Abstract
Three novel Ni complexes with the Schiff base ligand 2-methoxy-6-(E-2-pyridyliminomethyl)-phenol (L1) are described. In comparison with the similar 2-(pyridine-2-ylimino-methyl)phenol (pymp), the mode of coordination of L1 is altered due to the presence of methoxy substituent introducing the sterical hindrance. During the synthesis of the complexes, partial hydrolysis of the ligand was observed. Since such immediate hydrolysis of L1 was not detected during the reactions of Zn(II) and Cu(II) salts with L1, the DFT calculated structures of a series of similar Zn, Cu, Ni, and Co complexes with L1 are compared to account for the phenomenon.
1. Introduction
The first compounds that belong to imines or Schiff bases were synthesized by Hugo Schiff in 1864 [1]. They have now found a broad range of applications, e.g., as reagents in organic synthesis, chemosensors, dyes, and pigments, etc. Thanks to flexible structures and a variable number and character of donor atoms, Schiff bases have the ability to coordinate a wide range of metal ions including both main group and transition metals as well as lanthanides and actinides. The complexes were reported to possess electroluminescent, fluorescent and nonlinear optical properties, and biological activities (e.g., antiviral, antibacterial, antifungal, etc.) [2]. Reviews on the Schiff bases and their complexes are numerous and they usually focus on certain classes of Schiff bases such as derivatives of specific compounds, e.g., 4-aminoantipyrine [3] or equally specific applications such as cell imaging [4]. Compounds with certain coordination properties are often selected such as ligands of definite denticity [2].
We undertook a study of the coordination properties of relatively unexplored o-vanillin derivatives (Scheme 1). In 2019, we described their coordination properties towards copper(II) and zinc(II) as well as magnetic and catalytic properties of the obtained copper(II) complexes [5] and now, encouraged by potential catalytic properties [5,6,7,8], we initiated studies on the reactions of L1 with nickel(II) salts. Knowing that application of similar hemisalen ligands (Scheme 1) returned a variety of coordination modes with a wide choice of metal ions including nickel(II) [5,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26], we assumed that the synthesis would be simple and efficient. In this paper we describe the crystal structures of the obtained discrete and polynuclear products and unexpected synthetic problems that influenced the yield of the seemingly simple reaction.
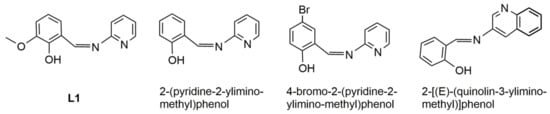
Scheme 1.
Formula of ligand L1 [5,27] and selected similar hemisalen ligands: 2-(pyridine-2-ylimino-methyl)phenol [9,10,11,12,13,14,15,16,17,19,23,24,25]; 4-bromo-2-(pyridine-2-yliminomethyl)phenol [21,22]; and 2-[(E)-(quinolin-3-yliminomethyl)]phenol [7].
2. Materials and Methods
2.1. Syntheses
Substrates: 2-hydroxy-3-methoxybenzaldehyde (o-vanillin, OV), Ni(NO3)2·6H2O, and aminopyridines (APs) were purchased from commercial sources. L1 was synthesized as described [27].
2.1.1. Tetranuclear Complex 1
The solution of L1 (0.114 g, 0.50 mmol) in 3.5 mL of methanol or ethanol was slowly added to the solution of Ni(NO3)2·6H2O (0.073 g, 0.25 mmol) in methanol (4.0 mL). Then, Et3N (0.15 mL, 1.00 mmol) was added to the reaction mixture. After 30 min, a solution of 2-AP (0.046 g, 0.50 mmol) in methanol (2 mL) was added. All operations were carried out at RT. Subsequently 15 min later, 100 µL H2O was introduced into the reaction mixture. Crystals for X-ray diffraction analysis were obtained at RT as a first crop of crystals appeared during the slow evaporation of the solvent. The synthesis was repeated at least five times to collect more crystals. Yield of a single attempt: 0.024 g (5.3%). Elemental analysis: anal. calcd. for C78H78N14Ni4O24 (we assumed the presence of the cluster (NO3)2(H2O)4 accompanying complex cation): C 51.18; H 4.30; and N 10.71, anal. found C 50.48; H: 4.35; and N: 10.79.
This procedure was also repeated several times, with the change of 2-AP to: 3-aminopyridine, 3-(aminomethyl)pyridine, pyridine, 2-hydroxy-6-methylpyridine, pyrazole, 2-methylimidazole, 4(5)-methylimidazole, 2-ethylimidazole, and diantipyrylmethane. Only in the case of pyridine did we obtain a product in which pyridine would partially replace the terminal 2-AP.
FT-IR: 3649(w), 3630(w), 3393(s), 3298(s), 3184(m), 3091(m), 3069(m), 2983(m), 2952(m), 2928(m), 2921(m), 2826(m), 2786(w), 1607(vs), 1605(vs), 1597(vs), 1587(vs), 1563(s), 1539(s), 1483(w), 1479(m), 1463(s), 1437(vs), 1433(vs), 1394(vs), 1374(s), 1366(s), 1330(vs), 1311(vs), 1293(m), 1279(m), 1254(m), 1215(vs), 1213(vs), 1194(vs), 1165(s), 1109(m), 1075(w), 1065(m), 1058(w), 1029(m), 991(m), 985(w), 981(m), 952(m), 882(w), 881(vw), 859(w), 847(w), 825(vw), 796(w), 771(w), 747(m), 731(s), 658(w), 637(w), 610(w), 581(vw), 550(w), 525(w), 518(w), 498(w), 464(vw), and 427(w) cm−1.
Evans μeff (DMSO-d6, 298 K): 6.62μB.
2.1.2. Trinuclear Complex 2
The methanolic solution (3.5 mL) of L1 (0.114 g, 0.50 mmol) was slowly added to the methanolic solution (4.0 mL) of Ni(NO3)2·6H2O (0.073 g, 0.25 mmol). Then, Et3N (0.15 mL, 1.00 mmol) was added to the reaction mixture. Crystals for X-ray diffraction analysis were obtained at 247 K. The synthesis was aimed at complex 1 but we also found complex 2 among the products. This was a side product, obtained only once and hence only characterized by X-ray diffraction.
2.1.3. Mononuclear Complex 3
The OV (0.076 g, 0.50 mmol) in methanol (2.0 mL) was added dropwise to the solution of 2-AP (0.023 g, 0.25 mmol) in methanol (2.0 mL). The mixture was introduced to the solution of Ni(NO3)2·6H2O (0.073 g, 0.25 mmol) in 2.0 mL of methanol/100 μL water. After 15 min of stirring, Et3N (0.07 mL, 0.50 mmol) was added to the clear willow-green solution. After ten minutes, a willow-green precipitate began to form. The precipitate was collected after 15 min of further stirring. Yield: 0.064 g (0.13 mmol, 54%). Elemental analysis: anal. calcd. for C21H22N2NiO7: C 53.31; H 4.69; and N 5.92, anal. found C 53.13; H 4.76; and N 6.01. Crystals for X-ray diffraction analysis were obtained at RT by slow evaporation of the remaining solvent.
