Abstract
Chimeric Antigen Receptor T-cell (CAR-T) immunotherapies are dependent upon designed transmembrane proteins to bind target antigens and stimulate an immune response. The success or failure of these CARs is only partially predictable, yet recent work has highlighted the importance of antigen binding scFvs driving distinct oligomerization states with varied CAR-T efficacy. Here, we sought to determine the extracellular structure of the anti-CD19 CAR 47G4-CD828Z. Unexpectedly, the resolved crystal structure revealed an IgVL homodimer bound along an inverted VL|VL interface. We found that the VL-VH linker, designed to be cleavage resistant, was cleaved, and the VH and CAR hinge domains were absent from the crystal structure lattice. Molecular Dynamics simulations revealed that the inverted VL|VL interface was more stable than the canonical VL|VL configuration. Our work substantiates the need to interrogate the scFv structure and CAR oligomerization state for optimal CAR-T design.
1. Introduction
Naturally occurring tumor-reactive lymphocytes (T-cells) have a remarkable capacity to react to pathogens [1], and a central direction in cancer immunotherapy has been to harness patients’ own (autologous) T-cells to fight their cancer through “Adoptive Cell Transfer” (ACT) [2,3]. One form of ACT, termed Chimeric Antigen Receptor (CAR) T-cell therapy, extends T-cells’ ability to bind specific target antigens expressed on the surface of tumor cells and initiate a cascade of cellular events that results in clearing tumors [4,5].
CAR T-cell therapy involves isolating T-cells from the patient’s blood (autologous T-cells) and genetically engineering them to express a CAR protein on its surface. These CAR-T cells (CAR-Ts) are expanded in large numbers in vitro and infused back into the patient to attack cancer cells. Moreover, as these CAR-Ts expand and persist in the patient they serve as a long-term “living drug” to keep the tumor in check [6].
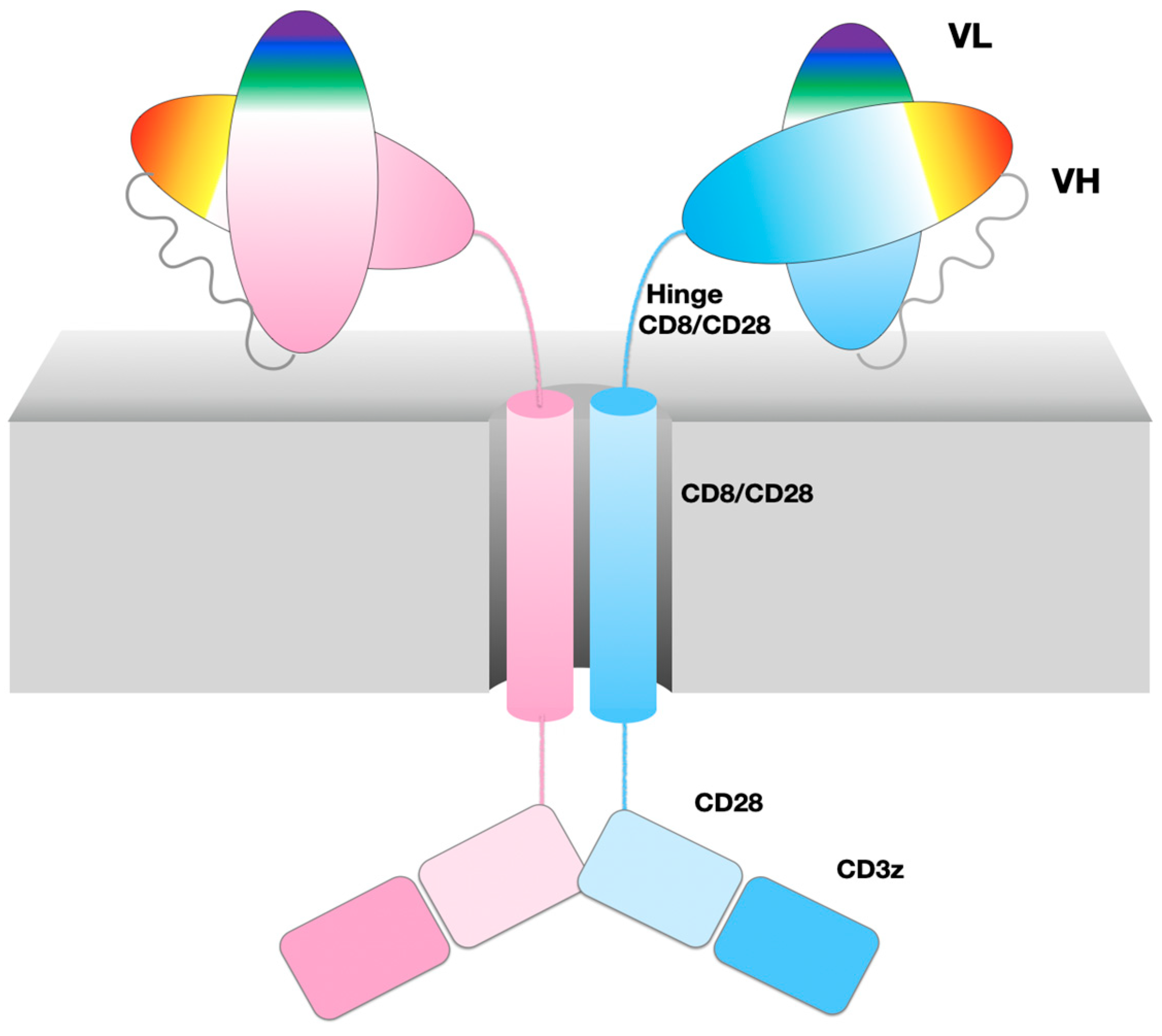
A chimeric antigen receptor (CAR) chains together different domains of native cell surface receptor proteins of the immune system to integrate a set of specific functions from binding a target cell surface antigen to signal transduction and T-cell stimulation. Current “second generation” CARs [4] are composed of antibody variable domains (scFv) targeting a specific antigen, followed by a hinge (also called spacer) connecting to a transmembrane helical domain, both typically extracted from a single pass transmembrane protein such as CD8 or CD28, followed by an intracellular costimulatory domain extracted from CD28 or 4-1BB, and lastly a signaling domain extracted from CD3z, as shown schematically in Figure 1. The optimization of CARs built from domains extracted from native cell surface receptors is a very active field of research [4,5].
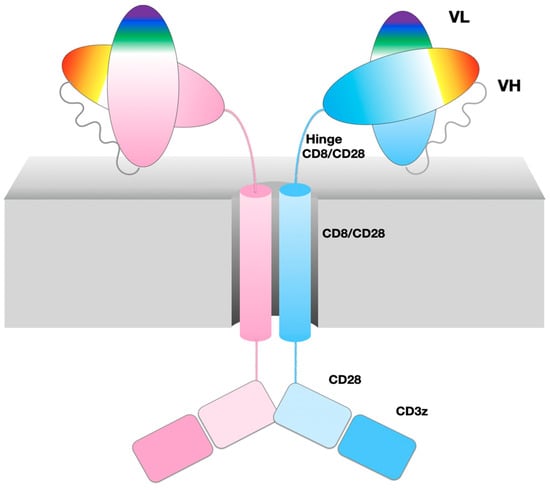
Figure 1.
47G4-CD828Z CAR domains and the VL-VL dimer. Cartoon of CAR domains, represented as an ideal dimer. A single chain is composed of two immunoglobulin domains VL and VH from the 47G4 antibody, chained together in that order as an scFv followed by the CD8a hinge and transmembrane regions, and costimulatory domains extracted from CD28 and CD3zeta. The CAR name describes its composition in abbreviated form 47G4-CD828Z.
