Biosynthesis of Silver Nanoparticles Using Bersama engleriana Fruits Extracts and Their Potential Inhibitory Effect on Resistant Bacteria
Abstract
:1. Introduction
2. Results and Discussion
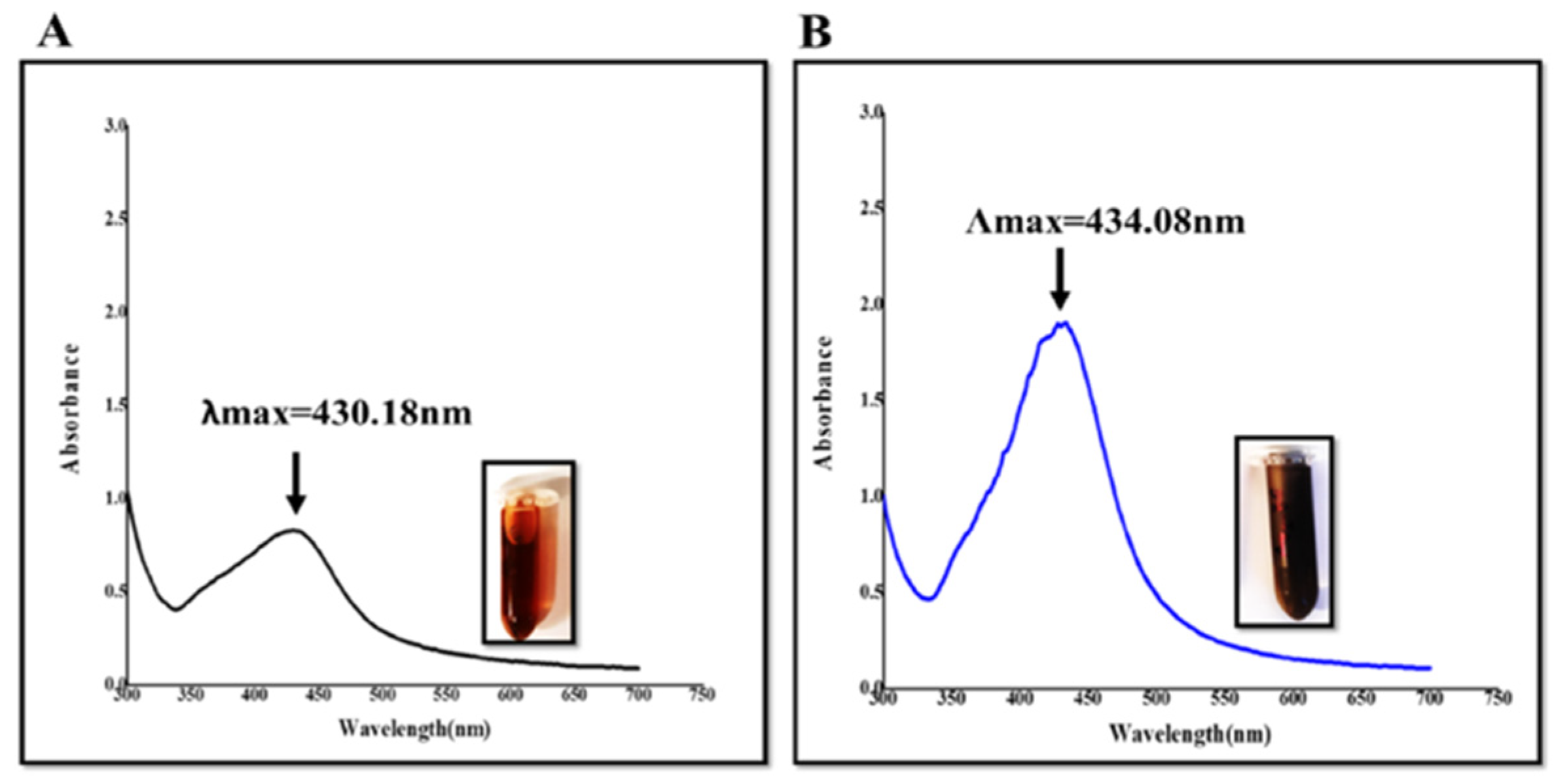
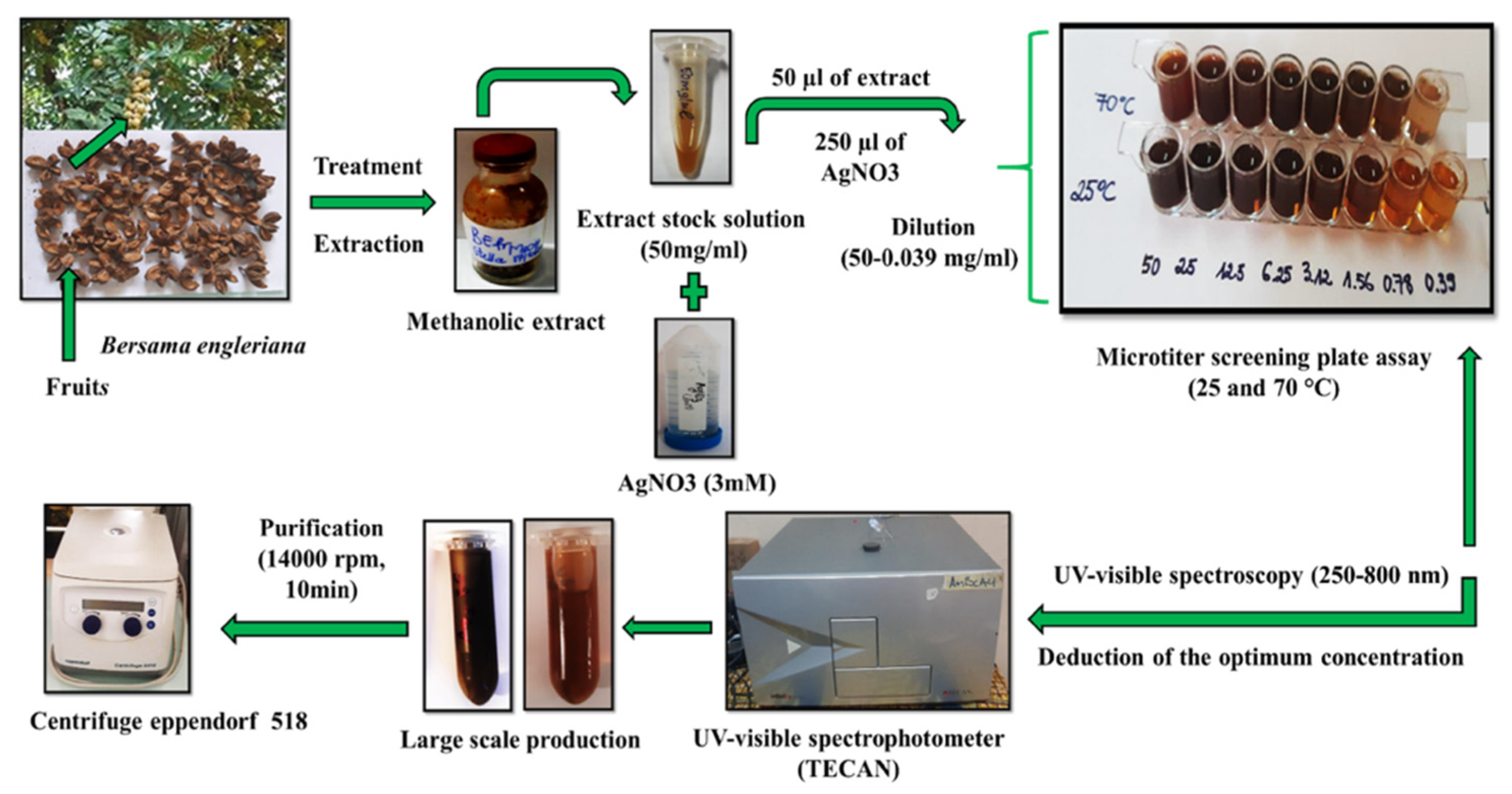
2.1. Green Synthesis and UV–Visible Spectroscopy Evaluation
2.2. Full Characterization of BEfr-SNPs
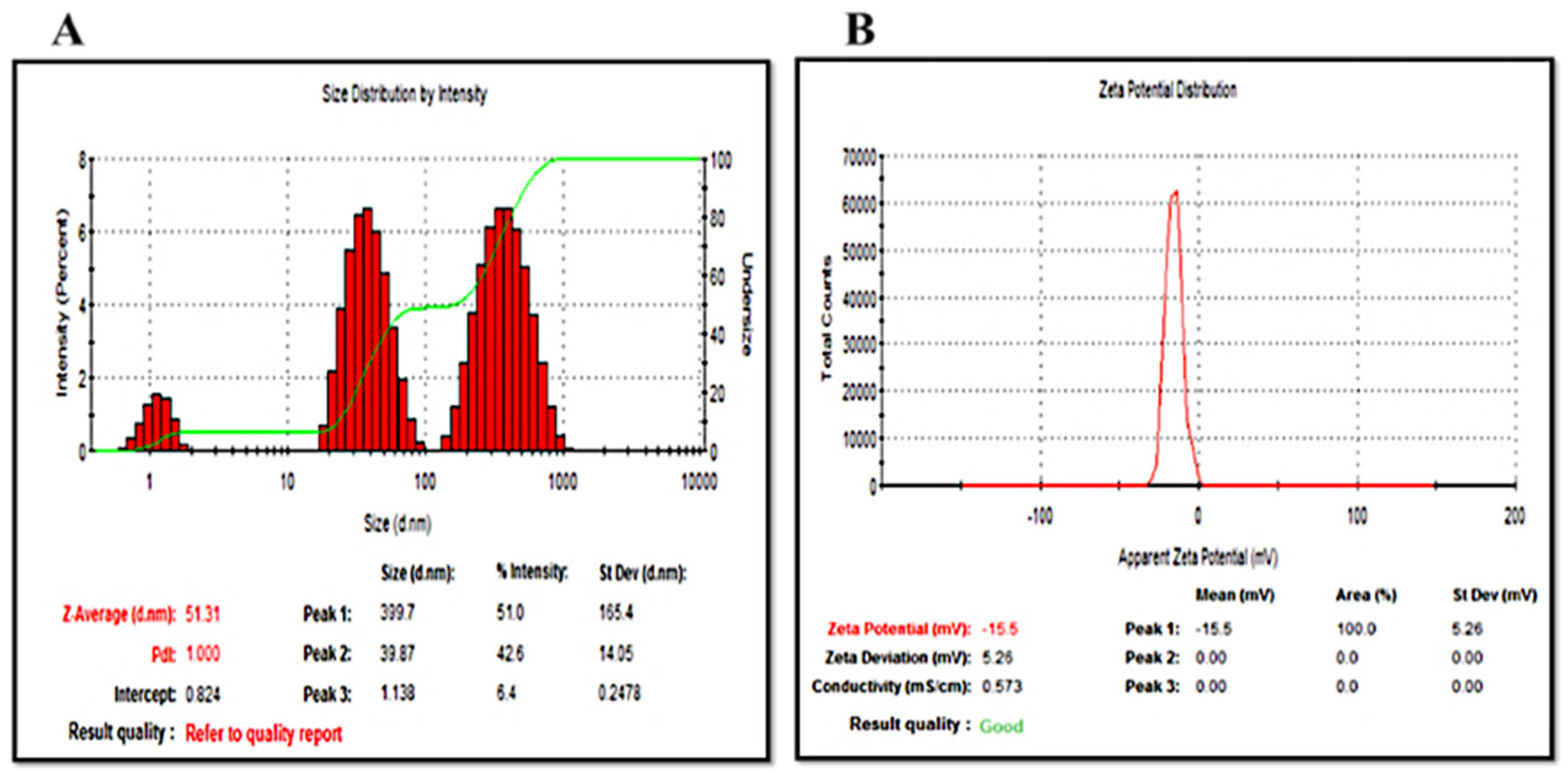
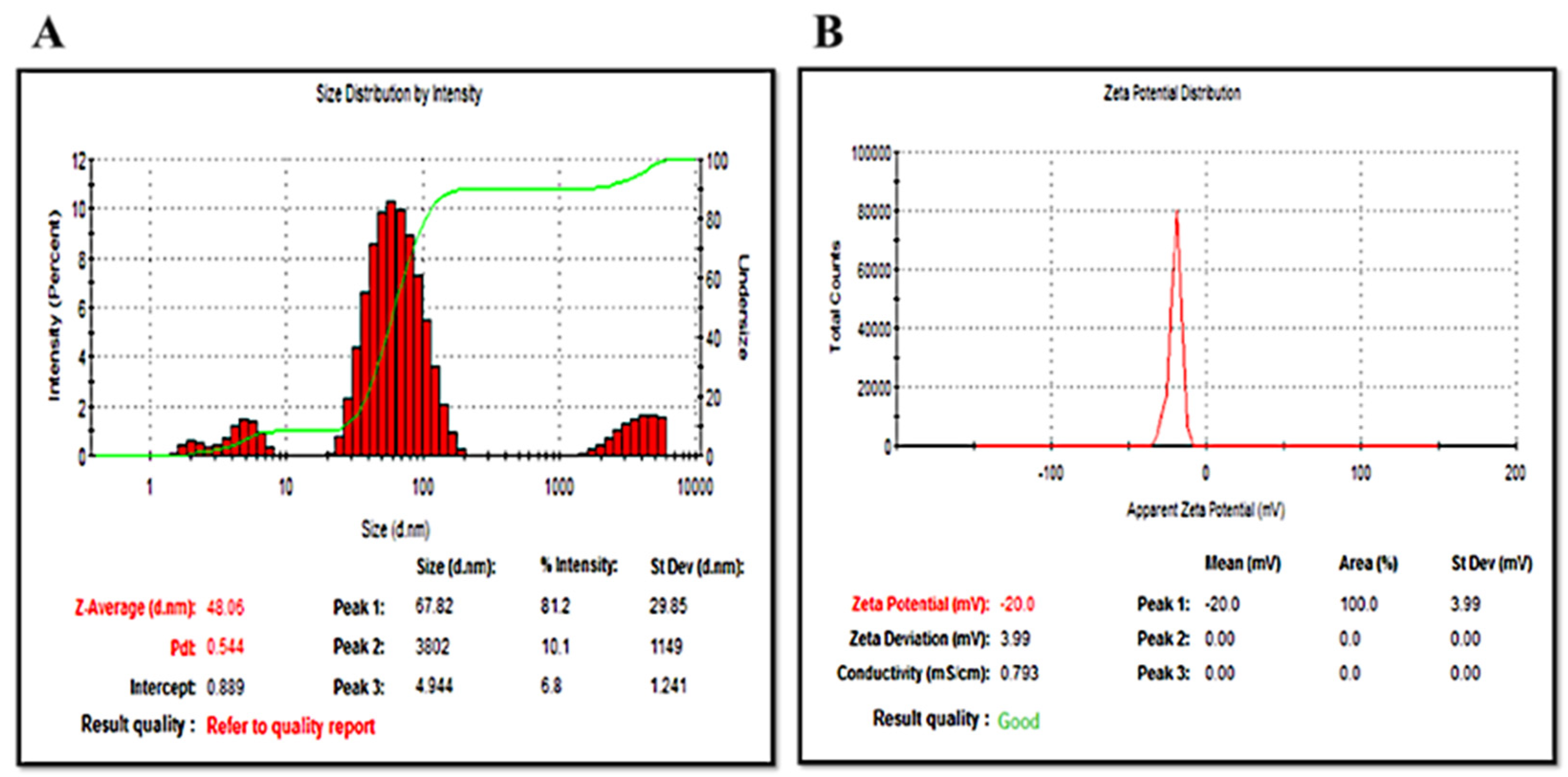
2.2.1. Dynamic Light Scattering (DLS) Evaluation
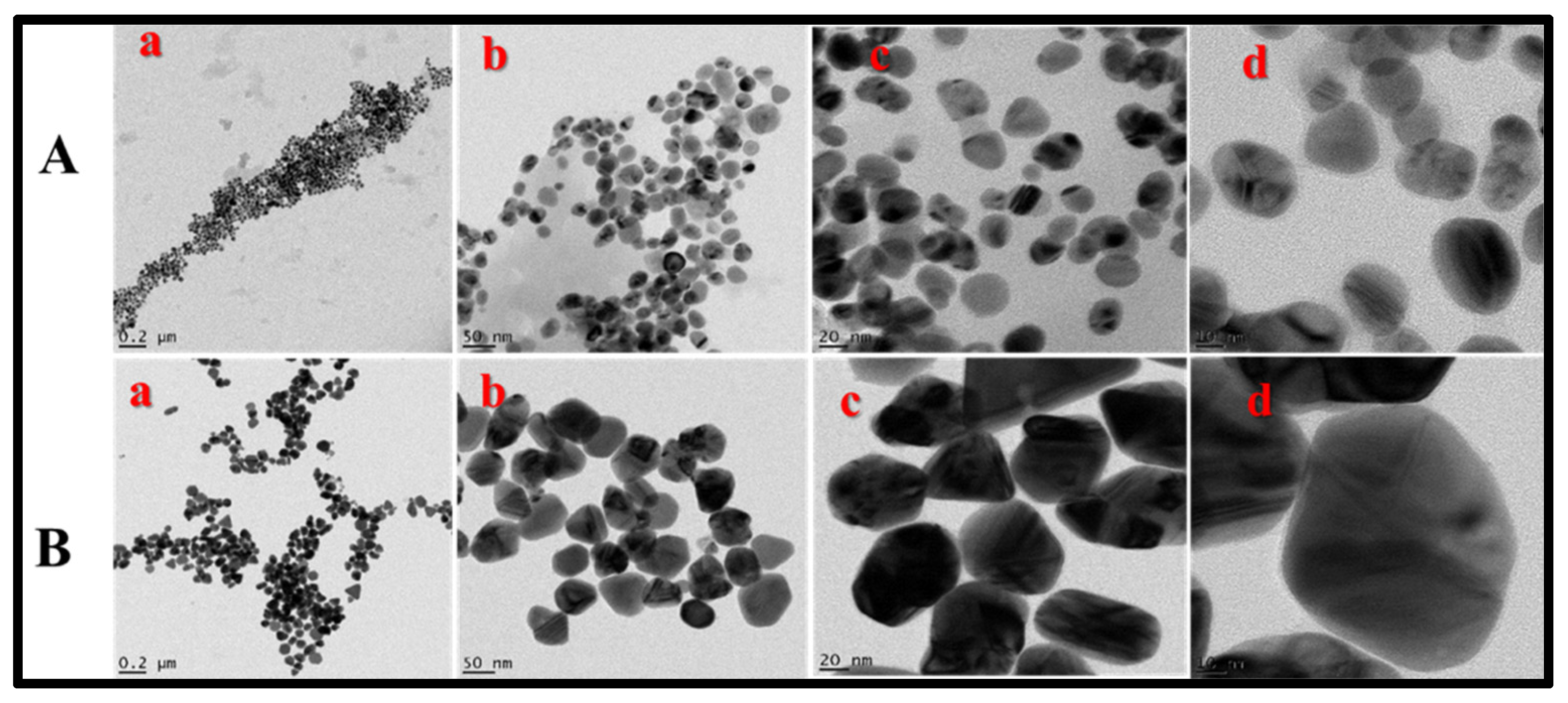
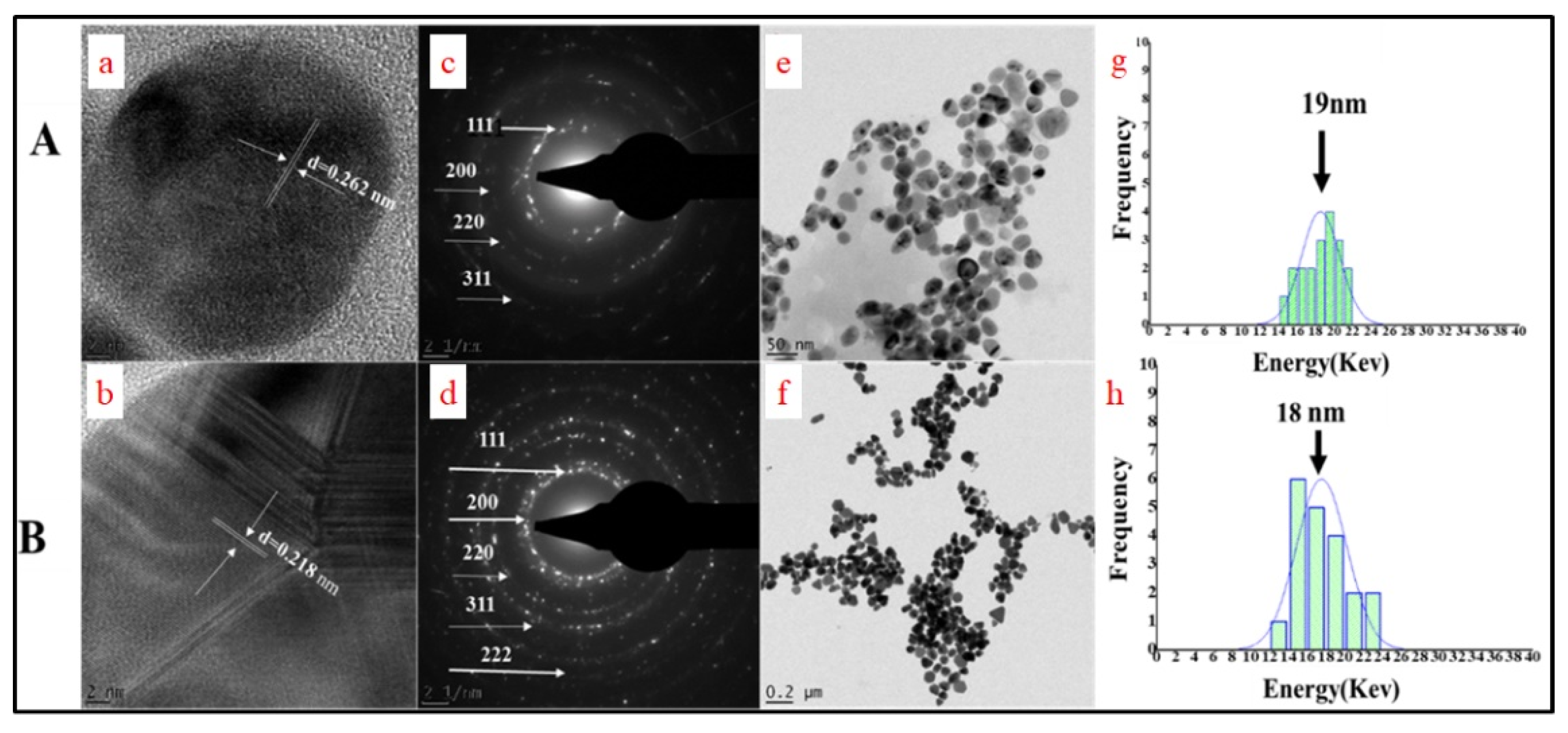
2.2.2. HRTEM Characterization of BEfr-SNPs
2.2.3. Size Distribution Frequency and Selected Area Diffraction Pattern
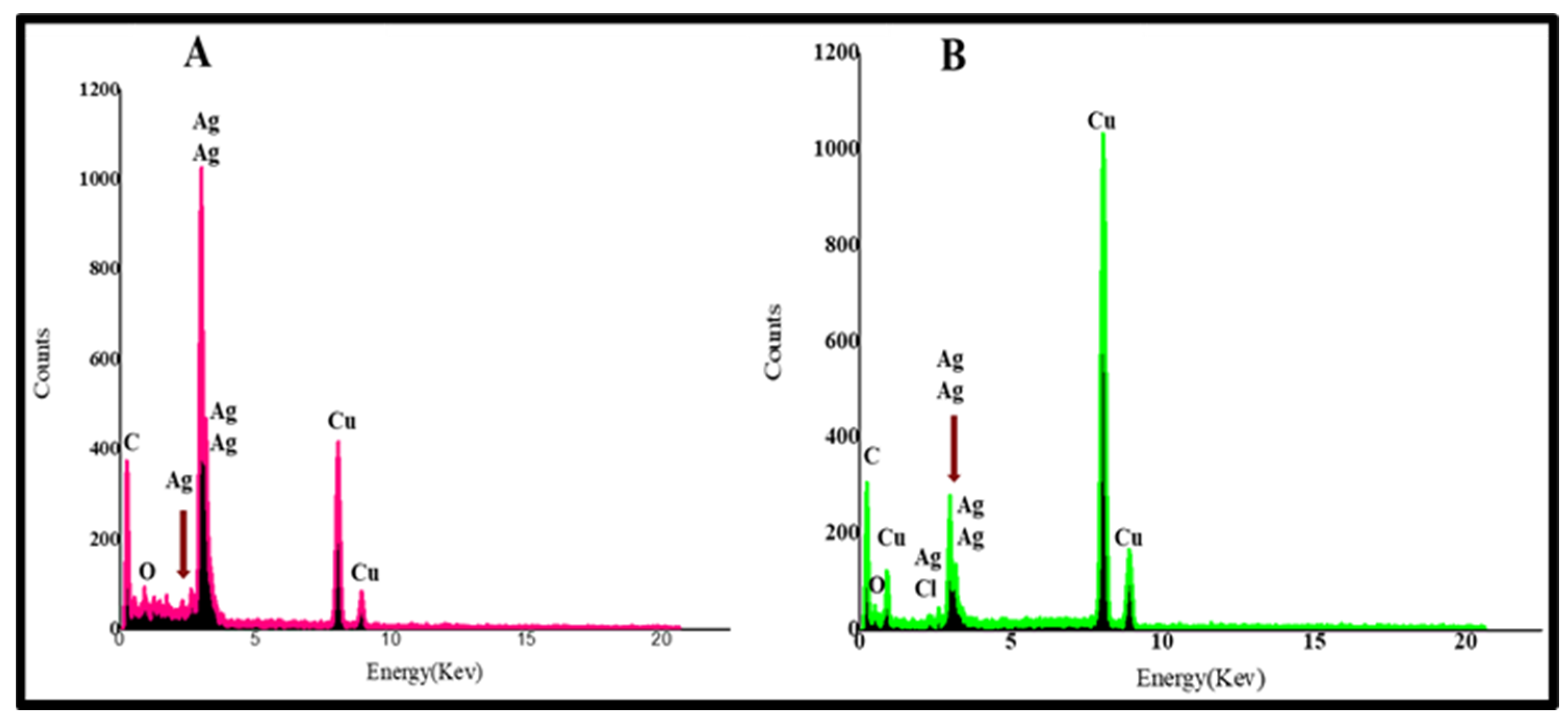
2.2.4. Energy Dispersive X-ray (EDX) Examination of BEfr-SNPs
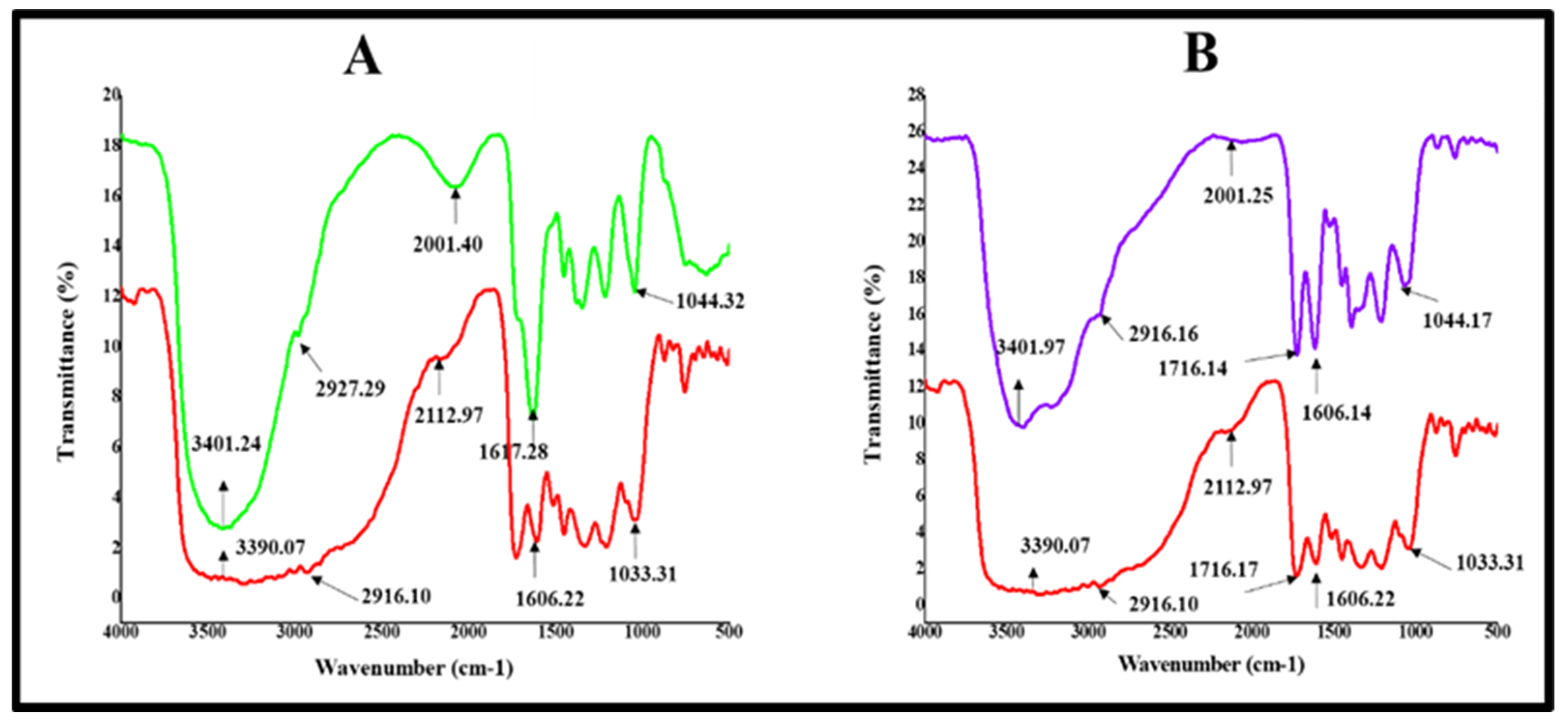
2.2.5. Fourier Transform Infrared Spectroscopy (FTIR) Analysis of BEfr-SNPs
2.3. Antibacterial Potential of Extracts and Derived Biogenic BEfr-SNPs
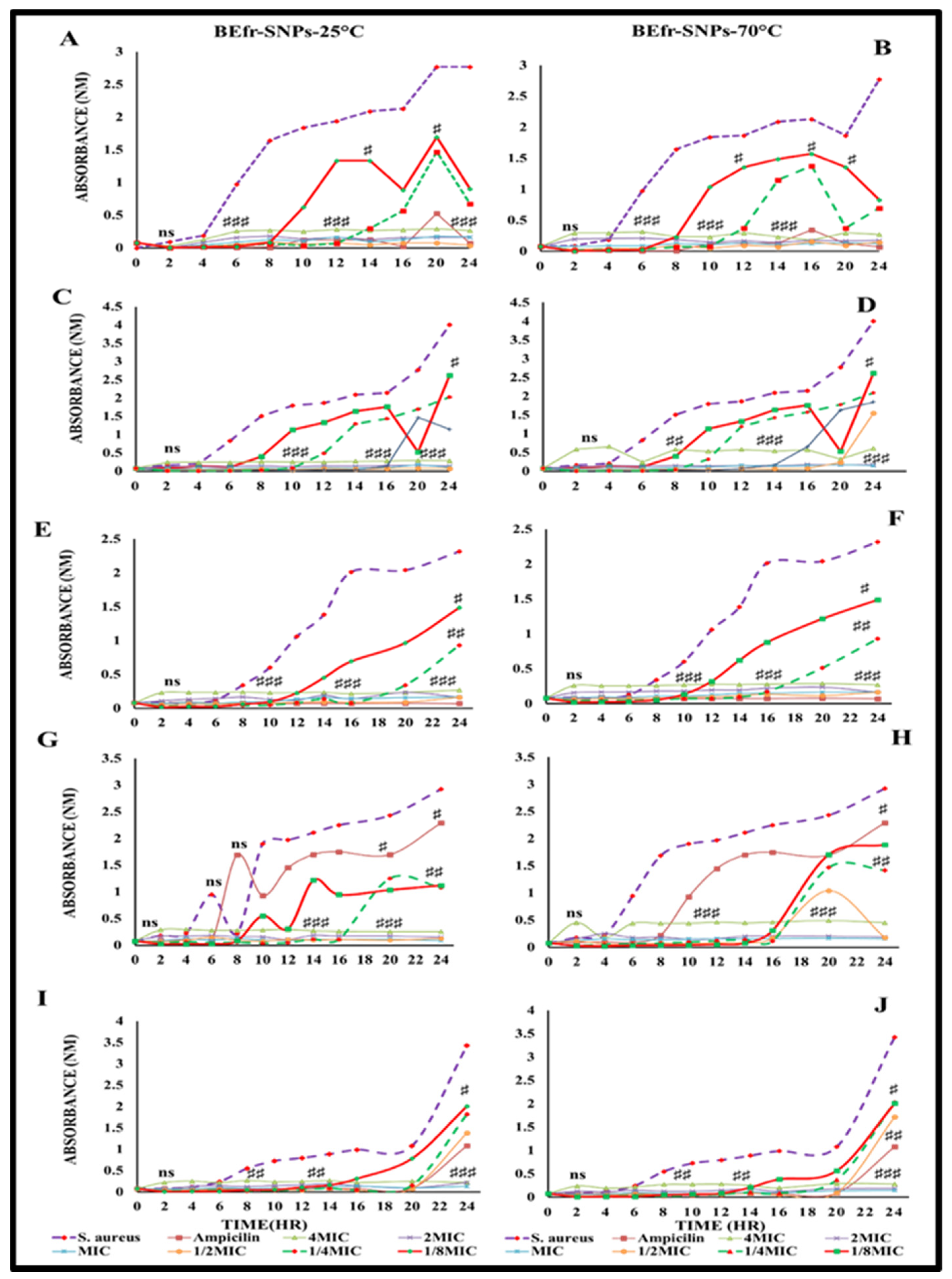
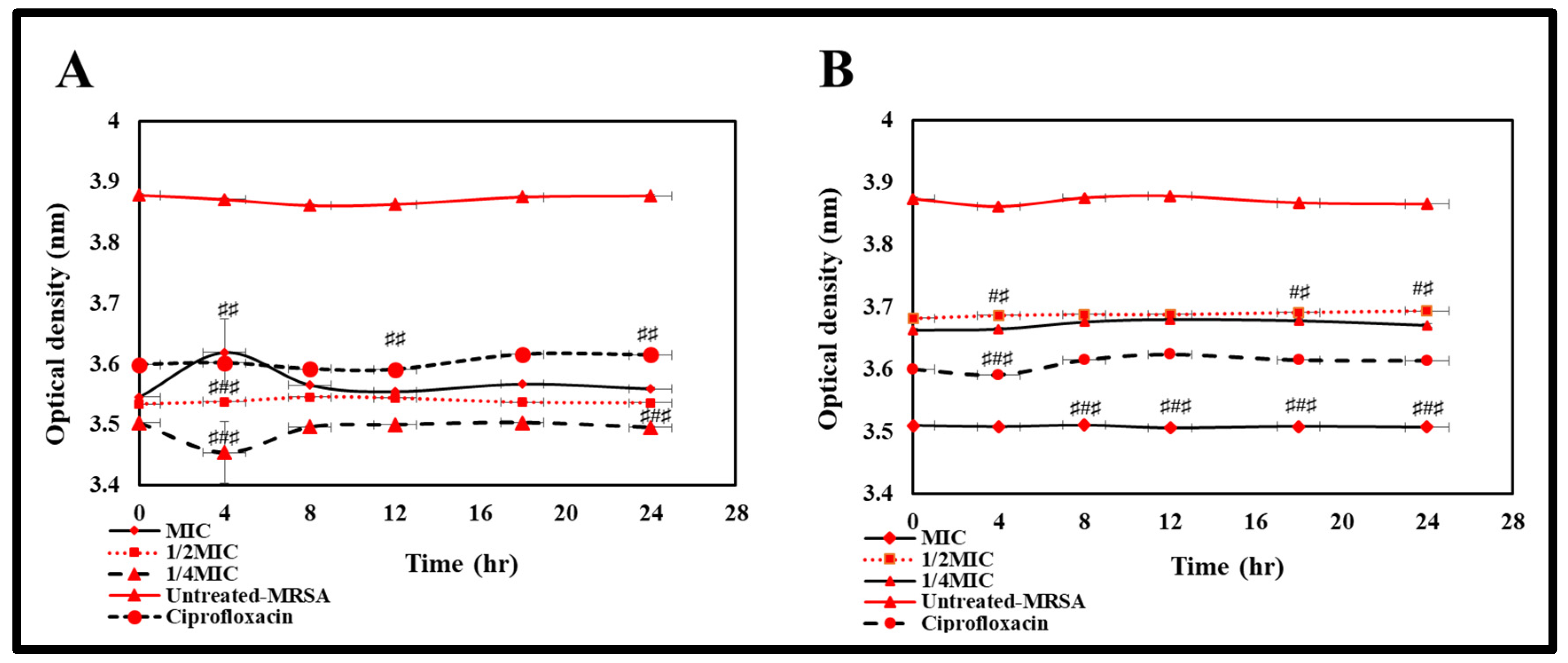
2.4. Evaluate the Time Kinetics Effect of Potential Silver Nanoparticles against the Susceptible Bacteria
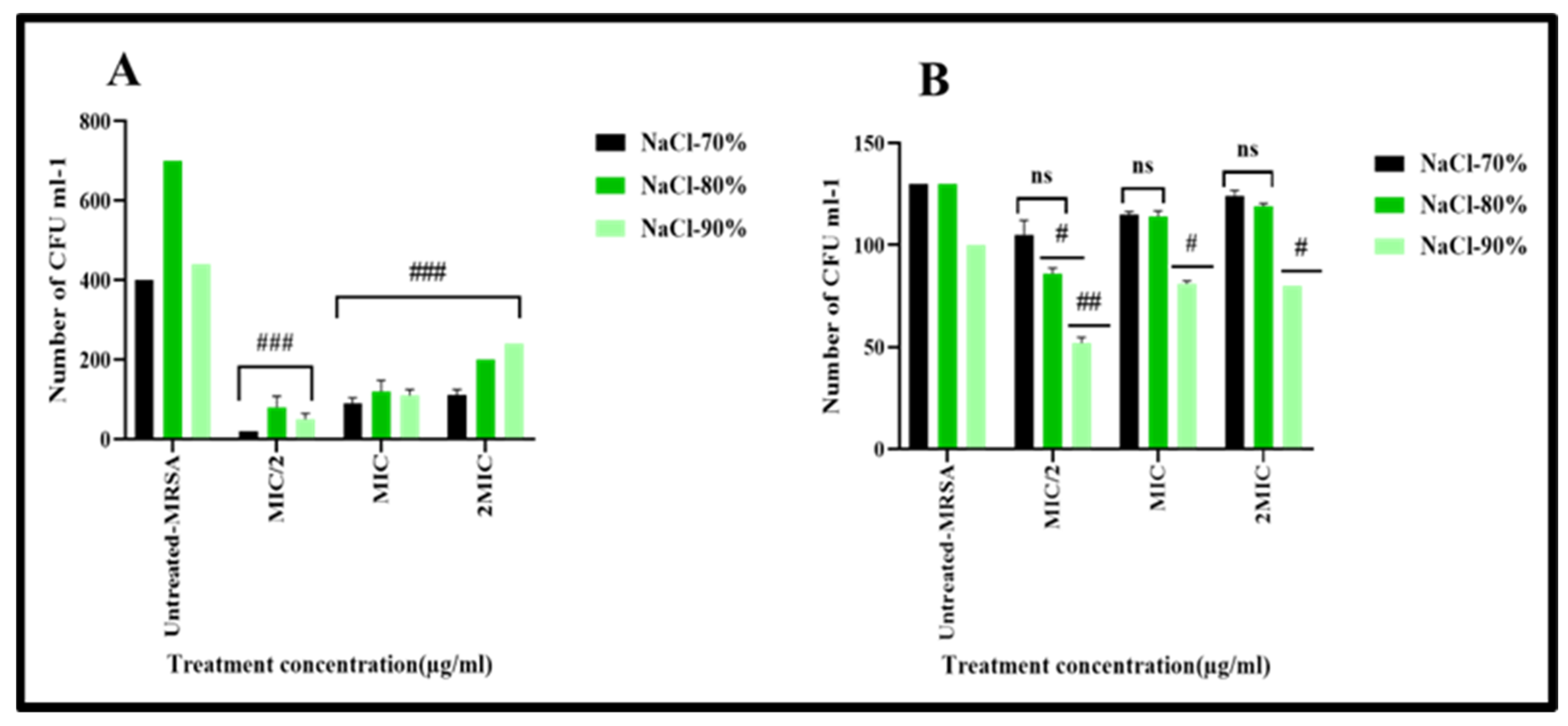
2.5. Evaluate the Effect of Biogenic BEfr-SNPs on Bacteria Cell Membrane of Methicillin-Resistant Staphylococcus aureus
2.6. Determination of the Effect of Bioactive BEfr-SNPs on Nucleotide Leakage
3. Materials and Methods
3.1. Plant Collection and Identification
3.2. Extraction Procedure
3.3. Biological Synthesis and Characterization of BEfr-SNPs from Bersama engleriana
3.4. Characterization of BEfr-SNPs
3.4.1. UV–Vis Spectrophotometry Analysis and Dynamic light Scattering (DLS) Analysis
3.4.2. HRTEM Characterization of BEfr-SNPs
3.4.3. Fourier Transform Infrared Spectroscopy (FTIR) Analysis of BEfr-SNPs
3.5. Assessment of In Vitro Antibacterial Effect of BEfr Extracts and BEfr-SNPs
3.5.1. Bacterial Strains
3.5.2. Antibacterial Screening of Extracts and Derived Silver Nanoparticles from Bersama engleriana
3.5.3. Evaluation of Minimal Inhibitory Concentration (MIC) of BEfr-SNPs
3.5.4. Study of Bacterial Time Kill Growth Inhibition of Active BEfr-SNPs
3.5.5. Analysis of the Effect of Extraction on Membrane Destabilization Using Loss of Salt Tolerance Assay
3.5.6. Analysis of the Effect of Bioactive BEfr-SNPs on Nucleotide Leakage