FT-IR: 3432(m), 3316(m), 3064(w), 3047(w), 3031(w), 2996(w), 2946(w), 2923(w), 2911(w), 2858(w), 2827(w), 2817(w), 2814(w), 1618(vs, sh), 1583(w), 1537(m), 1460(m), 1445(m), 1441(m), 1429(s, sh), 1408(m), 1398(w), 1367(w), 1365(w), 1335(w), 1334(m), 1264(vw), 1238(s, sh), 1205(s, sh), 1180(w), 1172(w), 1154(w), 1136(w), 1102(vw), 1072(w), 1046(w), 998(vw), 967(m, sh), 943(vw), 873(vw), 853(w), 769(vw), 761(w), 725(m, sh), 643(w), 574(vw), 562(vw), 539(vw), 509(vw), 498(vw), 435(w), and 425(vw) cm−1.
Evans μeff (DMSO-d6, 298 K): 3.33μB.
XRD powder diagram is presented in Supplementary Material as Figure S3.
2.1.4. Cubane 4
Since the compound was often described by other researchers [28,29,30,31,32], we present the synthesis and the experimental data obtained for 4 in Supplementary Materials as Table S1 and Figures S1–S3.
2.2. Physicochemical Measurements
2.2.1. FT-IR Measurements
FT-IR spectra were determined with a Nicolet iS50 FTIR spectrometer equipped with the Specac Quest single-reflection diamond attenuated total reflectance (ATR) accessory.
2.2.2. Elemental Analysis
Elemental analysis was performed at the Vario El Cube CHNS apparatus.
2.2.3. X-ray Diffraction
The crystal structure analyses were performed on an STOE IPDS II diffractometer using Mo Kα radiation of a microfocus X-ray source for complex 2 and Cu Kα radiation of a microfocus X-ray source for compounds 1 and 3. Crystals were cooled using a Cryostream 800 open flow nitrogen cryostat (Oxford Cryosystems). Data collection and image processing was performed with X-Area 1.75 (STOE & Cie GmbH, 2015) [33]. Intensity data were scaled with LANA (part of X-Area) in order to minimize differences in intensities of symmetry-equivalent reflections (multi-scan method). Structures were solved by direct methods and all non-hydrogen atoms were refined with anisotropic displacement parameters by full-matrix least squares procedure based on F2 using the SHELX–2014 program package [34]. The Olex [35] and Wingx [36] program suites were used to prepare the final version of CIF files. Olex [35] and Mercury [37] were used to prepare the figures. Hydrogen atoms were usually refined using the isotropic model with Uiso(H) values fixed to be 1.5 times Ueq of C atoms for –CH3 or 1.2 times Ueq for –CH2, –NH, and –CH groups. A disordered group of atoms consisting of nitrate anion and solvent molecules in the crystal structure of 1 was removed from the final refinement and solution with the program PLATON, and its SQUEEZE algorithm [38]. A summary of crystallographic data is shown in Table 1 and Table S1 in Supplementary Materials. CCDC 2063920–2063922 and 2072194, 2072195 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre.

Table 1.
Crystallographic data for complexes 1–3. Crystallographic data for cubane 4 are presented in Supplementary Materials in Table S1.
To confirm the identity of 3 and 4, which were also obtained as powders, we measured their XRD powder spectra in capillary on an STOE IPDS II diffractometer using Mo Kα radiation of a microfocus X-ray source. These results are presented in Supplementary Materials as Figure S3.
2.2.4. Magnetic Susceptibility Measurements in the Solution: Evans Method
The effective magnetic moment μeff of the samples obtained in the pure form (1 and 3) was determined in the solution by 1H NMR spectroscopy using the Evans method [39] with solvent (DMSO-d6) as internal reference and diamagnetic contributions according to the equations presented below [40]. A capillary with deuterated solvent was placed in the NMR tube containing a working solution prepared by dissolving the Ni compound 1 or 3 in DMSO-d6 (d = 1.190 g/mL at 25 °C). Deuterated solvent was used without further purification. 1H (internal standard Me4Si) spectra were recorded on a Bruker AV400 MHz spectrometer at 298 K and processed using Bruker’s Topspin 3.5 software:
where χmeas is total measured magnetic susceptibility (emu·mol−1); χp is the paramagnetic susceptibility (emu·mol−1); χd is the diamagnetic susceptibility (emu·mol−1); Δf is the chemical shift difference between the solvent in the presence of paramagnetic compound and in the pure solvent (Hz); F is the operating frequency of the NMR spectrometer (Hz); c is the concentration of the paramagnetic solution (mol·mL−1); M is the molar mass of the paramagnetic compound (emu·mol−1); T is the temperature during measurement (K); and μeff is the effective magnetic moment (μB). The ‘emu’ is the most widely used unit for the magnetic susceptibility (1 emu = 1 cm3 = 1 mL). Furthermore, π ≈ 3.14 was used in calculations.
2.2.5. Computational Details
Geometry optimizations were performed with the TPSSh density functional [41], which is known to provide accurate geometries and electronic structures for transition metal compounds [42,43,44]. Scalar relativistic effects were included with the zeroth-order regular approximation (ZORA) [45,46,47]. ZORA-recontracted [48] basis sets of the def2-TZVP family [49] were used for all atoms, with removal of the f polarization functions from main-group elements. All calculations were performed with ORCA [50]. Increased integration grids (Grid5, GridX7) and tight convergence criteria were applied throughout.
The Hirshfeld surfaces [51] and 2D fingerprint plots were generated with Crystal Explorer ver. 17.5 [52].
3. Results and Discussion
3.1. Synthesis and FT-IR Measurements
The initial reaction was carried out by simple mixing of the solutions of substrates: Ni(NO3)2 and imine L1 and triethylamine in methanol, which were allowed to react for a certain amount of time. In some of the attempts, 2-aminopyridine (2-AP) and o-vanillin (OV) were used separately instead of L1. As usual, we tried to isolate the products by crystallization; either the slow evaporation of solvent or lowering the temperature. During measurements of the FT-IR spectra of products isolated at various stages of evaporation of the solvent, we noticed that each time we obtained several different coordination complexes: 1, 2 and 3 (Figure 1). As revealed by X-ray measurements (see Section 3.2), all obtained complexes contained as ligands not only the imine L1 but also products of imine hydrolysis; either 2-AP or both 2-AP and OV. The formulas of the complexes are shown in Scheme 2. The presence of 2-AP and OV as ligands did not depend on the time of crystallization since we always obtained tetranuclear complex 1 as the first product with all of the ligands: L1, 2-AP, and OV. We tried to increase the yield of complexes 1 and 2 by adding 2-AP to the reaction mixture, but it did not work. Interestingly, the attempts to replace the terminal 2-AP with some other heterocyclic nitrogen bases were rather unsuccessful (despite pyridine). Therefore, we repeated the synthesis several times in order to collect enough 1 to analyze FT-IR spectra and EA. The elemental analysis was in accordance with its calculated composition (we assumed presence of nitrate ions and water molecules in the crystals on the basis of FT-IR spectra). From the same reaction mixture, we were able to obtain and identify by a single crystal diffraction complexes 2, 3, and cubane 4′; however, they always appeared together as a mixture during the evaporation of solvent. Each of the complexes contained as ligands the products of hydrolysis of L1 (see Scheme 2 and Supplementary Materials Figure S1).
