrossRef]
- Affan, M.A.; Foo, S.W.; Jusoh, I.; Hanapi, S.; Tiekink, E.R.T. Synthesis, characterization and biological studies of organotin(IV) complexes with hydrazone ligand. Inorg. Chim. Acta 2009, 362, 5031–5037. [Google Scholar] [CrossRef]
- Rapheal, P.F.; Manoj, E.; Kurup, M.R.P.; Suresh, E. Structural and spectral studies of novel Co(III) complexes of N(4)-substituted thiosemicarbazones derived from pyridine-2-carbaldehyde. Polyhedron 2007, 26, 607–616. [Google Scholar] [CrossRef]
- Barrios, V.A.E.; Méndez, J.R.R.; Aguilar, N.V.P.; Espinosa, G.A.; Rodríguez, J.L.D. FTIR–An Essential Characterization Technique for Polymeric Materials; InTech: Singapore, 2012; Volume 4. [Google Scholar]
- Vicente-Martínez, Y.; Caravaca, M.; Soto-Meca, A.; Martín-Pereira, M.; García-Onsurbe, M.d.C. Adsorption studies on magnetic nanoparticles functionalized with silver to remove nitrates from waters. Water 2021, 13, 1757. [Google Scholar]
- Trivedi, M.K.; Branton, A.; Trivedi, D.; Nayak, G.; Bairwa, K.; Jana, S. Spectroscopic characterization of disodium hydrogen orthophosphate and sodium nitrate after biofield treatment. J. Chromatogr. Sep. Tech. 2015, 6. [Google Scholar] [CrossRef]
- Parvathy, G.; Kaliammal, R.; Maheshwaran, G.; Devendran, P.; Kumar, M.K.; Sudhahar, S. Experimental and theoretical studies on 4-hydroxy-3-methoxybenzaldehyde nicotinamide organic co-crystal for third harmonic nonlinear optical applications. J. Mater. Sci. Mater. Electron. 2020, 31, 18937–18953. [Google Scholar] [CrossRef]
- Xavier, R.J.; Dinesh, P. Spectroscopic (FTIR, FT-Raman, 13C and 1H NMR) investigation, molecular electrostatic potential, polarizability and first-order hyperpolarizability, FMO and NBO analysis of 1-methyl-2-imidazolethiol. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2014, 118, 999–1011. [Google Scholar] [CrossRef] [PubMed]




| Complex No. | Layers | ||
|---|---|---|---|
| Lower | Middle | Upper | |
| 1 | Ni(NO3)2.6H2O (1 mmol) EtOH (5 mL) | EtOH (5 mL) | Phen (2 mmol) EtOH (7 mL) |
| 2 | NiCl2.6H2O (1 mmol) [EtOH + CHCl3 (1:1)] (5 mL) | [EtOH + CHCl3 (1:1)] (5 mL) | Phen (2 mmol) [EtOH + CHCl3 (1:1)] (7 mL) |
| 3 | Ni(NO3)2.6H2O (1 mmol) CH3CN (5 mL) | CH3CN (5 mL) | Phen (2 mmol) CH3CN (7 mL) |
| 4 | Ni(NO3)2.6H2O EtOH (5 mL) | EtOH (5 mL) | Phen (3 mmol) EtOH (7 mL) |
| Identification Code | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| CCDC Number | 1870131 | 1860200 | 1860201 | 1870142 |
| Chemical formula | C24H17N6NiO7 | C24H17Cl2N4NiO | C14H11N5NiO6 | C38H30N8NiO8 |
| Mr | 560.14 | 507.02 | 403.99 | 785.39 |