3.6. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nkengasong, J.N.; Tessema, S.K. Africa Needs a New Public Health Order to Tackle Infectious Disease Threats. Cell 2020, 183, 296–300. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E.; Stájer, A.; Baráth, Z. Antimicrobial Resistance in the Context of the Sustainable Development Goals: A Brief Review. Eur. J. Investig. Health Psychol. Educ. 2021, 11, 71–82. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). National Center for Emerging and Zoonotic Infectious Diseases (NCEZID) Progress Report, Protecting Health in 2020; CDC: Atlanta, GA, USA, 2020.
- Tincho, M.; Morris, T.; Meyer, M.; Pretorius, A. Antibacterial Activity of Rationally Designed Antimicrobial Peptides. Int. J. Microbiol. 2020, 2020, 2131535. [Google Scholar] [CrossRef] [Green Version]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [Green Version]
- Baptista, P.V.; McCusker, M.P.; Carvalho, A.; Ferreira, D.A.; Mohan, N.M.; Martins, M.; Fernanades, A.R. Nano-Strategies to Fight Multidrug Resistant Bacteria—“A Battle of the Titans”. Front. Microbiol. 2018, 9, 1441. [Google Scholar] [CrossRef] [Green Version]
- Majoumouo, M.S.; Sibuyi, N.R.S.; Tincho, M.B.; Mbekou, M.; Boyom, F.F.; Meyer, M. Enhanced Anti-Bacterial Activity of Biogenic Silver Nanoparticles Synthesized from Terminalia mantaly Extracts. Int. J. Nanomed. 2019, 14, 9031–9046. [Google Scholar] [CrossRef] [Green Version]
- Shehabeldine, A.M.; Elbahnasawy, M.A.; Hasaballah, A.I. Green Phytosynthesis of Silver Nanoparticles Using Echinochloa stagnina Extract with Reference to Their Antibacterial, Cytotoxic, and Larvicidal Activities. BioNanoScience 2021, 11, 526–538. [Google Scholar] [CrossRef]
- Singh, P.; Garg, A.; Pandit, S.; Mokkapati, V.R.S.S.; Mijakovic, I. Antimicrobial Effects of Biogenic Nanoparticles. Nanomaterials 2018, 8, 1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Backx, B.P.; dos Santos, M.S.; dos Santos, O.A.; Filho, S.A. The Role of Biosynthesized Silver Nanoparticles in Antimicrobial Mechanisms. Curr. Pharm. Biotechnol. 2021, 22, 762–772. [Google Scholar] [CrossRef]
- Shankar, S.S.; Rai, A.; Ankamwar, B.; Singh, A.; Ahmad, A.; Sastry, M. Biological synthesis of triangular gold nanoprisms. Nat. Mater. 2004, 3, 482–488. [Google Scholar] [CrossRef]