With the ability to recognize antigens expressed on the surface of tumor cells, anti-CD19 CAR-Ts target the cell surface CD19 molecule expressed on many B-lineage leukemias and lymphomas. They have been remarkably successful in treating relapsed/refractory disease, with two FDA approved therapies: CTL-019 (KYMRIAH, Novartis Pharmaceuticals Corp.) and axicabtagene ciloleucel (YESCARTA, Kite Pharma, Inc., Los Angeles, CA, USA) [4,7,8,9,10]. This success has inspired very strong interest in developing CARs against a broader range of cell-surface antigens, especially against solid tumors that represent 90% of cancers [11]. A total of 89 unique antigens (50 for hematological cancer, 57 for solid tumors, and 18 common to both) have been used as targets of CAR-Ts in over 800 ongoing clinical studies worldwide (https://clinicaltrials.gov/ct2/results/details?term=CAR-T+OR+%2528CAR-T+cell+therapy%2529&recrs=abdf, accessed on 19 April 2023), and more trials are projected in coming years [12].
While CAR-Ts hold great promise in the clinic, the biochemical and biophysical basis for what makes an effective CAR protein remains largely unknown. In fact, no intact structure of any single pass membrane protein on which these chimeric molecules are based has been determined to this day [13]. We sought initially to obtain a 3D structure of a CAR extracellular domain using X-ray crystallography.
2. Materials and Methods
2.1. Protein Purification and Crystallization
DNA coding for the extracellular region of the 47G4-CD828Z CAR was synthesized (Genscript, Piscataway, NJ, USA) and cloned into the mammalian cell expression vector pcDNA3.1(+) between the BamH1 and Xho1 sites of the polylinker under the control of the human cytomegalovirus (CMV) promoter. The construct began with a native CD8 leader sequence at the N-terminus to direct secretion and ended with TEV protease-cleavable octahistidine affinity tag at the C-terminus to aid in protein purification. The protein was expressed by transient transfection of Expi293 cells at high density using polyethylenimine [14]. Conditioned media containing secreted protein was harvested 4 days posttransfection, cleared of cells by centrifugation, and exchanged by cross-flow diafiltration with binding buffer containing 20 mM tris(hydroxymethyl)aminomethane (Tris) pH 7.5, 150 mM NaCl, and 40 mM imidazole. The 47G4-CD828Z CAR was purified with tandem nickel affinity and gel filtration chromatography using an AKTAexpress system (Cytiva, Marlborough, MA, USA). The elution buffer was of the same composition as the binding buffer except it contained 0.5 M imidazole and the gel filtration buffer contained 20 mM Tris pH 7.5 and 150 mM NaCl. Protein fractions were pooled and polyhistidine-tagged TEV protease [15] was used to cleave the C-terminal His8 tag from the CAR. The cleaved protein was passed through Ni-NTA agarose resin (Qiagen, Hilden, Germany) to remove the protease and tag. Protein purity and integrity was verified using SDS-PAGE prior to concentration for sparse-matrix crystallization screening. Sitting-drop vapor diffusion experiments were performed using the JCSG Core I–IV (Qiagen) and Wizard Precipitant Synergy (Molecular Dimensions, Sheffield, England) screens and slow-growing crystals nucleated in one condition after 3 weeks of incubation at room temperature. Crystallization was further optimized in the conditions given in Table 1.

Table 1.
Crystallization.
2.2. Data Collection and Processing, Structure Solution and Refinement
Crystals were flash frozen in liquid nitrogen after brief soaking in 3.4 M sodium malonate pH 8.5 (Hampton Research, Aliso Viejo, CA, USA) and screened in-house for diffraction. In order to improve diffraction, the crystals were annealed twice by blocking the cryostream for 1 s each annealing cycle prior to shipment to the synchrotron in the frozen state. Data were collected at beamline 19-ID (NYX) at the National Synchrotron Light Source II (Brookhaven National Laboratory, Upton, NY, USA), indexed and merged using HKL2000 [16], and converted to structure factors using CCP4 [17]. The structure was solved with molecular replacement using Phaser [18], searching independently with ensembles of isolated N- and C-terminal immunoglobulin domains from 8 different scFvs in the PDB (codes 6EJG, 4YJZ, 3AUV, 1KTR, 6I07, 1H8N, 5OGI, and 6J71). Two copies of only the N-terminal VL-derived domain of the extracellular domain of the 47G4-CD828Z CAR were found in the asymmetric unit. The model was automatically rebuilt using ARP/wARP [19] of the CCP4 package followed by manual model building in COOT [20] and refinement in Phenix [21]. Two molecules of PEG and a molecule of malonate were found ordered near crystal contacts. Data collection and processing, and structure solution and refinement statistics are listed in Table 2 and Table 3, respectively.

Table 2.
Data collection and processing. Values in parentheses are for outer shell.

Table 3.
Structure solution and refinement.
2.3. Structure Prediction
Alphafold2 predictions [22,23,24] were used to produce models of scFv monomers. Alphafold-multimer was used to produce models of scFv dimers and VL-VL conformations. The canonical VL-VL configuration structure was predicted by AlphaFold2 using a max template date of 10 May 2022. AlphaFold2 predicted 25 structural models; all but one (of low quality) were in the canonical VL-VL configuration (see Supplement File S3).
2.4. Molecular Dynamics Simulations
Both the inverted and canonical VL-VL dimer configurations were studied by MD simulation. The inverted VL-VL configuration structure was taken from the PDB crystal structure in this study (PDBid: 7JO8). The canonical VL-VL configuration structure was selected as the top-ranked AlphaFold2 model. MD simulations were conducted using NAMD2 following a similar protocol used previously [25]. Briefly, both systems were solvated in TIP3P water boxes with 100 mM of NaCl concentration. Protein parameters were assigned from the CHARMM36 force field. Production simulations consisted of 100 ns NPT equilibrium MD simulations. The trajectory Root Mean Square Deviation of the atomic positions (RMSD) was computed using Gromacs tools.
2.5. Steered Molecular Dynamics Simulations
Constant velocity SMD simulations were conducted using NAMD2 software. We ran 20 SMD replicas for each system represented by 20 structures taken every 2.5 ns across the last 50 ns of the equilibrium MD simulation. The chain A VL domain was held fixed by Cα atoms of residues 23, 34, 38, 48, 63, and 89, while the chain B VL domain was pulled by all heavy atoms at a constant velocity of 1 Å/ns with a pulling spring constant of 7 kcal/mol. The two VL-VL dimers were assumed to be unbound when the interaction energy reached 0 kcal/mol; we note that the same affinity trend existed when we defined the unbound state as an absence of native bound-state contacts (−64.9 ± 0.8 kcal/mol inverted vs. −46.4 ± 1.2 kcal/mol canonical).