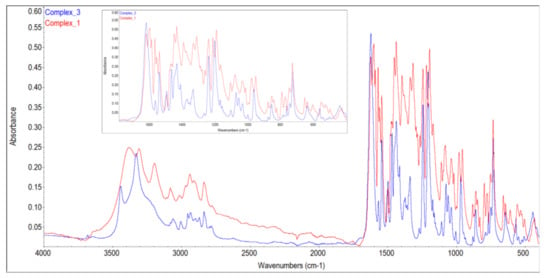
Figure 1.
FT-IR spectra of 1 and 3.
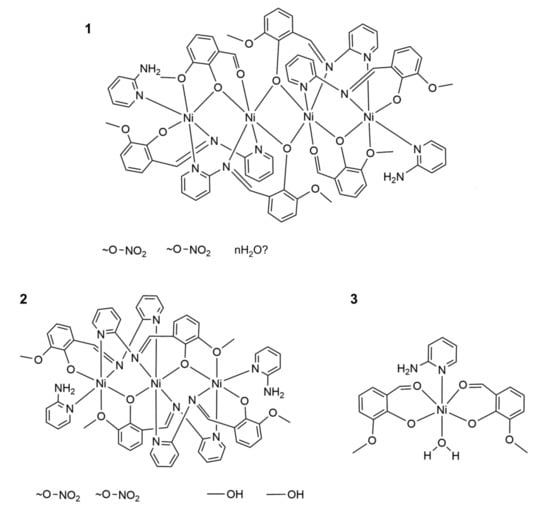
Scheme 2.
Formulas of the complexes 1–3 obtained as a result of the reaction between the nickel nitrate and ligand L1 in methanol.
We did not previously observe such immediate hydrolysis of ligand L1 during the synthesis of its zinc and copper(II) complexes [5], but a similar phenomenon has been reported for the reactions of nickel complexes with imines [53]. We tried to analyze the effect by the DFT calculations described in Section 3.4.
In order to obtain pure 3 and correlate the FT-IR spectrum with its molecular structure, we successfully carried out separate synthesis from Ni(NO3)2, 2-AP and OV in molar ratio 1:1:2, and triethylamine in methanol. It is important to preserve the described sequence of the addition of the reagents to produce 3 in this reaction (see Materials and Methods). Complex 3 could be easily distinguished from 1 by means of FT-IR spectroscopy and characteristic colors: olive for 1 and fluorescent willow-green for 3. The FT-IR spectrum of 1 features the presence of strong bands associated with the nitrate ion around 1350 cm−1, which is absent in the FT-IR spectrum of 3 (Figure 1). On the other hand, complexes 1 and 2 could not be differentiated by these simple tests because of their similar composition, and 2 could not be separated from 3 or 1, thus we neither obtained a FT-IR spectrum of pure 2, nor characterized it differently than an X-ray diffraction of a single crystal.
During this separate synthesis of 3 from nickel(II) nitrate, OV and 2-AP, the formation of different products was also observed. After isolation of the first crop of crystals of 3, we found crystals of the cubane 4 in the residual solids (see Supplementary Materials for details). Interestingly, cubane 4 can also form as the first product of the reaction between nickel(II) nitrate, OV, and 2-AP if the sequence of the addition of the reagents is altered (see Supplementary Materials). It has been the fourth product isolated within the system: nickel nitrate, OV, and AP, which points to the complexity of the equilibria in the studied reaction mixture. Moreover, there are even more possible reactions taking into account different complexes of nickel(II) and OV described in the literature; however, these were not isolated during our attempts [54].
3.2. Crystal Structures
Since the crystal structures can be measured for individual, small crystals we were able to characterize the structures of all products of the reaction between Ni(NO3)2 and imine L1, even if they were not obtained in larger amounts. The molecular structures of two polynuclear complexes 1 and 2 are shown in Figure 2 and Figure 3 and the analysis of Hirshfeld surface of complex 1 is shown in Figure 4. The mononuclear complex 3 that does not contain imine but only products of its hydrolysis, 2-AP and OV, is presented in Figure 5 and its Hirshfeld surface in Figure 6. In some of the syntheses carried out with the addition of certain heterocyclic nitrogen compounds we isolated previously known cubane 4 (see Supplementary Materials). Important bond lengths and angles are shown in the captions of Figure 2, Figure 3 and Figure 5 and crystallographic data are presented in Table 1 in the Materials and Methods.
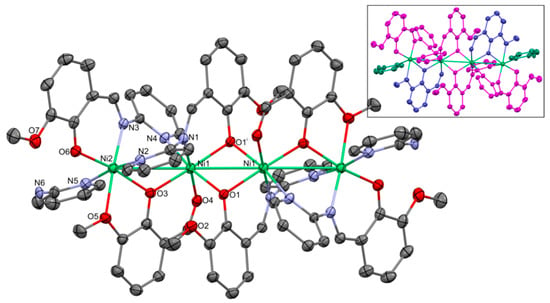
Figure 2.
Molecular structure of 1. Thermal ellipsoids at 30%. Numbering given for the Ni and heteroatoms of the independent part. In the insert the ligands are indicated with separate colors: L1—magenta; OV—blueish; and 2-AP—greenish. Important bond lengths and distances [Å]: Ni1–Ni2 3.406(2); Ni1–Ni1i 3.132(2); Ni1–O1 2.041(3); Ni1–O3 1.999(3); Ni1–O4 2.106(3); Ni1–N1 2.054(4); Ni1–N4 2.124(4); Ni2–O3 2.000(3); Ni2–O5 2.232(4); Ni2–O6 1.969(3); Ni2–N2 2.137(4); Ni2–N3 2.081(4); and Ni2–N5 2.140(4). Important angles [°]: O3–Ni1–O1 177.22(12); O3–Ni1–N1 93.38(14); O1–Ni1–N1 86.91(14); O3–Ni1–O4 84.15(13); O1–Ni1–O4 95.67(13); N1–Ni1–O4 176.66(14); O3–Ni1–N4 88.98(14); O1–Ni1–N4 88.24(14); N1–Ni1–N4 92.56(15); O1–Ni1–N4 168.94(13); O4–Ni1–N4 89.63(14); O6–Ni2–O3 168.19(13); O6–Ni2–N3 91.82(15); O3–Ni2–N3 99.26(14); O6–Ni2–N2 87.84(15); O3–Ni2–N2 87.22(14); N3–Ni2–N2 94.53(15); O6–Ni2–N5 93.49(15); O3–Ni2–N5 89.88(14); N3–Ni2–N5 93.79(15); N2–Ni2–N5 171.53(15); O6–Ni2–O5 93.73(14); O3–Ni2–O5 75.31(13); N3–Ni2–O5 174.26(14); N2–Ni2–O5 87.15(15); and N5–Ni2–O5 84.42(15). i: 1 − x, 1 − y, 1 − z.