- Singh, P.; Ahn, S.; Kang, J.-P.; Veronika, S.; Huo, Y.; Singh, H.; Chokkaligam, M.; Farh, M.E.-A.; Aceituno, V.C.; Kim, Y.J.; et al. In vitro anti-inflammatory activity of spherical silver nanoparticles and monodisperse hexagonal gold nanoparticles by fruit extract of Prunus serrulata: A green synthetic approach. Artif. Cells Nanomed. Biotechnol. 2018, 46, 2022–2032. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Jain, D.; Upadhyay, M.K.; Khandelwal, N.; Verma, H.N. Green Synthesis of Silver Nanoparticles Using Argemone Mexicana Leaf Extract and Evaluation of Their Anti-microbial Activities. Dig. J. Nanomater. Biostruct. 2010, 5, 483–489. [Google Scholar]
- Anuj, S.A.; Gajera, H.; Hirpara, D.G.; Golakiya, B.A. Bacterial membrane destabilization with cationic particles of nano-silver to combat efflux-mediated antibiotic resistance in Gram-negative bacteria. Life Sci. 2019, 230, 178–187. [Google Scholar] [CrossRef]
- Boateng, J.; Catanzano, O. Silver and Silver Nanoparticle-Based Antimicrobial Dressings. Therapeutic Dressings and Wound Healing Applications; John Wiley and Sons: Hoboken, NJ, USA, 2020; Chapter 8; pp. 157–184. [Google Scholar] [CrossRef] [Green Version]
- Burdușel, A.-C.; Gherasim, O.; Grumezescu, A.M.; Mogoantă, L.; Ficai, A.; Andronescu, E. Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview. Nanomaterials 2018, 8, 681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, G.; Shin, H.-S.; Kumar, A.; Vishnuprasad, C.N.; Patra, J.K. Photo-mediated optimized synthesis of silver nanoparticles using the extracts of outer shell fibre of Cocos nucifera L. fruit and detection of its antioxidant, cytotoxicity and antibacterial potential. Saudi J. Biol. Sci. 2020, 28, 980–987. [Google Scholar] [CrossRef] [PubMed]
- Dakshayani, S.S.; Marulasiddeshwara, M.B.; Sharath Kumar, M.N.; Ramesh, G.; Kumar, R.; Devaraja, S.; Hosamani, R. Antimicrobial, anticoagulant and antiplatelet activities of green synthesized silver nanoparticles using Selaginella (Sanjeevini) plant extract. Int. J. Biol. Macromol. 2019, 131, 787–797. [Google Scholar]
- Sengupta, A.; Sarkar, A. Synthesis and characterization of nanoparticles from neem leaves and banana peels: A green prospect for dye degradation in wastewater. Ecotoxicology 2021, 31, 537–548. [Google Scholar] [CrossRef]
- Hiemenz, P.C.; Rajagopalan, R. Principles of Colloid and Surface Chemistry, Revised and Expanded; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- El-Desouky, N.; Shoueir, K.R.; El-Mehasseb, I.; El-Kemary, M. Bio-inspired green manufacturing of plasmonic silver nanoparticles/Degussa using Banana Waste Peduncles: Photocatalytic, antimicrobial, and cytotoxicity evaluation. J. Mater. Res. Technol. 2020, 10, 671–686. [Google Scholar] [CrossRef]