| Crystal system, space group | Monoclinic, P21/n | Triclinic, P-1 | Monoclinic, P21/c | Triclinic, P-1 |
| Temperature (K) | 293 | 293 | 293 | 293 |
| a (Å) | 12.8433 (12) | 9.6283 (9), | 7.3081 (5) | 10.9832 (10) |
| b (Å) | 12.7849 (10) | 11.3616 (12) | 14.1152 (9) | 12.9050 (12), |
| c (Å) | 15.3264 (14) | 12.8315 (13) | 15.5505 (10) | 13.9599 (14) |
| α (°) | 65.101 (3) | 81.982 (3) | ||
| β (°) | 112.344 (3) | 84.546 (3) | 95.725 (2) | 76.578 (3) |
| γ (°) | 79.794 (3) | 67.807 (3) | ||
| V (Å3) | 2327.6 (4) | 1252.7 (2) | 1596.12 (18) | 1779.1 (3) |
| Z | 4 | 2 | 4 | 2 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα | Mo Kα |
| µ (mm−1) | 0.89 | 1.01 | 1.26 | 0.61 |
| Crystal size (mm) | 0.49 × 0.21 × 0.13 | 0.18 × 0.14 × 0.11 | 0.62 × 0.19 × 0.15 | 0.23 × 0.17 × 0.15 |
| Tmin, Tmax | 0.858, 0.906 | 0.785, 0.853 | 0.834, 0.867 | 0.834, 0.876 |
| No. of measured reflections | 66913 | 34226 | 41838 | 22519 |
| Independent reflections | 8530 | 9142 | 7770 | 8011 |
| Observed reflections [I > 2σ(I)] | 4055 | 4358 | 3360 | 4461 |
| Rint | 0.170 | 0.121 | 0.131 | 0.083 |
| R [F2 > 2σ(F2)] | 0.069 | 0.088 | 0.075 | 0.064 |
| wR (F2) | 0.155 | 0.293 | 0.158 | 0.195 |
| S | 1.02 | 1.09 | 1.01 | 1.00 |
| No. of reflections | 8530 | 9142 | 7770 | 8011 |
| No. of parameters | 347 | 293 | 247 | 482 |
| No. of restraints | 0 | 0 | 0 | 0 |
| H-atom treatment | a mixture of independent and constrained | constrained | ||
| Δρmax, Δρmin (e Å−3) | 0.51, −0.72 | 1.77, −0.77 | 0.52, −0.55 | 0.65, −0.53 |
| Complex 1 | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7W—H1OW···O5 | 0.65 (4) | 2.09 (4) | 2.693 (4) | 156 (5) |
| C21—H21A···O3i | 0.9300 | 2.5300 | 3.230 (5) | 132.00 |
| Symmetry codes: (i) x – ½, −y + 3/2, z – ½. | ||||
| Complex 2 | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1O1···Cl2i | 0.85 (10) | 2.37 (10) | 3.166 (5) | 157 (7) |
| C3—H3A···Cl1ii | 0.9300 | 2.7400 | 3.595 (6) | 153.00 |
| C15—H15A···Cl2iii | 0.9300 | 2.8200 | 3.714 (7) | 161.00 |
| Symmetry codes: (i) – x, −y + 2, −z + 1; (ii) −x + 1, −y + 1, −z + 2; (iii) −x, −y + 1, −z + 1. | ||||
| Complex 3 | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| C1—H1A···O2i | 0.9300 | 2.5200 | 3.064 (4) | 117.00 |
| C2—H2A···O2i | 0.9300 | 2.3000 | 2.949 (5) | 126.00 |
| C3—H3A···O3ii | 0.9300 | 2.5500 | 3.258 (3) | 133.00 |
| C8—H8A···O4iii | 0.9300 | 2.5100 | 3.332 (4) | 148.00 |
| Symmetry codes: (i) x, −y – 3/2, z – ½; (ii) −x + 1, −y – 1, −z – 1; (iii) −x + 2, y + ½, −z – ½. | ||||
| Complex 4 | ||||
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2X—H2XA···O7W | 0.9800 | 1.9400 | 2.814 (8) | 147.00 |