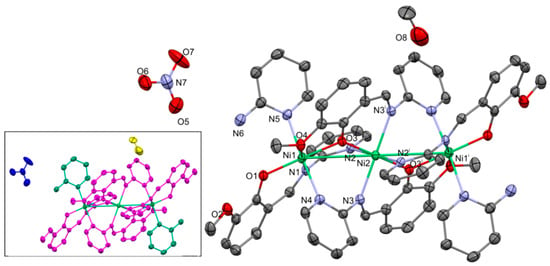
Figure 3.
Molecular structure of 2. Thermal ellipsoids at 30%. Numbering given mainly for the nickel and heteroatoms of the independent part. In the insert the ligands are indicated with the separate colors: L1—magenta; 2-AP—greenish; nitrate—blue; and methanol—yellow. Important bond lengths and distances [Å]: Ni1–Ni2 3.436(2); Ni1–O1 1.951(4); Ni1–O3 2.001(3); Ni1–O4 2.188(4); Ni1–N1 2.040(4); Ni1–N4i 2.178(5); Ni2–O3 1.973(4); Ni2–N2 2.222(4); and Ni2–N3 2.144(4). Important angles [°]: O3–Ni1–O1 165.05(15); O3–Ni1–N1 93.38(14); O1–Ni1–N1 92,69(16); O3–Ni1–O4 76.47(14); O1–Ni1–O4 89.60(15); N1–Ni1–O4 177.33(15); O3–Ni1–N4i 86.95(16); O1–Ni1–N4 86.91(16); N1–Ni1–N4i 91.83(17); O1–Ni1–N4i 86.91(16); O4–Ni1–N4i 86.90(16); O3–Ni2–O3i 180; O3–Ni2–N3 85.85(16); O3–Ni2–N2 90.37(15); and N3–Ni2–N2 94.21(16). i: 1 − x, 1 − y, 1 − z.
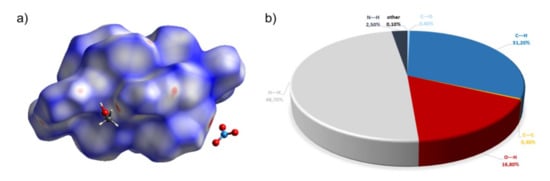
Figure 4.
(a) Hirshfeld surfaces of a complex 2. Red color: normalized contact distances dnorm shorter than the sum of van der Waals radii (dnorm = −0.414), white color: van der Waals contacts (dnorm = 0.502), and blue color: normalized contact distances exceeding the sum of van der Waals radii (dnorm = 1.406). (b) Hirshfeld surface fingerprint decompositions showing the main types of interactions for complex 2.
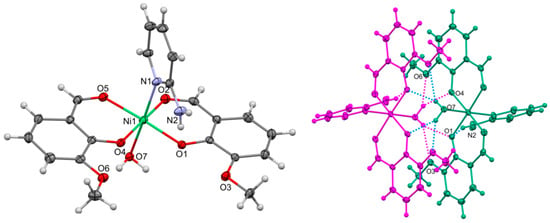
Figure 5.
Left side of the figure shows molecular structure of 3. Thermal ellipsoids at 30%. Numbering given mainly for the nickel and heteroatoms of the independent part. On the right side of the figure, hydrogen-bonded molecules within the dimer are indicated with separate colors. Important bond lengths and distances [Å]: Ni1–Ni2 3.436(2); Ni1–O1 1.995(2); Ni1–O2 2.044(3); Ni1–O4 2.001(4); Ni1–O5 2.049(2); and Ni1–O7 2.151(3). Important angles [°]: O1–Ni1–O4 89.19(10); O1–Ni1–O2 91.31(10); O4–Ni1–O2 176.45(10); O1–Ni1–O5 176.27(10); O4–Ni1–O5 90.63(10); O2–Ni1–O5 88.64(10); O1–Ni1–N1 93.72(10); O4–Ni1–N1 94.08(11); O2–Ni1–N1 89.39(11); O5–Ni1–N1 90.01(10); O1–Ni1–O7 88.64(10); O4–Ni1–O7 88.49(11); O2–Ni1–O7 88.02(11); O5–Ni1–O7 87.64(10); and N1–Ni1–O7 176.54(11).
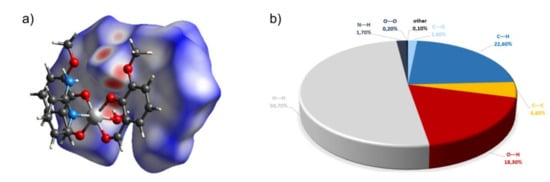
Figure 6.
(a) Hirshfeld surfaces of a complex 3. Red color: normalized contact distances dnorm shorter than the sum of van der Waals radii (dnorm = −0.557), white color: van der Waals contacts (dnorm = 0.431), and blue color: normalized contact distances exceeding the sum of van der Waals radii (dnorm = 1.303). (b) Hirshfeld surface fingerprint decompositions showing the main types of interactions for complex 3.
Polynuclear complexes 1 and 2 are centrosymmetric with the inversion center located between two inner Ni atoms in 1 and at the inner Ni atom in 2. The coordination geometry of both crystallographically independent Ni atoms within 1 and 2 is a slightly distorted octahedral. In the tetranuclear complex 1 nickel atoms are bridged by at least one phenoxide oxygen of imine L1 or OV. Two inner Ni1 atoms are connected by two phenoxide O atoms of L1 and their separation is shorter than that between the inner and terminal Ni atoms, which are linked by a single phenoxide of OV. Both L1 and OV utilize three donor atoms to coordinate to Ni, in the case of L1 these are one oxygen and two nitrogen atoms and OV utilizes three oxygen atoms to realize the bonding with Ni. The octahedral coordination of terminal Ni is completed with pyridine nitrogen of 2-AP. The inter-metal distances of over 3 Å do not suggest strong metal–metal interactions and are longer than these in complexes with ligands that impose close proximity of the metal atoms, e.g., pentanuclear Ni complexes of tripyridyldiamine [55] or tetranuclear Ni complexes with 2-(α-(5-phenyl)pyridylamino)-1,8-naphthyridine [56]. The Ni–Ni distances are also longer than in a tetranuclear Ni complex bridged by phenoxide atoms of N,N’-dimethyl-N,N’-bis(2-hydroxy-3-methoxy-5-methylbenzyl)ethylenediamine [57]. Contrary to the mentioned examples, the chain in 1 is not linear but bent with the Ni–Ni–Ni angles 149.66(3)°.
The tetranuclear complex cation in 1 is accompanied by the disordered nitrate anions and molecules of solvent. However, we were not able to build the correct model of disorder and therefore decided to remove the relevant electron densities from the refinement. Accordingly, we were not able to establish the exact nature of the interaction between these large complex cations and the anions. In the Supplementary Materials we illustrate the packing of the cation that shows the voids around Wyckoff’s special positions, which are occupied by the disordered anions and solvent molecules (Figure S4).