3. Results
3.1. CAR Extracellular Construct
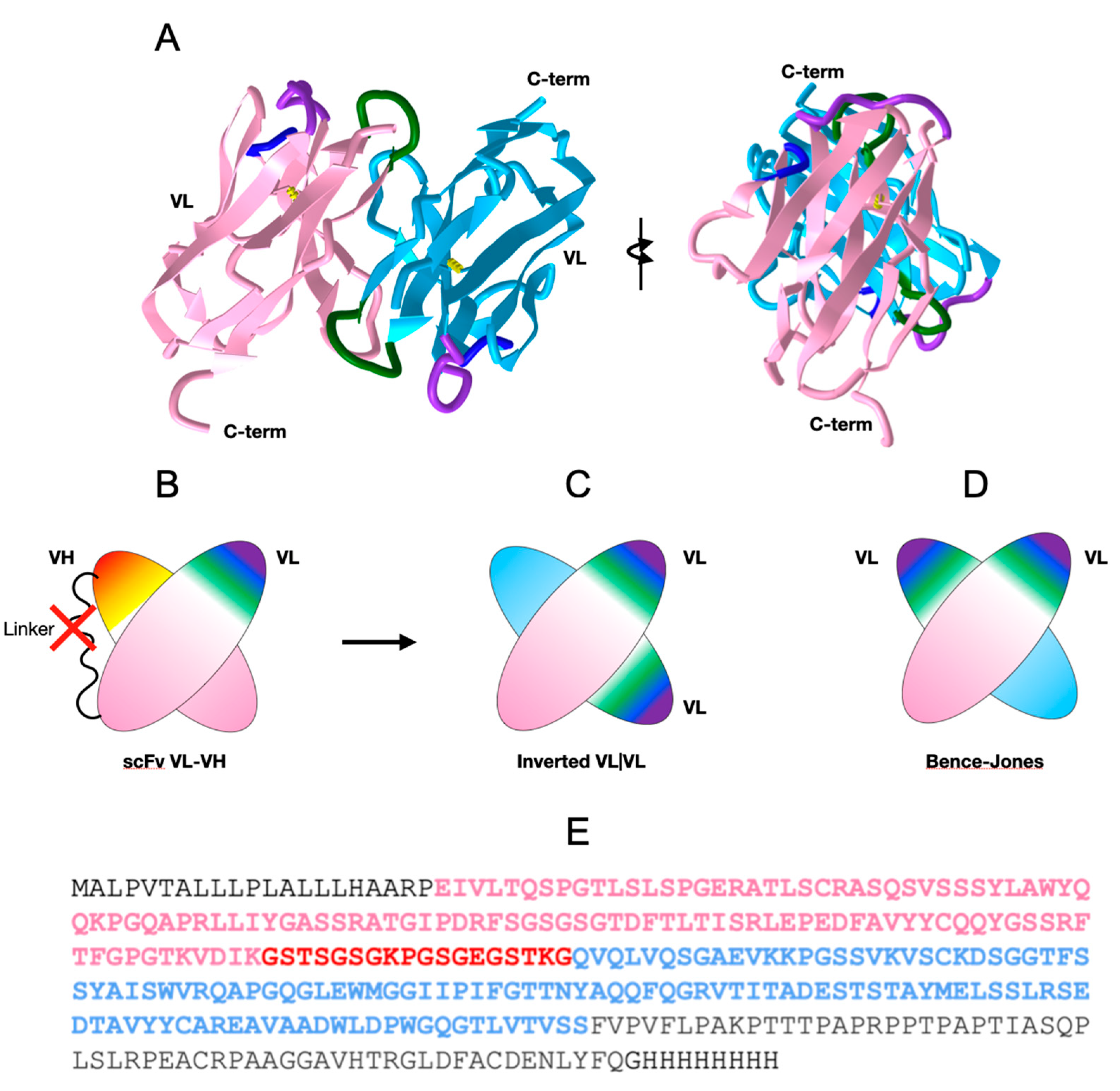
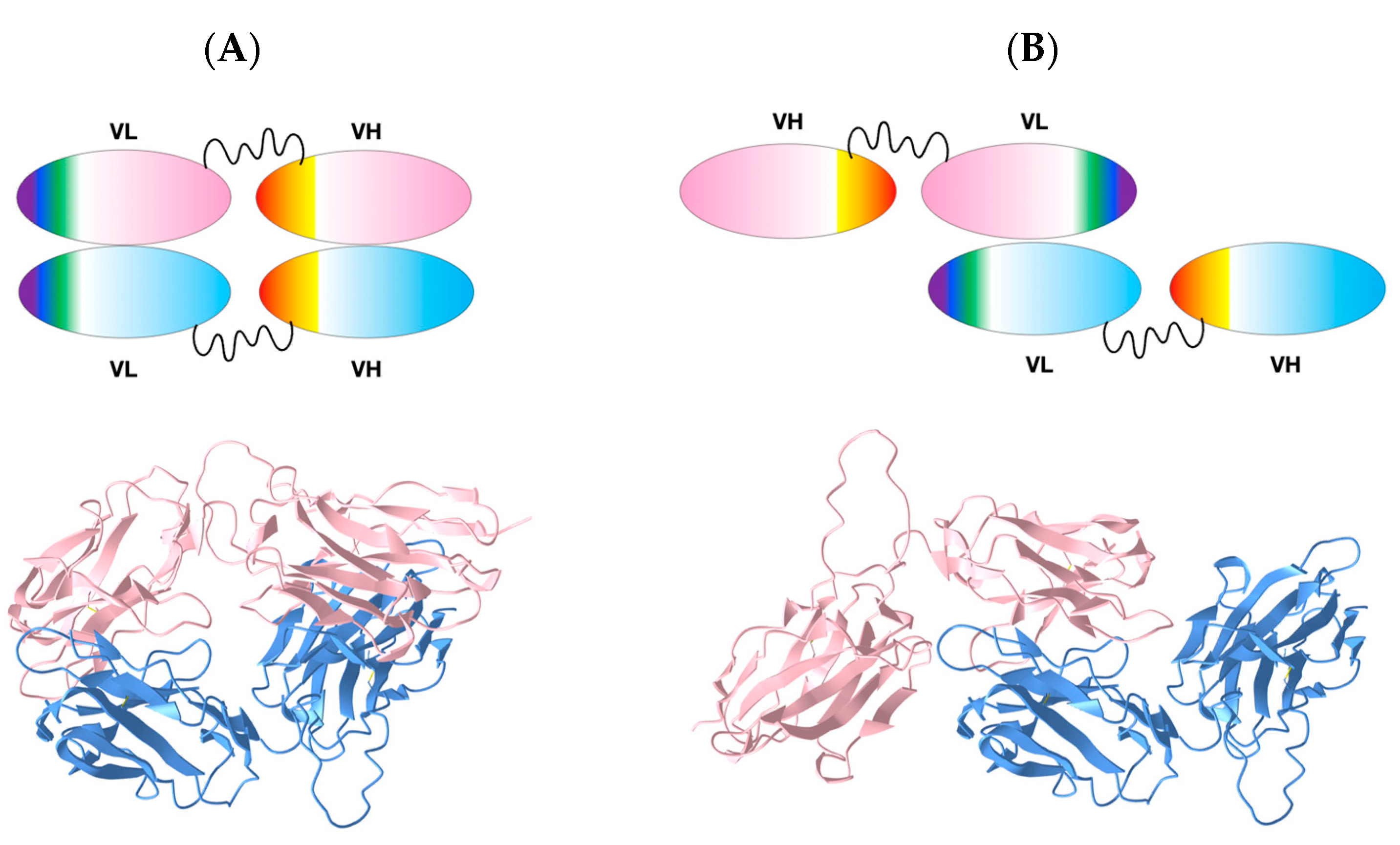
We generated constructs of the extracellular domains of the anti-human B-cell CD19 CAR 47G4-CD828Z [26] linking the VL and VH domains of the fully human anti-CD19 antibody [27], also called Hu19, to form an scFv in a VL to VH orientation connected by a linker peptide, followed by a CD8 hinge [10] to cover the entire extracellular region (See Figure 1 and Figure 2E). Importantly, the linker peptide (GSTSGSGKPGSGEGSTKG) connecting VL to VH was originally designed for “reduced aggregation and enhanced scFv proteolytic stability” [28]. This construct produced crystals suitable for diffraction analysis. The crystal structure obtained unexpectedly formed a VL-VL homodimer with an inverted (flipped) interface, unlike the well-known Bence-Jones protein (1REI) that forms as a VL-VL homodimer with a canonical IgV dimer interface (see Figure 2 and Figure 3 and Table S1 for sequence information).
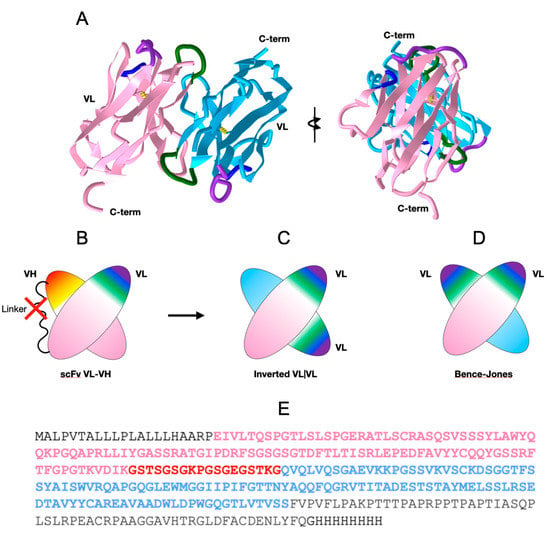
Figure 2.