- Ahmad, A.; Syed, F.; Shah, A.; Khan, Z.; Tahir, K.; Khan, A.U.; Yuan, Q. Silver and gold nanoparticles from Sargentodoxa cuneata: Synthesis, characterization and antileishmanial activity. RSC Adv. 2015, 5, 73793–73806. [Google Scholar] [CrossRef]
- Kumar, K.M.; Sinha, M.; Mandal, B.K.; Ghosh, A.R.; Kumar, K.S.; Reddy, P.S. Green synthesis of silver nanoparticles using Terminalia chebula extract at room temperature and their antimicrobial studies. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 91, 228–233. [Google Scholar] [CrossRef]
- Bagherzade, G.; Tavakoli, M.M.; Namaei, M.H. Green synthesis of silver nanoparticles using aqueous extract of saffron (Crocus sativus L.) wastages and its antibacterial activity against six bacteria. Asian Pac. J. Trop. Biomed. 2017, 7, 227–233. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, Q.; Wu, F.-G.; Dai, Y.; Chen, X. Polyphenol-Containing Nanoparticles: Synthesis, Properties, and Therapeutic Delivery. Adv. Mater. 2021, 33, 2007356. [Google Scholar] [CrossRef] [PubMed]
- Galvez, A.M.; Ramos, K.M.; Teja, A.J.; Baculi, R. Bacterial Exopolysaccharide-Mediated Synthesis of Silver Nanoparticles and Their Application on Bacterial Biofilms. J. Microbiol. Biotechnol. Food Sci. 2019, 8, 970–978. [Google Scholar] [CrossRef] [Green Version]
- Kuete, V.; Mbaveng, A.T.; Tsaffack, M.; Beng, V.P.; Etoa, F.-X.; Nkengfack, A.E.; Meyer, J.M.; Lall, N. Antitumor, antioxidant and antimicrobial activities of Bersama engleriana (Melianthaceae). J. Ethnopharmacol. 2008, 115, 494–501. [Google Scholar] [CrossRef]
- Okaiyeto, K.; Hoppe, H.; Okoh, A.I. Plant-Based Synthesis of Silver Nanoparticles Using Aqueous Leaf Extract of Salvia officinalis: Characterization and its Antiplasmodial Activity. J. Clust. Sci. 2020, 32, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Emani, S.; Gunjiganur, G.V.; Mehta, D.S. Determination of the antibacterial activity of simvastatin against periodontal pathogens, Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans: An in vitro study. Contemp. Clin. Dent. 2014, 5, 377–382. [Google Scholar] [CrossRef]
- Collins, T.L.; Markus, E.A.; Hassett, D.J.; Robinson, J.B. The Effect of a Cationic Porphyrin on Pseudomonas aeruginosa Biofilms. Curr. Microbiol. 2010, 61, 411–416. [Google Scholar] [CrossRef]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef] [Green Version]