The ligand composition of trinuclear 2 is different from 1 because the complex cation does not contain OV (Scheme 2, Figure 3). It contains only four anions of deprotonated imine L1 and 2 molecules of 2-AP. Therefore, we think that it must form independently when the concentrations of ligands change as a result of evaporation of the solvent. Complex 2 is centrosymmetric with the inversion center located at the central Ni, therefore it is linear. Every two nickel atoms are triply bridged by imine molecules. Likewise, in 1, the distances between nickel atoms do not indicate metal–metal interactions. Although the composition of 2 is very similar to the formerly described (Ni3(pymp)4(pya)2)(NO3)2(CH3OH)2 [12] and other trinuclear complexes with this ligand [19,25], it features a different coordination pattern of the central and terminal nickel atoms. The presence of the additional methoxy substituent in L1 “inverts” the position of the ligand within the complex and phenoxide oxygen coordinates to the terminal Ni (and forms bridges), whereas pyridyl rings are bonded to the inner Ni atom. Therefore, instead of a N2O4 coordination mode of the inner metal atom described for trinuclear pymp complexes of transition metals [12,19,25], the coordination of the inner Ni atom in 2 is N4O2. The change in the coordination mode of the imine must have an influence on the Ni–Ni distances which are longer in 2 compared with the pymp complex.
The positions of nitrate anion and methanol in 2 are well established and in Figure S5, which illustrates crystal packing, we can see that the clusters of nitrate anions and methanol molecules do occupy special positions within the unit cell as was also suggested for 1. Nevertheless, the analysis of Hirshfeld surface presented in Figure 4 indicates that the most important interactions between molecules in complex 2 are van der Waals forces. The decomposed fingerprint plot shows that the hydrophobic H···H (48.7%) interactions are the major factor in the crystal packing with C···H (31.2%) interactions representing the next highest contribution. Red areas on the Hirshfeld surface correspond to strong interactions between N6–H···O5 and weaker C18–H···O8.
One more product of the reaction(s) was a mononuclear complex 3 with OV, 2-AP, and water illustrated in Figure 5. The non-centrosymmetric arrangement of OV ligands in this octahedral complex is probably imposed by the intramolecular interactions of the water molecule. In the given conformation of ligands, it forms two bifurcated hydrogen bonds with the OV ligands of the adjacent molecule complex. The hydrogen bonded dimers are kept together by four bifurcated H-bonds. There is one intramolecular H-bond within the complex and one rather weak bond judging from the donor acceptor distance between the dimers. The hydrogen-bonded dimer of 3 is shown in Figure 5. On the Hirshfeld surface of 3 shown in Figure 6, we see the red spots that represent the above-mentioned strong bifurcate O7–H···O1, O7–H···O4 contacts between dimers, while the blue regions correspond to weak interactions such as C—H···H contacts. The decomposed fingerprint plot shows that similar to compound 2, the H···H (50.7%) interactions are the major factor in the crystal packing with C···H (22.6%) and hydrogen bonds O···H (18.3%) as second and third highest contributions, respectively.
3.3. Magnetic Properties by Evans Method
Our final attempt to characterize the obtained complexes experimentally employed 1H NMR measurements of the effective magnetic moment μeff of the pure, crystalline 1 and 3 redissolved in DMSO-d6 using the Evans method (see Materials and Methods). The NMR spectra for complexes 1 and 3 are presented in Supplementary Materials as Figures S6–S9. The obtained values of magnetic moments μeff for complexes 1 and 3 are 6.62 μB and 3.33 μB, respectively (Table 2). As we see in Table 2, the experimental magnetic moment for 1 does not exactly agree with the value predicted for typical octahedral tetra-nickel(II) systems with S = 4 (8.94 μB, 8 unpaired electrons). The observed reduction of the magnetic moment resulted either from the coupling between bonded Ni atoms, or from the formation of a mixture of complexes in the prepared solution. On the other hand, the value of the magnetic moment established for complex 3 seemed to be in good accordance with the anticipated magnetic moment that equals 2.83 μB for Ni(II) systems with S = 1. It is also comparable to the available experimental data for similar octahedral Ni(II) complexes, which may exhibit magnetic moments in a range of 3.15–3.60 μB [58,59].

Table 2.
Effective magnetic moments μeff of crystalline 1 and 3 redissolved in DMSO-d6 determined by 1H NMR spectroscopy using the Evans method.
3.4. DFT Calculations
To identify the possible electronic origins of the divergent reactivity observed for the different metal ions, we performed density functional theory calculations with the TPSSh functional [41] and scalar relativistic corrections for the series of Co, Ni, Cu, and Zn complexes with ligand L1. Geometry optimizations of the M(L1)2 complexes in various conformations of the ligand resulted in a unique favored geometry for each metal ion. The calculated geometries are illustrated in Figure 7. The optimized geometries and corresponding electronic structures reveal that the Zn and Cu complexes have distinct coordination preferences from the Ni and Co analogues. In the zinc complex, the closed-shell Zn(II) ion adopts a geometry that is almost perfectly tetrahedral, with the chelating planes of the two ligands (as defined by the two O–Zn–N angles) arranged almost perpendicularly to each other (twist angle of 86° in the optimized structure). In the copper complex, the Cu(II) is also highly symmetric, but the ligands are now rotated towards coplanarity (twist angle of 41°). The end result is a coordination geometry intermediate between tetrahedral and square planar, a feature that is frequently encountered in Cu(II) chelates [60]. The single unpaired electron occupies a molecular orbital that is identifiable as the dx2−y2 orbital of Cu with σ-antibonding character with respect to the four ligating atoms. The singly occupied frontier orbitals of Cu, Ni, and Co complexes are depicted in the Supplementary Materials in Figure S10.
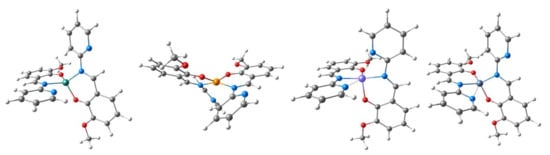
Figure 7.
DFT-optimized structures of M(L1)2 models. From left to right: M = Zn, Cu, Ni, and Co.
For the Zn and Cu complexes, no interaction is observed between the metal and the pyridine nitrogens. In contrast, the optimized structures of the Co and Ni complexes feature a distortion of either one (Co) or both (Ni) of the L1 ligands to bring the pyridine nitrogen(s) in close proximity to the metal ion. In the case of the cobalt complex, this results in a quasi-5-coordinate (distorted square pyramidal) arrangement, with one close Co–N(pyridine) distance of 2.53 Å. Regarding the nickel complex, both pyridine nitrogens have Ni–N distances of 2.49 Å, leading to a distorted octahedral coordination sphere where both ligands function approximately as tridentate fac-O,N,N-donors. Although the short distances between metal and pyridine nitrogens are ca. 0.5 Å longer than the two other metal–N distances, they clearly represent weak bonding interactions, particularly in the case of the Ni complex where the Mayer bond orders for the Ni–pyridine interactions are greater than 25% of the bond order of the two other Ni–N or Ni–O bonds. Presumably, it is only the highly unfavorable energetics of the four-membered ring that prevent any closer interaction of the pyridine nitrogen with the metal. This weak interaction is accompanied by the distortion of the ligand compared to the conformation adopted in the genuine 4-coordinate Cu and Zn complexes: the pyridine bends toward the metal by ca. 10° away from its non-interacting conformation, while the imine C=N bond is shortened by up to 0.05 Å to facilitate the interaction. The above observations strongly suggest that M(L1)2 complexes of Co and Ni are coordinatively unsaturated systems, in contrast to the Cu and Zn complexes. The strong propensity of Ni(II) to adopt an octahedral coordination sphere and the inability of L1 to act as an efficient tridentate ligand to a single metal site are consistent with the observed formation of oligonuclear species. At the same time, M–O/N Mayer bond orders explicitly adopt the lowest values for Ni complexes consistent with the increased coordination number as illustrated in Table 3.