47G4-CD828Z CAR domains and the VL-VL dimer. (A) VL-VL homodimer observed in the crystal with an inverted configuration. VL domains’ CDR1 colored in purple, CDR2 in blue, and CDR3 in green. (PDB:7JO8 3D visualization link: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?mMva3qCc76kn11LG6, accessed on 19 April 2023). (B) Cartoon of the scFv moiety, formed by linking a VL and a VH domain with a canonical VL|VH interface between the two Ig domains (C) Cartoon of the 47G4 VL-VL homodimer formed in the crystal with an inverted configuration. (D) Cartoon of a Bence-Jones VL-VL homodimer with a canonical interface (similar to a VL|VH heterodimer) for comparison (see Figure 3 for details). (E) Protein sequence of the extracellular region of the of the anti-CD19 47G4 CAR with the CD8 secretion signal at the N terminus (black), the VL domain (pink), the linker (red), the VH domain (blue), followed by the CD8a hinge (black) and the His8-tag.
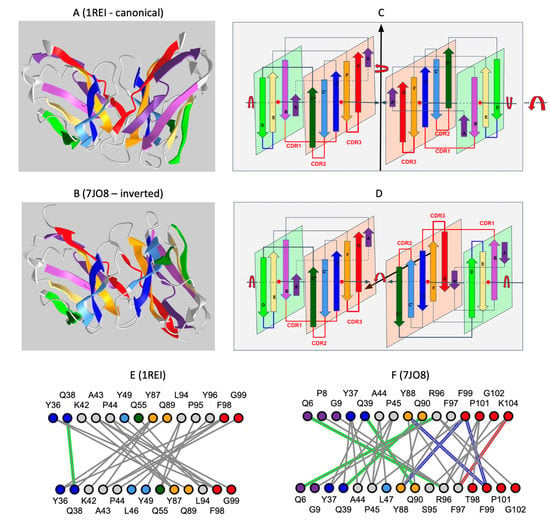
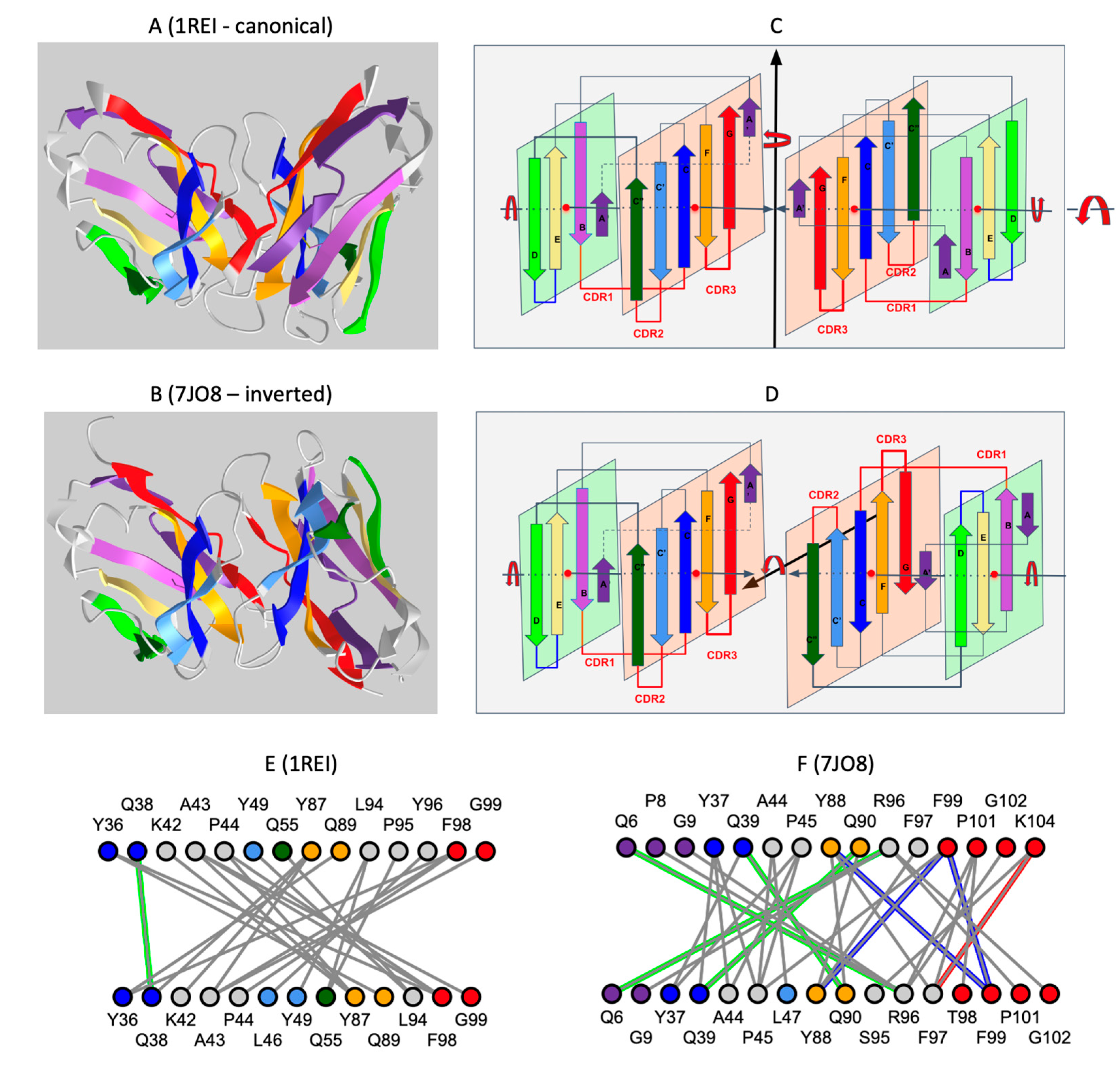
Figure 3.
Comparison of the inverted VL-VL dimer vs. the Bence-Jones VL-VL dimer. (A) Canonical dimer interface using the GFCC’ sheet observed in the Bence-Jones protein (1REI): The observed quaternary axis of symmetry is vertical. This is the classical interface of antibody VH-VL domains with the CDRs on the same side. It is also found in the Bence-Jones protein as a VL-VL: 3D visualization of the Bence-Jones protein with a canonical interface iCn3D-1REI homodimer (VL-VL): https://structure.ncbi.nlm.nih.gov/icn3d/share.html?RK2mWw4w3BfULb2H6, (accessed on 19 April 2023). (B) Inverted dimer interface using the GFCC’ sheet observed in the CAR VL-VL dimer (7JO8): The observed quaternary axis of symmetry is horizontal coming out of the page plane. It corresponds to a flip (180 degrees rotation) of one VL domain vs. the other. In this case, CDRs are on opposite sides; the C terminus G strands are pointed in opposite directions. The figure shows the structure of a VL-VL dimer interacting through the GFCC’ sheet in antiparallel iCn3D-7JO8 inverted homodimer: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?eGmk1SeSjLyok3p37, (accessed on 19 April 2023). The second row (C,D) represents a schematic representation of the variable domains in both dimers. The third row (E,F) compares the interactions between the GFCC’C” sheet between the two dimers. The Bence-Jones protein symmetric dimer interface is similar to a canonical pseudosymmetric VH-VL interface, forming in particular H-bonds between residues Q38 in symmetric positions. The inverted VL-VL dimer interface forms a tighter symmetric interface with 4 residues forming H-bonds (green) (Q6-R96 Q39-Q90), aromatic interactions (blue) (Y88-F99, F99-F99), and aromatic cation interaction (red) (F97-K104). See 3D visualization: comparison of the two VLVL dimer interfaces: https://structure.ncbi.nlm.nih.gov/icn3d/share.html?EqpkRWuB1bCaBXTc9, (accessed on 19 April 2023). Coloring of Ig strands using the full rainbow spectrum (iCn3D: strands A/A′: dark violet, strand B: light violet, strand C: dark blue, strand C′: light blue, strand C″: dark green, strand D light green, strand E: yellow, strand F: orange, strand G: red. Loops in grey.