- Van De Vel, E.; Sampers, I.; Raes, K. A review on influencing factors on the minimum inhibitory concentration of essential oils. Crit. Rev. Food Sci. Nutr. 2017, 59, 357–378. [Google Scholar] [CrossRef]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.-H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.-Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Chung, I.-M.; Park, I.; Seung-Hyun, K.; Thiruvengadam, M.; Rajakumar, G. Plant-Mediated Synthesis of Silver Nanoparticles: Their Characteristic Properties and Therapeutic Applications. Nanoscale Res. Lett. 2016, 11, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumarasinghe, K.G.U.R.; Silva, W.C.H.; Fernando, M.D.A.; Palliyaguru, L.; Jayawardena, P.S.; Shimomura, M.; Fernando, S.S.N.; Gunasekara, T.D.C.P.; Jayaweera, P.M. One-Pot Reducing Agent-Free Synthesis of Silver Nanoparticles/Nitrocellulose Composite Surface Coating with Antimicrobial and Antibiofilm Activities. BioMed Res. Int. 2021, 2021, 6666642. [Google Scholar] [CrossRef] [PubMed]
- Permana, A.D.; Anjani, Q.K.; Sartini; Utomo, E.; Volpe-Zanutto, F.; Paredes, A.J.; Evary, Y.M.; Mardikasari, S.A.; Pratama, M.R.; Tuany, I.N.; et al. Selective delivery of silver nanoparticles for improved treatment of biofilm skin infection using bacte-ria-responsive microparticles loaded into dissolving microneedles. Mater. Sci. Eng. C 2021, 120, 111786. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [Green Version]
- Kvitek, D.J.; Will, J.L.; Gasch, A.P. Variations in Stress Sensitivity and Genomic Expression in Diverse S. cerevisiae Isolates. PLoS Genet. 2008, 4, e1000223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aljabali, A.A.A.; Akkam, Y.; Al Zoubi, M.S.; Al-Batayneh, K.M.; Al-Trad, B.; Alrob, O.A.; Alkilany, A.M.; Benamara, M.; Evans, D.J. Synthesis of Gold Nanoparticles Using Leaf Extract of Ziziphus zizyphus and their Antimicrobial Activity. Nanomaterials 2018, 8, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labruère, R.; Sona, A.J.; Turos, E. Anti–Methicillin-Resistant Staphylococcus aureus Nanoantibiotics. Front. Pharmacol. 2019, 10, 1121. [Google Scholar] [CrossRef]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The Effect of Charge at the Surface of Silver Nanoparticles on Antimicrobial Activity against Gram-Positive and Gram-Negative Bacteria: A Preliminary Study. J. Nanomater. 2015, 2015, 53. [Google Scholar] [CrossRef] [Green Version]
- Nalwade, A.R.; Jadhav, A.A. Biosynthesis of silver nanoparticles using leaf extract of Daturaalba Nees. and evaluation of their antibacterial activity. Arch. Appl. Sci. Res. 2013, 5, 45–49. [Google Scholar]