Table 3.
Calculated Mayer bond orders for complexes M(L1)2.
4. Conclusions
We have found that in the presence of nickel cations the imine ligand L1 immediately begins to hydrolyze, which results in the formation of various heteroligand nickel complexes. The difference between the reaction of nickel(II) and previously described reactions of copper(II) and zinc(II) with L1 [5] is ascribed to the coordinative unsaturation of stoichiometric Ni(L1)2 complexes, indicated by DFT calculations.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cryst11050512/s1. Synthesis of Cubane 4. Table S1. Crystallographic data for cubane 4 and 4′. Figure S1: Molecular structures of 4. Numbering given for the independent part. Important bond lengths/distances [Å]: Ni1–O1 2.0059(19); Ni1–O2 2.028(2); Ni1–O4 2.0355(19); Ni1–O4i 2.0406(19); Ni1–O4ii 2.0593(19); Ni1–O5 2.096(2); Ni1–Ni1i 3.0659(6); Ni1–Ni1ii 3.0872(6); i y + 1/4, − x + 3/4, − z + 3/4; ii − x + 1, − y + 1/2, z. Figure S2: FT-IR spectrum of 4. Figure S3. XRD powder spectra of (a) 3 and (b) 4. Simulated spectra were calculated with Mercury and re-calculated to Mo Kα using the Bragg equation (the Default calculation in Mercury returns the values for Cu Kα). Figure S4: Crystal packing of 1. Figure S5: Crystal packing of 2. Darker blue—methanol, lighter blue—nitrate. 1H NMR DATA of isolated Ni complexes. Figure S6: 1H NMR of 1 at room temperature in DMSO-d6. Abbreviations in the figure: g—grease, h—H2O in deuterated solvent DMSO-d6, s—residual signal of deuterated solvent DMSO-d6. Figure S7: 1H NMR of 3 at room temperature in DMSO-d6. Abbreviations in the figure: g—grease, h—H2O in deuterated solvent DMSO-d6, m—methanol, s—residual signal of deuterated solvent DMSO-d6, ●—unidentified compound. 1H NMR spectra for the effective magnetic moment determination in solution (Evans method). Figure S8: 1H NMR of DMSO-d6 in the presence of paramagnetic compound 1 solution in DMSO-d6 at room temperature. Abbreviations in the figure: out—outer tube, in—inner tube/capillary. Figure S9: 1H NMR of DMSO-d6 in the presence of paramagnetic compound 3 solution in DMSO-d6 at room temperature. Abbreviations in the figure: out—outer tube, in—inner tube/capillary. Figure S10: Singly occupied orbitals of the M(L1)2 models of (a) Cu, (b) Ni, and (c) Co.
Author Contributions
Conceptualization, A.D.; investigation, M.S., D.A.P., J.B., K.K.-L. and A.D.; methodology M.S., D.A.P., K.K.-L. and A.D.; visualization, M.S., D.A.P., K.K.-L. and A.D.; writing—original draft, M.S., D.A.P., K.K.-L. and A.D.; writing—review & editing, K.K.-L. and A.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available within the manuscripts, Supplementary Materials and from The Cambridge Crystallographic Data Centre as CCDC 2063920–2063922 and 2072194, 2072195.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schiff, H. Mittheilungen aus dem Universitätslaboratorium in Pisa: Eine neue Raiche organischer Basen. Justus Liebigs Ann. Chem. 1864, 131, 118–119. [Google Scholar] [CrossRef]
- Liu, X.; Hamon, J.-R. Recent developments in penta-, hexa- and heptadentate Schiff base ligands and their metal complexes. Coord. Chem. Rev. 2019, 389, 94–118. [Google Scholar] [CrossRef]
- Sakthivel, A.; Jeyasubramanian, K.; Thangagiri, B.; Dhaveethu Raja, J. Recent advances in Schiff base metal complexes derived from 4-aminoantipyrine derivatives and their potential applications. J. Mol. Struct. 2020, 1222, 128885. [Google Scholar] [CrossRef]
- Udhayakumari, D.; Inbaraj, V. A Review on Schiff Base Fluorescent Chemosensors for Cell Imaging Applications. J. Fluoresc. 2020, 30, 1203–1223. [Google Scholar] [CrossRef]
- Mielcarek, A.; Bieńko, A.; Saramak, P.; Jezierska, J.; Dołęga, A. Cu/Zn heterometallic complex with solvent-binding cavity, catalytic activity for oxidation of 1-phenylethanol and unusual magnetic properties. Dalton Trans. 2019, 48, 17780–17791. [Google Scholar] [CrossRef]
- Chang, F.; Zhang, D.; Xu, G.; Yang, H.; Li, J.; Song, H.; Sun, W.-H. Synthesis and characterization of new bis(1-aryliminomethylenylnaphthalen-2-oxy)nickel complexes and their catalytic behavior for vinyl polymerization of norbornene. J. Organomet. Chem. 2004, 689, 936–946. [Google Scholar] [CrossRef]
- Ikram, M.; Rehman, S.U.; Rehman, S.; Baker, R.J.; Schulzke, C. Synthesis, characterization and distinct butyrylcholinesterase activities of transition metal complexes of 2-[(E)-(quinolin-3-ylimino)methyl]phenol. Inorg. Chim. Acta 2012, 390, 210–216. [Google Scholar] [CrossRef]
- Kuchtanin, V.; Kleščíková, L.; Šoral, M.; Fischer, R.; Růžičková, Z.; Rakovský, E.; Moncoľ, J.; Segľa, P. Nickel(II) Schiff base complexes: Synthesis, characterization and catalytic activity in Kumada–Corriu cross-coupling reactions. Polyhedron 2016, 117, 90–96. [Google Scholar] [CrossRef]
- Drummond, J.; Wood, J.S. Crystal and Molecular Structure and Magnetic Properties of a Tetrameric Copper Complex formed by the Terdentate Ligand N-2-Pyridylsalicylaldimine. J. Chem. Soc. Dalton Trans. 1972, 365–369. [Google Scholar] [CrossRef]
- Shen, Y.-Z.; Gu, H.; Pan, Y.; Dong, G.; Wu, T.; Jin, X.-P.; Huang, X.-Y.; Hu, H. Synthesis and characterization of dialkylgallium (dialkylindium) complexes of N-salicylidene 2-aminopyridine and N-salicylidene 2-methoxyaniline: Crystal structure of dimethyl[N-salicylidene 2-aminopyridine]gallium. J. Organomet. Chem. 2000, 605, 234–238. [Google Scholar] [CrossRef]