The intact extracellular region was purified and used in crystallization; it is apparent that the flexible linker joining the VL and VH domains of the scFv portion of the CAR was proteolyzed during crystallization, despite its intended design [28], since the unit cell dimensions and lattice do not provide the space for the other regions of the extracellular domain to be present if they had been disordered. Subsequent sparse-matrix crystallization screening of purified protein supplemented with protease inhibitors failed to yield other crystallization hits.
The observed VL-VL structure, obtained after proteolysis of the linker, raises questions surrounding scFv linkers, variable domain swapping, and scFv oligomerization, and, therefore, regarding CAR oligomerization made possible through their IgV domains rearrangements. While the significance of this observed structure in the context of a full CAR, especially in vivo, has to be established, it points to possible domain swaps in scFv dimers.
3.2. scFv Linkers and Oligomerization through Domain Swapping
CAR designs to date have assumed that scFvs are in a monomeric form within a single CAR chain in order to bind one target. Yet, scFvs are prone to dimerization and aggregation, and the linker length between the VH and VL domains impacts scFv formation and, consequently, CAR formation. scFv oligomerization states are very sensitive to VL-VH linkers and can dimerize to form diabodies. Reducing the scFv linker length to 3–5 residues can lead to the formation of (dimeric) diabodies vs. (monomeric) scFvs through domain swapping [29,30,31] to form two canonical VL|VH interfaces to bind their targets. In the context of CAR-Ts, a recent study has demonstrated the importance of the scFv linker length in anti-CD22 CARs [32,33], differing only in the linker length with a drastic effect on the clinical outcomes [32,33]. The two CARs derived from the m971 anti-CD22 monoclonal antibody [34,35] differ in linker length, and it is known from X-ray crystallography that the short (5 residue) linker forms a diabody [36]. Short linkers are sometimes called Winter linkers [30].
We performed a size exclusion chromatography (AnSEC) study of the extracellular region 47G4-CD828Z CAR (Figure 2E) that confirms the formation of an oligomer, running primarily as a dimer with a small peak as a tetramer (see Figure S1). Yet, given that our crystals led to the formation of a single domain VL-VL dimer after cleavage of the linker, we are left with the question regarding the mode of dimerization for the 47G4 VL-linker-VH scFv.
3.3. Predicted scFv Dimers in Domain-Swapped or Strand-Swapped Forms
In the absence of an experimental structure of an intact CAR or simply of an intact CAR extracellular region, we can interrogate monomeric or dimeric forms through either structure prediction and/or homology model building. Alphafold2 predictions [22,23,24] show swapped diabody forms of the scFvs, regardless of the linker length. These are canonical (parallel) IgV|IgV interfaces and are plausible from a structure prediction viewpoint. However, our observed crystal structure shows an inverted VL-VL interface (Figure 2).
On the other hand, the PDB database offers a variety of IgV interfaces, from a canonical parallel domain interface [37,38] to an inverted (antiparallel/flipped) interface [39,40], partially matching the one we observe in the present structure. A list of PDB files containing a variety of VL-VL interfaces observed in crystal structures is available in the Supplementary Material (File S1).
Alphafold2 models of scFv dimers of current CARs, especially with the 47G4 sequence, show the formation of swapped VH and VL domains in dimeric forms with long linkers and short linkers, yet long linkers allow for a variety of domain arrangements. In the case of the 47G4 scFv, the VL-to-VH sequence (Figure 2E) with a long linker (Figure 2E) leads to swapped domain models (see Supplement Files S2). This is consistent with observations that scFvs in VL-to-VH arrangements form diabodies with linkers longer than 5 residues while exhibiting higher flexibility [41] than in the VH-to-VL order. For the 47G4 sequence, however, four out of five Alphafold2 predictions exhibit a G-strand swapping of VH domains (https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?type=icn3dpng&url=https://zenodo.org/api/files/4c8a1713-cd80-4ee5-9e3c-93782c4b3193/5models_Hu19_scFv_AF2_icn3d_loadable.png, accessed on 19 April 2023). The highest-ranking model presents an antiparallel VL-VL arrangement, albeit not with the interface observed in the X-ray structure observed in this study (Figure 3). This strand swapping is sequence-dependent and does not happen in Alphafold2 models with all VH|VL sequence combinations. G-strand swapping may represent an aggregation mechanism and may point to the importance of pairing VL and VH domains to maximize their interface stability.
3.4. The Observed VL-VL Homodimer Interface
VL-VL homodimers have been observed originally in Bence-Jones proteins [42]. Unlike the Bence-Jones structure (PDBid:1REI), the 47G4-VLVL structure in this study exhibits an inverted, antiparallel VL-VL interface (see Figure 2). In fact, it has been shown that a single residue mutation in a VL domain can induce a rotation [43] or inversion [44,45] of the VL-VL domain interfaces (3D link showing an example of inversion due to a single mutation: 1LVE vs. 5LVE), and a previous survey in the literature found 61 VL-VL dimers in the PDB database [40] among which only nine formed a noncanonical quaternary arrangement. All these dimer complexes possess exact C2 symmetry (except for one with partial dissymmetry) but differ in the rotation angle and translational displacement between monomers. The 47G4-VL-VL dimer exhibits C2 symmetry. (A survey of the current PDB database for structures exhibiting significant VL-VL contacts in crystal structures can be found in Supplement Figure S1).
3.5. The Modeled vs. the Observed VL-VL Homodimer Interface: Binding Affinities
We hypothesized that preference for different VL-VL dimer configurations is due to more favorable binding affinity. To test this hypothesis, we conducted free energy calculations of VL-VL dimer formation for the solved inverted crystal structure, as well as the canonical interface of the same VL domain as predicted by AlphaFold2 [22,23]. As shown in Figure 4A, AlphaFold2 predicted the canonical interface for all 25 structural models but one model of low quality. It does not capture the inverted interface. This is surprising, as in many cases AlphaFold2 predicts oligomeric complexes with high accuracy [22,46].