- Bapat, R.A.; Chaubal, T.V.; Joshi, C.P.; Bapat, P.R.; Choudhury, H.; Pandey, M.; Gorain, B.; Kesharwani, P. An overview of application of silver nanoparticles for biomaterials in dentistry. Mater. Sci. Eng. C 2018, 91, 881–898. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The Antibacterial Mechanism of Silver Nanoparticles and Its Application in Dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majoumouo, M.S.; Tincho, M.B.; Morris, T.; Hiss, D.C.; Boyom, F.F.; Mandal, C. Antiproliferative potential of methanolic and aqueous extracts and their methanolic fractions derived from fruits of Bersama engleriana against a panel of four cancer cell lines. Cogent Biol. 2020, 6, 1727636. [Google Scholar] [CrossRef]
- Song, H.Y.; Ko, K.K.; Oh, I.H.; Lee, B.T. Fabrication of Silver Nanoparticles and Their Antimicrobial Mechanisms. Eur. Cells Mater. 2006, 11, 58–59. [Google Scholar]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions onEscherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Kumar, N.; Das, S.; Jyoti, A.; Kaushik, S. Synergistic effect of silver nanoparticles with doxycycline against Klebsiella pneumoniae. Int. J. Pharm. Pharm. Sci. 2016, 8, 183–186. [Google Scholar]
- Majoumouo, M.S.; Sharma, J.R.; Sibuyi, N.R.S.; Tincho, M.B.; Boyom, F.F.; Meyer, M. Synthesis of Biogenic Gold Nanoparticles from Terminalia mantaly Extracts and the Evaluation of Their In Vitro Cytotoxic Effects in Cancer Cells. Molecules 2020, 25, 4469. [Google Scholar] [CrossRef]
- Clinical Laboratory Standard Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard-Ninth Edition Document, Replaces M07-A8; Clinical Laboratory Standard Institute: Wayne, PA, USA, 2012; Volume 29, p. 2. [Google Scholar]
- Ho, P.L.; Cao, L.; Dai, C.; Li, Z.; Fan, Z.; Song, Y.; Wu, Y.; Cao, Z.; Li, W. Antibacterial Activity and Mechanism of a Scorpion Venom Peptide Derivative In Vitro and In Vivo. PLoS ONE 2012, 7, e40135. [Google Scholar]
- Etame, R.E.; Mouokeu, R.S.; Pouaha, C.L.C.; Kenfack, I.V.; Tchientcheu, R.; Assam, J.P.A.; Poundeu, F.S.M.; Tiabou, A.T.; Etoa, F.X.; Kuiate, J.R.; et al. Effect of Fractioning on Antibacterial Activity of Enantia chlorantha Oliver (Annonaceae) Methanol Extract and Mode of Action. Evid. Based Complement. Altern. Med. 2018, 2018, 4831593. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, D.M.; Melo, F.G.; Balogun, S.O.; Flach, A.; de Souza, E.C.A.; de Souza, G.P.; Rocha, I.D.N.A.; da Costa, L.A.M.A.; Soares, I.M.; da Silva, L.I.; et al. Antibacterial mode of action of the hydroethanolic extract of Leonotis nepetifolia (L.) R. Br. involves bacterial membrane perturbations. J. Ethnopharmacol. 2015, 172, 356–363. [Google Scholar] [CrossRef]