| C1—H1A···O7Wi | 0.9500 | 2.5200 | 3.310 (7) | 141.00 |
| C2X—H2XC···O3 | 0.9800 | 1.9200 | 2.806 (9) | 149.00 |
| C3—H3A···O5ii | 0.9500 | 2.2700 | 3.148 (9) | 153.00 |
| C5—H5A···O4ii | 0.9500 | 2.5900 | 3.498 (8) | 160.00 |
| C8—H8A···O1iii | 0.9500 | 2.5700 | 3.457 (9) | 155.00 |
| C9—H9A···O1iv | 0.9500 | 2.5200 | 3.463 (8) | 174.00 |
| C13—H13A···O6v | 0.9500 | 2.5000 | 3.223 (7) | 133.00 |
| C18—H18A···O4vi | 0.9500 | 2.4400 | 3.319 (9) | 153.00 |
| C20—H20A···O7Wvi | 0.9500 | 2.5400 | 3.379 (9) | 147.00 |
| C22—H22A···O5i | 0.9500 | 2.3800 | 3.068 (9) | 129.00 |
| C22—H22A···O6i | 0.9500 | 2.5800 | 3.349 (7) | 138.00 |
| C27—H27A···O6vii | 0.9500 | 2.4300 | 3.256 (6) | 145.00 |
| C30—H30A···O1viii | 0.9500 | 2.4800 | 3.345 (9) | 152.00 |
| C33—H33A···O3ii | 0.9500 | 2.5600 | 3.407 (7) | 149.00 |
| Symmetry codes: (i) −x + 2, −y + 1, −z + 1; (ii) x + 1, y – 1, z; (iii) x, y – 1, z; (iv) −x + 2, −y + 1, −z; (v) −x + 1, −y + 1, −z + 1; | ||||
| (vi) x + 1, y, z; (vii) x + 1, y, z – 1; (viii) −x + 3, −y + 1, −z. | ||||
| Crystal | Degr. Steps | TGA (°C) | DTA Peak (°C) (Process) | Mass Loss Exp. (%) | Mass-Loss Calc. (%) | Assig. Process, Product |
|---|---|---|---|---|---|---|
| 1 | 1 | 30–106.7 (75.1) | 92.9 (exo) | 12.9 | 14.28 | NO3 + H2O |
| 2 | 106.7–158.5 (333.9) | 131.5 (exo) | ||||
| 3 | 283.7–342.5 (762.2) | 351.2 (endo) | 34.9 | 32.2 | phen | |
| 4 | 342.5–517.5 | 502.3 (endo) | 10.8 | 11.06 | NO3 | |
| 5 | 517.5–789.7 | 713.2 (endo) | 9.7 | 9.5 | C3H3N | |
| residue | Ni, NiO, carbonaceous products (26.9%) | |||||
| 2 | 1 | 30–190.4 | 119.2 (exo) | 14.5 | 13.98 | 2Cl |
| 2 | 190.4–557.6 | 472.1 (exo) | 27 | 26.2 | C8H5N, H2O | |
| 3 | 557.6—870 | 786.9 (endo) | 13.5 | 12.8 | C4H3N | |
| residue | Ni, NiO, carbonaceous products (42.2%) | |||||
| 3 | 1 | 30–149.7 | 125.9 (exo) | 6 | 7.67 | CH3, ½ O2 |
| 2 | 294.7–400.0 | 390.4 (endo) | 37.9 | 41.1 | C12H8N | |
| 3 | 400–518.4 | 505.7 (endo) | 7.0 | 6.4 | CN | |
| 4 | 518.4–830.2 | 732.8 (endo) | 14.0 | 13.1 | C3H3N | |
| residue | Ni, NiO, carbonaceous products (31.6%) | |||||
| 4 | 1 | 30–190.4 | - | 6.2 | 5.9 | C2H5OH |
| 2 | 190.4–237.7 | 233.0 (exo) | 7.1 | 7.89 | NO3 | |
| 3 | 237.7–422.4 | 384.3 (endo) | 27.5 | 25.2 | Phen + H2O | |
| 4 | 422.4–517.4 | 502.4 (endo) | 6.2 | 5.9 | NO2 | |
| 5 | 517.4–850.2 | 767.1 (endo) | 21.4 | 22.9 | phen | |
| residue | Ni, NiO, carbonaceous products (27.5%) | |||||
| 1 | 2 | ||||
| Bond Lengths/Angles | Exp | M06-2X | Bond Lengths/Angles | Exp | M06-2X |
| Ni1---N2 | 2.07 | 1.96 | Ni1---Cl2 | 2.36 | 2.24 |