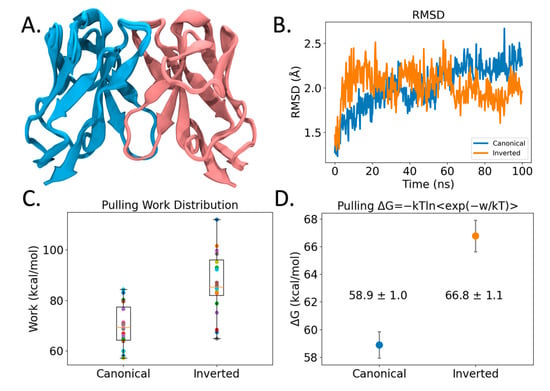
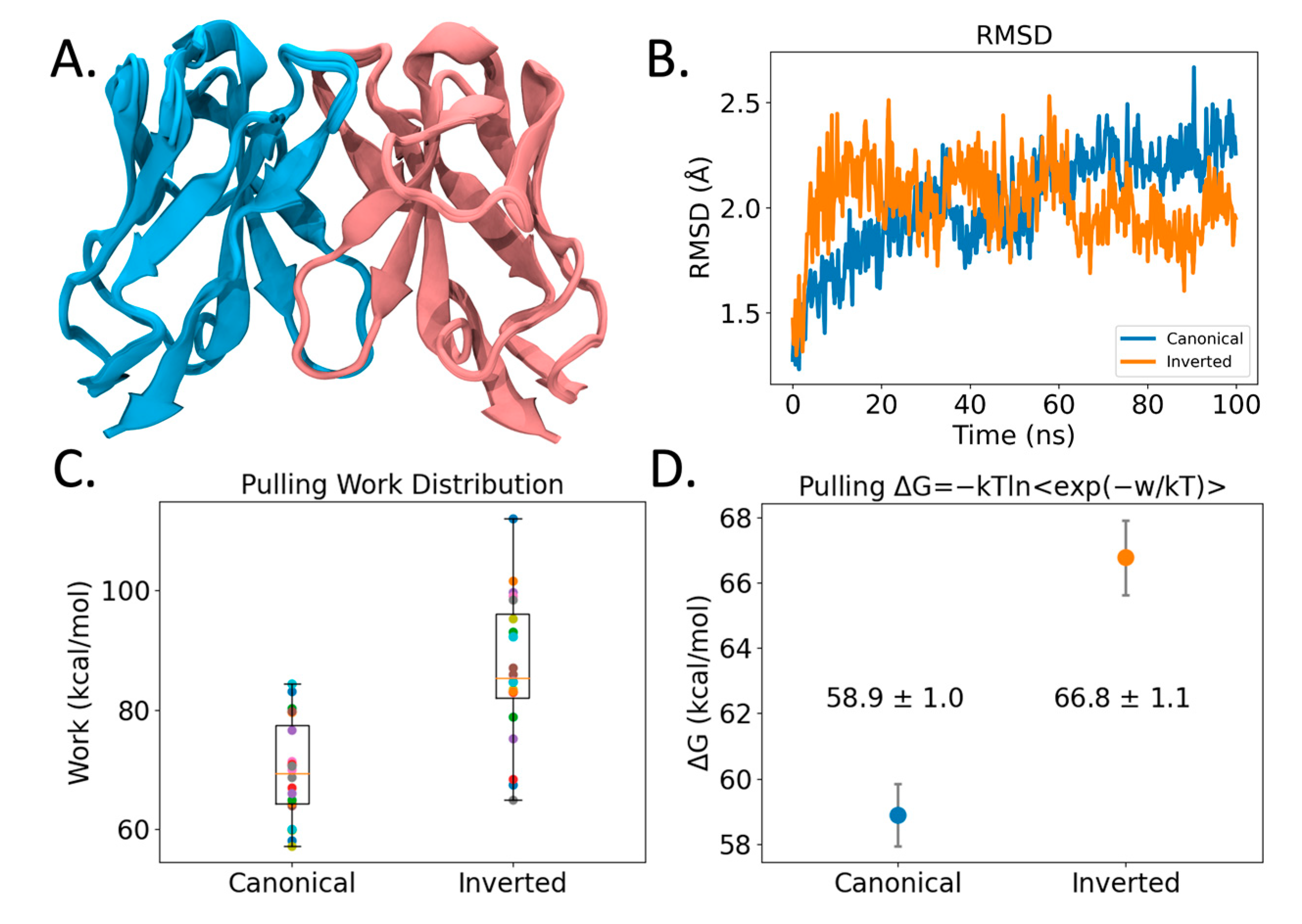
Figure 4.
Modeling of VL-VL binding interface. (A) Structural overlay of 25 VL-VL binding complex structures predicted by AlphaFold2 in the canonical configuration. Blue and pink coloring identifies different VL Ig domains. (B) RMSD profile of VL-VL complex showing stability across the Molecular Dynamics (MD) simulation. (C) Pulling work required to pull VL-VL complexes apart using Steered Molecular Dynamics (SMD). Shown are boxplots and points of all 20 replicas in different colors. (D) VL-VL binding free energies of SMD simulations from (C) computed using Jarzynski’s inequality. Coloring denotes Canonical (blue) and Inverted (orange) conformation binding free energies. Error bars represent 95% CI.
However, here, we are asking AlphaFold2 to predict VL-VL homodimerization along two competitive interfaces, with one interface (canonical) being highly populated by sequence and structure libraries, that AlphaFold2 heavily relies upon for prediction. Reports in the literature have emerged that give a varying view of the ability of Alphafold2 to predict conformational diversity and/or quaternary configurations. Some studies show that the program can predict different monomeric vs. dimeric conformational states [47]. Other studies show that it does not accurately reproduce competing structures of fold-switching proteins [48] arguing that the lack of conformational diversity is due to its deep learning algorithm depending on pattern recognition rather than pure protein biophysics. Yet, the very success of AlphaFold2 lies in balancing deep learning and intrinsic knowledge of protein tertiary structure [23], but to address the complexity of quaternary interactions more biophysical knowledge is required.
We resorted to Molecular Dynamics (MD) methods to further study the observed dimeric interface. From 100 ns simulations (see Figure 4B), both the observed inverted crystal structure interface as well as the predicted canonical dimer interface remain bound, arguing that there is stability in both the predicted and the observed VL-VL interfaces. However, steered MD simulations (SMD) (see Methods) measuring the work required to pull the VL domains apart revealed that the inverted VL-VL dimer interface is more stable for the 47G4 VL domain (see Figure 4C). Likewise, when we use Jarzynski’s equality to convert the pulling work to free energy values [49], we find that the inverted VL-VL dimer interface has a more favorable binding affinity than the canonical VL-VL interface: −66.8 ± 1.1 kcal/mol vs. −58.9 ± 1.0 kcal/mol (±95% CI); although, from the SMD method we stress the relative qualitative trend rather than exact quantitative values. Ultimately, these modeling findings support our hypothesis that different VL-VL dimer configurations occur due to more favorable binding affinity.
3.6. Hypothetical Models of Canonical and Inverted VL-VL Dimers of scFvs
While the inverted VL-VL structure obtained during crystallization can be considered an anecdote, we interrogated 47G4 scFv dimerization through an Alphafold-multimer (v2.2) [22]. The predicted models are essentially G-strand-swapped dimeric structures (see above), while the original version (Alphafold2.0) produced top-ranking models without dimerization, except for an unusual diabody pairing VL-VL and VH-VH domains; both domains have a canonical interface (Figure 5A and Supplement Files S4). Surprisingly, no classical VLVH diabody was predicted. In contrast, building a full-length dimerized scFv model, linking VL and VH domains based on the inverted VL-VL structure, would produce quasi free VH domains (Figure 5B). Machine Learning (ML) methods are still evolving, and oligomeric models can only be taken as suggestions at this stage. In this case, they only suggest possible mechanisms of oligomerization or aggregation. New functional chain architectures could be imagined using VL domains for dimerization, as long as VH domains would not aggregate and could bind antigen targets as single Ig domains, similar to nanobody VHH domains [50].
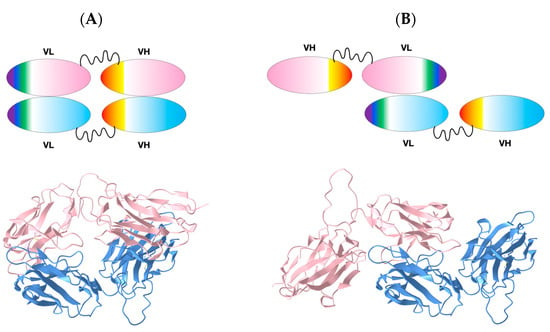
Figure 5.
Comparison of VL-VL based scFv dimer. (A) VL-VL diabody: a canonical VL-VL dimer interface leads to a pairing of VH domains. Top: Cartoon, Bottom: AlphaFold2-predicted structure. (B) Inverted VL-VL diabody model leading to free VH domains. Top: Cartoon, Bottom: structural model built on the basis of the observed inverted VL-VL structure. Blue and pink coloring identifies different scFv domains. Models are available as supplemental data (Supplement Files S4). 3D models comparison link: https://www.ncbi.nlm.nih.gov/Structure/icn3d/full.html?type=icn3dpng&url=https://zenodo.org/api/files/4c8a1713-cd80-4ee5-9e3c-93782c4b3193/VLVLDIAB_VLINVDIAB_icn3d_loadable.png, accessed on 19 April 2023).