| Codes | Dynamic Light Scattering Parameters | |||||
|---|---|---|---|---|---|---|
| Optimum Concentration (mg/mL) | λmax (nm) | Average Diameter (nm) | Average Zeta Potential (mV) | Deviation/Conductivity (ms/cm) | Pdi | |
| BeFr-SNPs-25 °C | 0.39 | 434.08 | 51.31 | −17.30 ± 1.28 | 5.26/0.573 | 0.480 |
| BeFr-SNP-70 °C | 0.78 | 430.18 | 48.06 | −20.83 ± 0.59 | 7.40/10.788 | 0.665 |
| Peak Position in BEfr) (cm−1) | Peak Position in SNPs at 25 °C (cm−1) | Shift in Position (cm−1) | Peak Position in SNPs at 70 °C (cm−1) | Shift in Position (cm−1) | Type of Chemicals Groups |
|---|---|---|---|---|---|
| 1033.31 | 1044.32 | +11.03 | 1044.17 | +10.86 | C-O carboxylic acids, esters, ethers |
| 1606.22 | 1617.28 | +11.06 | 1606.14 | −0.08 | –C=C–stretch alkenes |
| 1716.17 | -------------- | -------- | 1716.14 | −0.03 | C=O Anhydrides |
| 2916.10 | 2927.29 | +11.19 | 2916.16 | +0.06 | H–C=O: C–H stretch aldehydes |
| 3390.07 | 3401.24 | +11.17 | 3401.97 | +11.9 | O-H, Alcohol, phenol |
| Extracts | Microorganism Acronyms | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | |
| BEfrAQ | 500 | 500 | 500 | 500 | 500 | 500 | 500 | 500 |
| BEfrMeOH | 500 | 500 | 500 | 500 | 500 | 500 | 125 | 125 |
| BEfeAQ | 500 | 500 | 500 | 500 | 500 | 500 | 500 | >500 |
| BEfeMeOH | 500 | 500 | 500 | 500 | 500 | 500 | 500 | >500 |
| BEeAQ | >500 | >500 | >500 | >500 | >500 | >500 | >500 | >500 |
| BEeMeOH | 500 | >500 | 500 | 500 | 500 | 500 | 500 | 500 |
| Ampicilin | 62.5 | >62.5 | >62.5 | >62.5 | 15.625 | 62.5 | 3.901 | 7.8125 |
| Acronym | MIC (µg/mL) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacterial Strains | |||||||||||||
| A | B | C | D | E | F | G | H | I | K | L | M | N | |
| BEfrMeOH | 500 | 500 | 500 | 500 | 500 | >500 | 125 | 125 | >500 | >500 | >500 | >500 | >500 |
| BEfr-SNPs-25 °C | >50 | >50 | >50 | >50 | >50 | 0.3906 | 6.25 | 0.7815 | 0.7812 | 0.3906 | 0.3906 | >50 | 0.3906 |
| BEfr-SNPs-70 °C | > 50 | > 50 | > 50 | > 50 | > 50 | 3.125 | > 50 | 6.25 | 3.125 | 1.5612 | 3.125 | > 50 | >50 |
| Ampicilin | 62.5 | >62.5 | >62.5 | >62.5 | 15.625 | 62.5 | 3.901 | 7.8125 | 7.8125 | 7.8125 | 3.9062 | >62.5 | 62.50 |