- Zhu, H.-L.; Liu, X.-Y. Syntheses and Crystal Structures of a Pair of Trinuclear Cobalt(II) Complexes with the Cations [Co3L4(APYH)2] and [Co3L4(APY)2] (APY = 2-aminopyridine, L = N-salicylaldehyde-2-iminopyridine). Synth. React. Inorg. Met. Org. Nano Metal. Chem. 2005, 35, 193–196. [Google Scholar] [CrossRef]
- Ran, J.-W.; Zhang, S.-Y.; Hu, B.; Xu, B.; Li, Y. Trinuclear and mononuclear nickel(II) complexes incorporating tridentate 2-[(pyridine-2-ylimine)methyl]phenol ligand: Syntheses, crystal structures and magnetic properties. Inorg. Chem. Commun. 2008, 11, 1474–1477. [Google Scholar] [CrossRef]
- Miao, J.; Zhao, Z.; Chen, H.; Wang, D.; Nie, Y. Bis{2-[(2-pyridyl)iminomethyl]-phenolato}copper(II). Acta Crystallogr. Sect. E Struct. Rep. Online 2009, E65, m904. [Google Scholar] [CrossRef]
- Zhao, Q.-H.; Chen, H.-Y.; Li, L.-N.; Xie, M.-J. Tris [2-(2-pyridylimino-methyl)phenolato(0.67)]-europium(III) nitrate. Acta Crystallogr. Sect. E Struct. Rep. Online 2009, E65, m697. [Google Scholar] [CrossRef] [PubMed]
- Robert, F.; Tinant, B.; Clérac, R.; Jacquemin, P.-L.; Garcia, Y. Self-assembly of asymmetric tetranuclear Cu(II) [2×2] grid-like complexes and of a dinuclear Ni(II) complex from pyridyl-phenol Schiff base ligands. Polyhedron 2010, 29, 2739–2746. [Google Scholar] [CrossRef]
- Chang, H.-C.; Cole, J.M.; Lin, T.-C.; Sylvester, S.O.; Waddell, P.G. Bis{l-2-[(pyridin-2-yl)iminomethyl]-phenolato}bis[(2-formylphenolato)-copper(II)]. Acta Crystallogr. Sect. E Struct. Rep. Online 2011, E67, m690. [Google Scholar] [CrossRef]
- Giri, S.; Biswas, S.; Drew, M.G.B.; Ghosh, A.; Saha, S.K. Structure and magnetic properties of a tetranuclear Cu(II) complex containing the 2-(pyridine-2-yliminomethyl)-phenol ligand. Inorg. Chim. Acta 2011, 368, 152–156. [Google Scholar] [CrossRef]
- Dame, A.N.; Bharara, M.S.; Barnes, C.L.; Walensky, J.R. Synthesis of Thorium(IV) and Uranium(IV) Salicylaldiminate Pseudo-Halide Complexes. Eur. J. Inorg. Chem. 2015, 2996–3005. [Google Scholar] [CrossRef]
- Zhang, Y.-Z.; Brown, A.J.; Meng, Y.-S.; Sun, H.-L.; Gao, S. Linear trinuclear cobalt(II) single molecule magnet. Dalton Trans. 2015, 44, 2865–2870. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.; Mongal, B.N.; Dutta, A.; Mondal, P.; Lewis, W.; Saba, N.; Naskar, S. Complexation study of Schiff base ligand: Pyridin-2-ylimino methyl naphthanol with Co2+, Mn2+ and Ni2+ ions in solid and solution phase. J. Coord. Chem. 2016, 69, 2364–2376. [Google Scholar] [CrossRef]
- Jia, W.-G.; Zhang, H.; Zhang, T.; Xie, D.; Ling, S.; Sheng, E.-H. Half-Sandwich Ruthenium Complexes with Schiff-Base Ligands: Syntheses, Characterization, and Catalytic Activities for the Reduction of Nitroarenes. Organometallics 2016, 35, 503–512. [Google Scholar] [CrossRef]
- Jafari, M.; Salehi, M.; Kubicki, M.; Arab, A.; Khaleghian, A. DFT studies and antioxidant activity of Schiff base metal complexes of 2-aminopyridine. Crystal structures of cobalt(II) and zinc(II) complexes. Inorg. Chim. Acta 2017, 462, 329–335. [Google Scholar] [CrossRef]
- Enamullah, M.; Vasylyeva, V.; Abdul Quddus, M.; Islam, M.K.; Höfert, S.-P.; Janiak, C. Spontaneous resolution of a Δ/Λ-chiral-at-metal pseudo-tetrahedral Schiff-base zinc complex to a racemic conglomerate with C–H⋯O organized 41- and 43-helices. CrystEngComm 2018, 20, 4724–4734. [Google Scholar] [CrossRef]
- Enamullah, M.; Islam, M.A.; Kautz, A.C.; Janiak, C. Synthesis, spectroscopy, electrochemistry, and molecular structure of tetrakis{(E)-2-((pyridin-2-ylimino)methyl)phenolato}(hydroxido)0.5(nitrato)1.5-tetracopper(II) nitrate hydroxide. J. Coord. Chem. 2018, 71, 2557–2568. [Google Scholar] [CrossRef]
- Liu, S.; Deng, Y.-F.; Li, C.A.; Chang, X.; Zhang, Y.-Z. A linear trinuclear ferrous single molecule magnet. Dalton Trans. 2018, 47, 16704–16708. [Google Scholar] [CrossRef]
- Pradhan, K.C.; Barik, S.; Singh, B.C.; Mohapatra, P.; Kisan, H.K.; Pal, S. Synthesis, characterisation and theoretical studies of a series of Iridium(III) heteroleptic complexes with Schiff base ligands. J. Mol. Struct. 2020, 1211, 128058. [Google Scholar] [CrossRef]
- Mielcarek, A.; Wiśniewska, A.; Dołęga, A. Unassisted formation of hemiaminal ether from 4-aminopyridine and o-vanillin—experimental and theoretical study. Struct. Chem. 2018, 29, 1189–1200. [Google Scholar] [CrossRef]
- Ayikoe, K.; Butcher, R.J.; Gultneh, Y. Tetra-μ3-methanolato-tetrakis[(2-formyl-6-methoxyphenolato)methanolnickel(II)]. Acta Crystallogr. Sect. E Struct. Rep. Online 2010, E66, m1487–m1488. [Google Scholar] [CrossRef]
- Yu, G.-M.; Zhao, L.; Zou, L.-F.; Guo, Y.-N.; Xu, G.-F.; Li, Y.-H.; Tang, J. A Tetranuclear Nickel(II) Cubane Complex with O-Vanillin Ligand. J. Chem. Crystallogr. 2011, 41, 606–609. [Google Scholar] [CrossRef]
- Habib, F.; Cook, C.; Korobkov, I.; Murugesu, M. Novel in situ manganese-promoted double-aldol addition. Inorg. Chim. Acta 2012, 380, 378–385. [Google Scholar] [CrossRef]
- Costes, J.-P.; Novitchi, G.; Vendier, L.; Pilet, G.; Luneau, D. Magnetic ordering of NiII4 Cubane complexes through hydrogen bonds. C. R. Chim. 2012, 15, 849–855. [Google Scholar] [CrossRef]
- Zhang, S.-H.; Zhang, Y.D.; Zou, H.H.; Guo, J.J.; Li, H.P.; Song, Y.; Liang, H. A family of cubane cobalt and nickel clusters: Syntheses, structures and magnetic properties. Inorg. Chim. Acta 2013, 396, 119–125. [Google Scholar] [CrossRef]
- X-Area 1.75, STOE & Cie GmbH, Software Package for Collecting Single-Crystal Data on STOE Area-Detector Diffractometers, for Image Processing, Scaling Reflection Intensities and for Outlier Rejection; STOE: Darmstadt, Germany, 2015.