4. Discussion
Our structural knowledge of single-pass transmembrane receptors is limited [13]. With the development of single-pass transmembrane chimeric antigen receptors for CAR-T cell immunotherapies, it is imperative that we gain knowledge of their structure for accurate rational design. Recent studies and clinical trials show the need to understand CAR oligomerization and aggregation on the T-cell surface to optimize T-cell stimulation and limit toxicities. These works point to CAR oligomerization and functional dependence on scFv constructs, especially the VH-VL linker and VH-VL stability, yet the determinants of scFv-driven CAR oligomerization have not been identified [33]. This study sought to determine the extracellular structure of the anti-CD19 CAR 47G4-CD828Z. We unexpectedly captured a homodimer of a single IgVL domain with an inverted VL-VL interface. Further investigation revealed that the VL-VH linker was cleaved in the crystal, despite its designed cleavage resistance, with the VH and CAR Hinge domains absent from the crystal structure lattice. Molecular Dynamics simulations support the finding that the inverted VL-VL interface is more stable (enhanced binding affinity) than the canonical IgV interface predicted by Alphafold, as in the Bence-Jones protein. We find that IgV domain pairing and interface stability is an important and nontrivial parameter in scFv structure and, consequently, CAR oligomerization and CAR-T cell function. We conclude that it is important to interrogate the pairing and stability of VH-VL interfaces in single-chain antibody fragments when optimizing scFvs for use as binding moieties of CARs.
While we are still left with the question surrounding the extracellular structure of the intact CAR including the VL-VH and hinge domains of CAR-T-cells in biological conditions, this work substantiates the need to interrogate the scFv structure and CAR oligomerization state for optimal CAR-T design and calls for further structural studies to reveal the full picture of CARs expressed at the surface of T-cells.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cryst13040710/s1. Supplemental files can be found in the Zenodo repository: https://doi.org/10.5281/zenodo.7809232 (accessed on 19 April 2023) [51]. Any file in repository can be accessed as an iCn3D [52] from the Zenodo API record record: https://zenodo.org/api/records/7809232 (accessed on 19 April 2023) [53].
Author Contributions
Conceptualization: P.Y., J.N.K. and W.A.H.; funding acquisition: J.N.K. and P.Y.; crystallization and structure determination: J.C. and S.W.; in silico structural analysis: P.Y.; structure predictions: D.R.B. and P.Y.; MD simulations: D.R.B.; visualization: P.Y. and D.R.B.; writing of manuscript: P.Y., J.C. and D.R.B.; review and editing: D.R.B., J.N.K., W.A.H. and P.Y.; supervision: W.A.H. and P.Y.; project administration: W.A.H. and P.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the NIH intramural program and through a research contract #HHSN261201800773P. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Structural Data was deposited to the Protein Data Bank (PDB) under the PDBid 7JO8. Alphafold2 models are available in the Zenodo repository: https://doi.org/10.5281/zenodo.7809232 (accessed on 19 April 2023) [51]. Any file in repository can be accessed as an iCn3D [52] URL from the Zenodo API record: https://zenodo.org/api/records/7809232 (accessed on 19 April 2023) [53].
Acknowledgments
We thank our collaborators from the Frederick National Center for Cancer Research (FNLCR): Dominic Esposito and Jane Jones for protein expression and size exclusion chromatography experiments, and Raul Cachau for useful discussions. We thank our collaborators from NCBI Jiyao Wang for iCn3D developments allowing the sharing of 3D visualizations and structural analyses through lifelong links used in this paper, James Song for improved topological labeling of Conserved Domains (CDD) of Immunoglobulin domains, and Tom Madej for performing structural database searches of quaternary structural homologs of Immunoglobulin variable domains VL-VL dimers. We also thank David Goldstein and Mariam Malik for setting up extramural collaborations with the NY Structural Biology Center.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, N.; Bevan, M.J. CD8+ T cells: Foot soldiers of the immune system. Immunity 2011, 35, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Restifo, N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 2015, 348, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, S.A.; Dudley, M.E. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Curr. Opin. Immunol. 2009, 21, 233–240. [Google Scholar] [CrossRef]
- Kochenderfer, J.N.; Rosenberg, S.A. Treating B-cell cancer with T cells expressing anti-CD19 chimeric antigen receptors. Nat. Rev. Clin. Oncol. 2013, 10, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Brudno, J.N.; Lam, N.; Vanasse, D.; Shen, Y.-w.; Rose, J.J.; Rossi, J.; Xue, A.; Bot, A.; Scholler, N.; Mikkilineni, L.; et al. Safety and feasibility of anti-CD19 CAR T cells with fully humanbinding domains in patients with B-cell lymphoma. Nat. Med. 2020, 26, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Fraietta, J.A.; June, C.H.; Xu, Z.; Joseph Melenhorst, J.; Lacey, S.F. Engineered T Cell Therapies from a Drug Development Viewpoint. Engineering 2019, 5, 140–149. [Google Scholar] [CrossRef]
- Maude, S.L.; Laetsch, T.W.; Buechner, J.; Rives, S.; Boyer, M.; Bittencourt, H.; Bader, P.; Verneris, M.R.; Stefanski, H.E.; Myers, G.D.; et al. Tisagenlecleucel in Children and Young Adults with B-Cell Lymphoblastic Leukemia. N. Engl. J. Med. 2018, 378, 439–448. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Han, W.; Zhang, Y. Tisagenlecleucel, an approved anti-CD19 chimeric antigen receptor T-cell therapy for the treatment of leukemia. Drugs Today 2017, 53, 597–608. [Google Scholar] [CrossRef]
- Gill, S.; June, C.H. Going viral: Chimeric antigen receptor T-cell therapy for hematological malignancies. Immunol. Rev. 2015, 263, 68–89. [Google Scholar] [CrossRef]
- Kochenderfer, J.N. Chimeric Antigen Receptors Targeting CD-19. US10287350B2, 14 May 2019. [Google Scholar]
- Yong, C.S.M.; Dardalhon, V.; Devaud, C.; Taylor, N.; Darcy, P.K.; Kershaw, M.H. CAR T-cell therapy of solid tumors. Immunol. Cell Biol. 2017, 95, 356–363. [Google Scholar] [CrossRef]
- Moreno-Cortes, E.; Forero-Forero, J.V.; Lengerke-Diaz, P.A.; Castro, J.E. Chimeric antigen receptor T cell therapy in oncology—Pipeline at a glance: Analysis of the ClinicalTrials.gov database. Crit. Rev. Oncol. Hematol. 2021, 159, 103239. [Google Scholar] [CrossRef]
- Bugge, K.; Lindorff-Larsen, K.; Kragelund, B.B. Understanding single-pass transmembrane receptor signaling from a structural viewpoint-what are we missing? FEBS J. 2016, 283, 4424–4451. [Google Scholar] [CrossRef]
- Backliwal, G.; Hildinger, M.; Hasija, V.; Wurm, F.M. High-density transfection with HEK-293 cells allows doubling of transient titers and removes need for a priori DNA complex formation with PEI. Biotechnol. Bioeng. 2008, 99, 721–727. [Google Scholar] [CrossRef]