| Ciprofloxacin | 0.468 | 0.468 | 0.234 | 0.468 | 0.234 | 0.234 | 0.234 | 0.234 | 0.234 | 0.468 | 0.234 | 0.468 | 0.468 |
| Bacterial Strains | Acronym | Reference No. | Supplier |
|---|---|---|---|
| Pseudomonas aeruginosa | P. aeruginosa | NR-48982 | BEI resources |
| Streptococcus pneumonia | S. pneumonia | ATCC 49619 | ATCC |
| Salmonella enterica typhimurium | S. enterica | NR-13555 | BEI resources |
| Klebsiella pneumoniae | K. pneumoniae | ATCC 700603 | ATCC |
| Staphylococcus aureus | S. aureus | ATCC 43300 | ATCC |
| Eschericha coli | E. coli | ATCC 25922 | ATCC |
| Staphylococcus aureus | S. aureus | NR-45003 | BEI resources |
| Salmonela enterica | S. enterica | NR-4311 | BEI resources |
| Methicilin resistant Staphylococcus aureus | MRS. aureus | ATCC 33591 | ATCC |
| Klebsiella pneumoniae | K. pneumoniae | ATCC 13883 | ATCC |
| Staphylococcusaureus | S. aureus | NR-48374 | BEI resources |
| Staphylococcus epidermidis | S. epidermidis | ATCC 12228 | ATCC |
| Streptococcus pyogenes | S. pyogenes | ATCC 19615 | ATCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majoumouo, M.S.; Tincho, M.B.; Yimta, Y.D.; Adekiya, T.A.; Aruleba, R.T.; Ayawei, N.; Boyom, F.F.; Morris, T. Biosynthesis of Silver Nanoparticles Using Bersama engleriana Fruits Extracts and Their Potential Inhibitory Effect on Resistant Bacteria. Crystals 2022, 12, 1010. https://doi.org/10.3390/cryst12071010
Majoumouo MS, Tincho MB, Yimta YD, Adekiya TA, Aruleba RT, Ayawei N, Boyom FF, Morris T. Biosynthesis of Silver Nanoparticles Using Bersama engleriana Fruits Extracts and Their Potential Inhibitory Effect on Resistant Bacteria. Crystals. 2022; 12(7):1010. https://doi.org/10.3390/cryst12071010
Chicago/Turabian StyleMajoumouo, Michele Stella, Marius Belmondo Tincho, Youmbi Diane Yimta, Tayo Alex Adekiya, Raphael Taiwo Aruleba, Nimibofa Ayawei, Fabrice Fekam Boyom, and Thureyah Morris. 2022. "Biosynthesis of Silver Nanoparticles Using Bersama engleriana Fruits Extracts and Their Potential Inhibitory Effect on Resistant Bacteria" Crystals 12, no. 7: 1010. https://doi.org/10.3390/cryst12071010
APA StyleMajoumouo, M. S., Tincho, M. B., Yimta, Y. D., Adekiya, T. A., Aruleba, R. T., Ayawei, N., Boyom, F. F., & Morris, T. (2022). Biosynthesis of Silver Nanoparticles Using Bersama engleriana Fruits Extracts and Their Potential Inhibitory Effect on Resistant Bacteria. Crystals, 12(7), 1010. https://doi.org/10.3390/cryst12071010