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A Complete Structure Solution, Refinement and Analysis Program. J. Appl. Crystallogr. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Farrugia, L.J. WinGX and ORTEP for Windows: An update. J. Appl. Crystallogr. 2012, 45, 849–854. [Google Scholar] [CrossRef]
- Macrae, C.F.; Sovago, I.; Cottrell, S.J.; Galek, P.; McCabe, P.; Pidcock, E.; Platings, M.; Shields, G.P.; Stevens, J.S.; Towler, M.; et al. Mercury 4.0: From visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 53, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Crystallogr. Sect. C Cryst. Struct. Commun. 2015, C71, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Evans, D.F. The Determination of the Paramagnetic Susceptibility of Substances in Solution by Nuclear Magnetic Resonance. J. Chem. Soc. 1959, 2003–2005. [Google Scholar] [CrossRef]
- Bain, G.A.; Berry, J.F. Diamagnetic Corrections and Pascal’s Constants. J. Chem. Educ. 2008, 85, 532–536. [Google Scholar] [CrossRef]
- Staroverov, V.N.; Scuseria, G.E.; Tao, J.; Perdew, J.P. Comparative Assessment of a New Nonempirical Density Functional: Molecules and Hydrogen-Bonded Complexes. J. Chem. Phys. 2003, 119, 12129–12137. [Google Scholar] [CrossRef]
- Bühl, M.; Kabrede, H. Geometries of Transition-Metal Complexes from Density-Functional Theory. J. Chem. Theory Comput. 2006, 2, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Bühl, M.; Reimann, C.; Pantazis, D.A.; Bredow, T.; Neese, F. Geometries of third-row transition-metal complexes from density-functional theory. J. Chem. Theory Comput. 2008, 4, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.P. Bioinorganic Chemistry Modeled with the TPSSh Density Functional. Inorg. Chem. 2008, 47, 10357–10365. [Google Scholar] [CrossRef]
- Van Lenthe, E.; Baerends, E.J.; Snijders, J.G. Relativistic Regular Two-component Hamiltonians. J. Chem. Phys. 1993, 99, 4597–4610. [Google Scholar] [CrossRef]
- Van Lenthe, E.; Baerends, E.J.; Snijders, J.G. Relativistic Total-Energy Using Regular Approximations. J. Chem. Phys. 1994, 101, 9783–9792. [Google Scholar] [CrossRef]
- Van Leeuwen, R.; van Lenthe, E.; Baerends, E.J.; Snijders, J.G. Exact solutions of regular approximate relativistic wave equations for hydrogen-like atoms. J. Chem. Phys. 1994, 101, 1272–1281. [Google Scholar] [CrossRef]
- Pantazis, D.A.; Chen, X.Y.; Landis, C.R.; Neese, F. All-electron scalar relativistic basis sets for third-row transition metal atoms. J. Chem. Theory Comput. 2008, 4, 908–919. [Google Scholar] [CrossRef] [PubMed]
- Weigend, F.; Ahlrichs, R. Balanced Basis Sets of Split Valence, Triple Zeta Valence and Quadruple Zeta Valence Quality for H to Rn: Design and Assessment of Accuracy. Phys. Chem. Chem. Phys. 2005, 7, 3297–3305. [Google Scholar] [CrossRef]
- Neese, F.; Wennmohs, F.; Becker, U.; Riplinger, C. The ORCA quantum chemistry program package. J. Chem. Phys. 2020, 152, 224108. [Google Scholar] [CrossRef]
- Spackman, M.A.; Jayatilaka, D. Hirshfeld Surface Analysis. CrystEngComm. 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer 17; University of Western Australia: Crawley, Australia, 2017. [Google Scholar]
- Pait, M.; Sarkar, A.; Colacio, E.; Ray, D. Hydrolysis on Di-Schiff Base Ligand During Dinuclear Ni(II) Complex Formation: Synthesis, Crystal Structures and Magneto-Structural Correlation Studies. Proc. Natl. Acad. Sci. India Sect. A Phys. Sci. 2014, 84, 189–196. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, D.-Q. Aqua-bis(o-vanillinato-κ2O,O’)nickel(II). Acta Crystallogr. Sect. E Struct. Rep. Online 2008, E64, m298. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Lo, W.-C.; Chou, C.-C.; Lee, G.-H.; Chen, J.-M.; Peng, S.-M. Synthesis, Crystal Structures, and Magnetic Properties of a Series of Linear Pentanickel(II) Complexes: [Ni5(μ5-tpda)4 × 2] (X = Cl−, CN−, N3−, NCS−) and [Ni5(μ5-tpda)4(CH3CN)2](PF6)2 (tpda2− = the Tripyridyldiamido Dianion). Inorg. Chem. 1998, 37, 4059–4065. [Google Scholar] [CrossRef] [PubMed]
- Tsou, L.-H.; Sigrist, M.; Chiang, M.-H.; Horng, E.-C.; Chen, C.-H.; Huang, S.-L.; Lee, G.-H.; Peng, S.-M. Asymmetric tetranuclear nickel chains with unidirectionally ordered 2-(α-(5-phenyl)pyridylamino)-1,8-naphthyridine ligands. Dalton Trans. 2016, 45, 17281–17289. [Google Scholar] [CrossRef] [PubMed]
- Botana, L.; Ruiz, J.; Mota, A.J.; Rodríguez-Diéguez, A.; Seco, J.M.; Oyarzabal, I.; Colacio, E. Anion controlled structural and magnetic diversity in unusual mixed-bridged polynuclear NiII complexes with a versatile bis(2-methoxy phenol)diamine hexadentate ligand. An experimental and theoretical magneto-structural study. Dalton Trans. 2014, 43, 13509–13524. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Mandal, D.; Ghosh, D.; Goldberg, I.; Chaudhury, M. Equilibrium Studies in Solution Involving Nickel(II) Complexes of Flexidentate Schiff Base Ligands: Isolation and Structural Characterization of the Planar Red and Octahedral Green Species Involved in the Equilibrium. Inorg. Chem. 2003, 42, 8439–8445. [Google Scholar] [CrossRef]
- Salem, N.M.H.; Rashad, A.R.; El Sayed, L.; Haase, W.; Iskander, M.F. Synthesis, characterization, molecular and supramolecular structures of nickel(II) complexes derived from α-diketone and α-ketoaldehyde bisaroylhydrazones. Polyhedron 2009, 28, 2137–2148. [Google Scholar] [CrossRef]
- Stamos, N.-A.; Ferentinos, E.; Chrysina, M.; Raptopoulou, C.P.; Psycharis, V.; Sanakis, Y.; Pantazis, D.A.; Kyritsis, P.; Mitrikas, G. Unusual 31P Hyperfine Strain Effects in a Conformationally Flexible Cu(II) Complex Revealed by Two-Dimensional Pulse EPR Spectroscopy. Inorg. Chem. 2020, 59, 3666–3676. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).