| Ni1---N3 | 2.08 | 1.99 | Ni1---O3 | 2.09 | 2.37 |
| Ni1---N4 | 2.08 | 1.99 | Ni1---N4 | 2.09 | 1.98 |
| Ni1---O5 | 2.07 | 1.89 | Ni1---N5 | 2.12 | 2.00 |
| Ni1---N6 | 2.08 | 2.20 | Ni1---N6 | 2.09 | 2.16 |
| Ni1---O7 | 2.11 | 2.22 | Ni1---N7 | 2.09 | 2.19 |
| N2—Ni1—N3 | 98 | 84 | Cl2—Ni1—O3 | 90 | 102 |
| N2—Ni1—O5 | 91 | 90 | Cl2—Ni1—N4 | 93 | 94 |
| N2—Ni1—O7 | 90 | 89 | Cl2—Ni1—N6 | 95 | 90 |
| N2—Ni1—N6 | 78 | 106 | Cl2—Ni1—N7 | 94 | 97 |
| O7—Ni1—N3 | 86 | 77 | O3—Ni1—N4 | 95 | 89 |
| O7—Ni1—N4 | 91 | 90 | O3—Ni1—N5 | 86 | 74 |
| O7—Ni1—O5 | 93 | 113 | O3—Ni1—N6 | 92 | 96 |
| 3 | 4 | ||||
| Bond lengths/angles | Exp | M06-2X | Bond lengths/angles | Exp | M06-2X |
| Ni1---N2 | 1.94 | 1.98 | Ni1---N2 | 2.10 | 2.27 |
| Ni1---N3 | 2.03 | 2.06 | Ni1---N3 | 2.08 | 1.96 |
| Ni1---N4 | 1.97 | 2.03 | Ni1---N4 | 2.10 | 1.98 |
| Ni1---O5 | 2.19 | 1.97 | Ni1---N5 | 2.09 | 2.26 |
| Ni1---O6 | 2.06 | 1.98 | Ni1---N6 | 2.08 | 1.96 |
| N2—Ni1—N3 | 99 | 154 | Ni1---N7 | 2.10 | 2.01 |
| N2—Ni1—N4 | 172 | 130 | N2—Ni1—N3 | 93 | 102 |
| N3—Ni1—N4 | 77 | 76 | N2—Ni1—N4 | 92 | 92 |
| N3—Ni1—O5 | 94 | 89 | N2—Ni1—N6 | 95 | 97 |
| N4—Ni1—O5 | 83 | 92 | N2—Ni1—N7 | 79 | 75 |
| N2—Ni1—O5 | 91 | 92 | N3—Ni1—N4 | 79 | 84 |
| N2—Ni1—O6 | 86 | 90 | N3—Ni1—N5 | 94 | 88 |
| -- | -- | -- | N3—Ni1—N6 | 92 | 91 |
| Complexes | EHOMO (eV) | ELUMO (eV) | Egap (eV) |
|---|---|---|---|
| 1 | −7.11 | −1.47 | 5.63 |
| 2 | −6.51 | −1.33 | 5.18 |
| 3 | −8.19 | −1.25 | 6.94 |
| 4 | −6.44 | −1.70 | 4.74 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alramadhan, S.A.; Hammud, H.H.; Ali, B.F.; Ghabbour, H.A.; Sarfaraz, S.; Ayub, K. Structures, Characterization and DFT Studies of Four Novel Nickel Phenanthroline Complexes. Crystals 2023, 13, 738. https://doi.org/10.3390/cryst13050738
Alramadhan SA, Hammud HH, Ali BF, Ghabbour HA, Sarfaraz S, Ayub K. Structures, Characterization and DFT Studies of Four Novel Nickel Phenanthroline Complexes. Crystals. 2023; 13(5):738. https://doi.org/10.3390/cryst13050738
Chicago/Turabian StyleAlramadhan, Safiah A., Hassan H. Hammud, Basem F. Ali, Hazem A. Ghabbour, Sehrish Sarfaraz, and Khurshid Ayub. 2023. "Structures, Characterization and DFT Studies of Four Novel Nickel Phenanthroline Complexes" Crystals 13, no. 5: 738. https://doi.org/10.3390/cryst13050738
APA StyleAlramadhan, S. A., Hammud, H. H., Ali, B. F., Ghabbour, H. A., Sarfaraz, S., & Ayub, K. (2023). Structures, Characterization and DFT Studies of Four Novel Nickel Phenanthroline Complexes. Crystals, 13(5), 738. https://doi.org/10.3390/cryst13050738