- Blommel, P.G.; Fox, B.G. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr. Purif. 2007, 55, 53–68. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef]
- Langer, G.; Cohen, S.X.; Lamzin, V.S.; Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008, 3, 1171–1179. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Žídek, A.; Bates, R.; Blackwell, S.; Yim, J.; et al. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Bell, D.R. Evolution of Thyroglobulin Loop Kinetics in EpCAM. Life 2021, 11, 915. [Google Scholar] [CrossRef] [PubMed]
- Alabanza, L.; Pegues, M.; Geldres, C.; Shi, V.; Wiltzius, J.J.W.; Sievers, S.A.; Yang, S.; Kochenderfer, J.N. Function of Novel Anti-CD19 Chimeric Antigen Receptors with Human Variable Regions Is Affected by Hinge and Transmembrane Domains. Mol. Ther. J. Am. Soc. Gene Ther. 2017, 25, 2452–2465. [Google Scholar] [CrossRef]
- King, D.J.; Rao-Naik, C.; Pan, C.; Cardarelli, J.; Blanset, D. Human Antibodies that Bind CD19 and Uses Thereof. US20100104509A1, 29 April 2010. [Google Scholar]
- Whitlow, M.; Bell, B.A.; Feng, S.L.; Filpula, D.; Hardman, K.D.; Hubert, S.L.; Rollence, M.L.; Wood, J.F.; Schott, M.E.; Milenic, D.E.; et al. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 1993, 6, 989–995. [Google Scholar] [CrossRef]
- Poljak, R.J. Production and structure of diabodies. Structure 1994, 2, 1121–1123. [Google Scholar] [CrossRef]
- Holliger, P.; Prospero, T.; Winter, G. “Diabodies”: Small bivalent and bispecific antibody fragments. Proc. Natl. Acad. Sci. USA 1993, 90, 6444–6448. [Google Scholar] [CrossRef]
- Le Gall, F.; Reusch, U.; Little, M.; Kipriyanov, S.M. Effect of linker sequences between the antibody variable domains on the formation, stability and biological activity of a bispecific tandem diabody. Protein Eng. Des. Sel. 2004, 17, 357–366. [Google Scholar] [CrossRef]
- Singh, N.; Frey, N.V.; Engels, B.; Barrett, D.M.; Shestova, O.; Ravikumar, P.; Shyu, A.; Highfill, S.; Zhao, L.; Peng, L.; et al. Single Chain Variable Fragment Linker Length Regulates CAR Biology and T Cell Efficacy. Blood 2019, 134, 247. [Google Scholar] [CrossRef]
- Singh, N.; Frey, N.V.; Engels, B.; Barrett, D.M.; Shestova, O.; Ravikumar, P.; Cummins, K.D.; Lee, Y.G.; Pajarillo, R.; Chun, I.; et al. Antigen-independent activation enhances the efficacy of 4-1BB-costimulated CD22 CAR T cells. Nat. Med. 2021, 27, 842–850. [Google Scholar] [CrossRef]
- Xiao, X.; Ho, M.; Zhu, Z.; Pastan, I.; Dimitrov, D.S. Identification and characterization of fully human anti-CD22 monoclonal antibodies. mAbs 2009, 1, 297–303. [Google Scholar] [CrossRef]
- Haso, W.; Lee, D.W.; Shah, N.N.; Stetler-Stevenson, M.; Yuan, C.M.; Pastan, I.H.; Dimitrov, D.S.; Morgan, R.A.; FitzGerald, D.J.; Barrett, D.M.; et al. Anti-CD22-chimeric antigen receptors targeting B-cell precursor acute lymphoblastic leukemia. Blood 2013, 121, 1165–1174. [Google Scholar] [CrossRef]
- Ereño-Orbea, J.; Liu, X.; Sicard, T.; Kucharska, I.; Li, W.; Borovsky, D.; Cui, H.; Feng, Y.; Dimitrov, D.S.; Julien, J.P. Structural details of monoclonal antibody m971 recognition of the membrane-proximal domain of CD22. J. Biol. Chem. 2021, 297, 100966. [Google Scholar] [CrossRef]
- Novotný, J.; Haber, E. Structural invariants of antigen binding: Comparison of immunoglobulin VL-VH and VL-VL domain dimers. Proc. Natl. Acad. Sci. USA 1985, 82, 4592–4596. [Google Scholar] [CrossRef]
- Chothia, C.; Novotný, J.; Bruccoleri, R.; Karplus, M. Domain association in immunoglobulin molecules. The packing of variable domains. J. Mol. Biol. 1985, 186, 651–663. [Google Scholar] [CrossRef]
- Youkharibache, P. Topological and Structural Plasticity of the Single Ig Fold and the Double Ig Fold Present in CD19. Biomolecules 2021, 11, 1290. [Google Scholar] [CrossRef]
- Szent-Gyorgyi, C.; Stanfield, R.L.; Andreko, S.; Dempsey, A.; Ahmed, M.; Capek, S.; Waggoner, A.S.; Wilson, I.A.; Bruchez, M.P. Malachite green mediates homodimerization of antibody VL domains to form a fluorescent ternary complex with singular symmetric interfaces. J. Mol. Biol. 2013, 425, 4595–4613. [Google Scholar] [CrossRef]
- Carmichael, J.A.; Power, B.E.; Garrett, T.P.; Yazaki, P.J.; Shively, J.E.; Raubischek, A.A.; Wu, A.M.; Hudson, P.J. The crystal structure of an anti-CEA scFv diabody assembled from T84.66 scFvs in V(L)-to-V(H) orientation: Implications for diabody flexibility. J. Mol. Biol. 2003, 326, 341–351. [Google Scholar] [CrossRef]
- Epp, O.; Lattman, E.E.; Schiffer, M.; Huber, R.; Palm, W. The molecular structure of a dimer composed of the variable portions of the Bence-Jones protein REI refined at 2.0-A resolution. Biochemistry 1975, 14, 4943–4952. [Google Scholar] [CrossRef]
- Huang, D.B.; Chang, C.H.; Ainsworth, C.; Brünger, A.T.; Eulitz, M.; Solomon, A.; Stevens, F.J.; Schiffer, M. Comparison of crystal structures of two homologous proteins: Structural origin of altered domain interactions in immunoglobulin light-chain dimers. Biochemistry 1994, 33, 14848–14857. [Google Scholar] [CrossRef] [PubMed]
- Pokkuluri, P.R.; Huang, D.B.; Raffen, R.; Cai, X.; Johnson, G.; Stevens, P.W.; Stevens, F.J.; Schiffer, M. A domain flip as a result of a single amino-acid substitution. Structure 1998, 6, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Peterson, F.C.; Baden, E.M.; Owen, B.A.; Volkman, B.F.; Ramirez-Alvarado, M. A single mutation promotes amyloidogenicity through a highly promiscuous dimer interface. Structure 2010, 18, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Feng, B.Y.; Varshney, A.; Pierce, B.G. Benchmarking AlphaFold for protein complex modeling reveals accuracy determinants. Protein Sci. 2022, 31, e4379. [Google Scholar] [CrossRef] [PubMed]
- Cummins, M.C.; Jacobs, T.M.; Teets, F.D.; DiMaio, F.; Tripathy, A.; Kuhlman, B. AlphaFold accurately predicts distinct conformations based on the oligomeric state of a de novo designed protein. Protein Sci. 2022, 31, e4368. [Google Scholar] [CrossRef]
- Chakravarty, D.; Porter, L.L. AlphaFold2 fails to predict protein fold switching. Protein Sci. 2022, 31, e4353. [Google Scholar] [CrossRef]
- Jarzynski, C. Equilibrium free-energy differences from nonequilibrium measurements: A master-equation approach. Phys. Rev. E 1997, 56, 5018–5035. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural Single-Domain Antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- Zenodo Repository. Available online: https://doi.org/10.5281/zenodo.7809232 (accessed on 19 April 2023). [CrossRef]
- Wang, J.; Youkharibache, P.; Zhang, D.; Lanczycki, C.J.; Geer, R.C.; Madej, T.; Phan, L.; Ward, M.; Lu, S.; Marchler, G.H.; et al. iCn3D, a Web-Based 3D Viewer for Sharing 1D/2D/3D Representations of Biomolecular Structures. Bioinformatics 2020, 36, 131–135. [Google Scholar] [CrossRef]
- Available online: https://zenodo.org/api/records/7809232 (accessed on 19 April 2023).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).