2.2.1. Identification of Ricin and RCA120 by Tryptic Fingerprinting and Sequencing
The aim of the LC-ESI MS/MS analysis (this section) and the MALDI-TOF MS/MS analysis (
Section 2.2.2) was to unambiguously identify ricin and RCA120 in the protein preparations prepared from
R. communis and to thoroughly check for potential cross-contaminations with other
R. communis proteins. To this end, both preparations were reduced, alkylated, and underwent tryptic digestion. The tryptic fragments were analyzed and sequenced by LC-ESI MS/MS, focusing on identification of the isoforms of ricin D, ricin E, and their corresponding A and B chains, as well as RCA120.
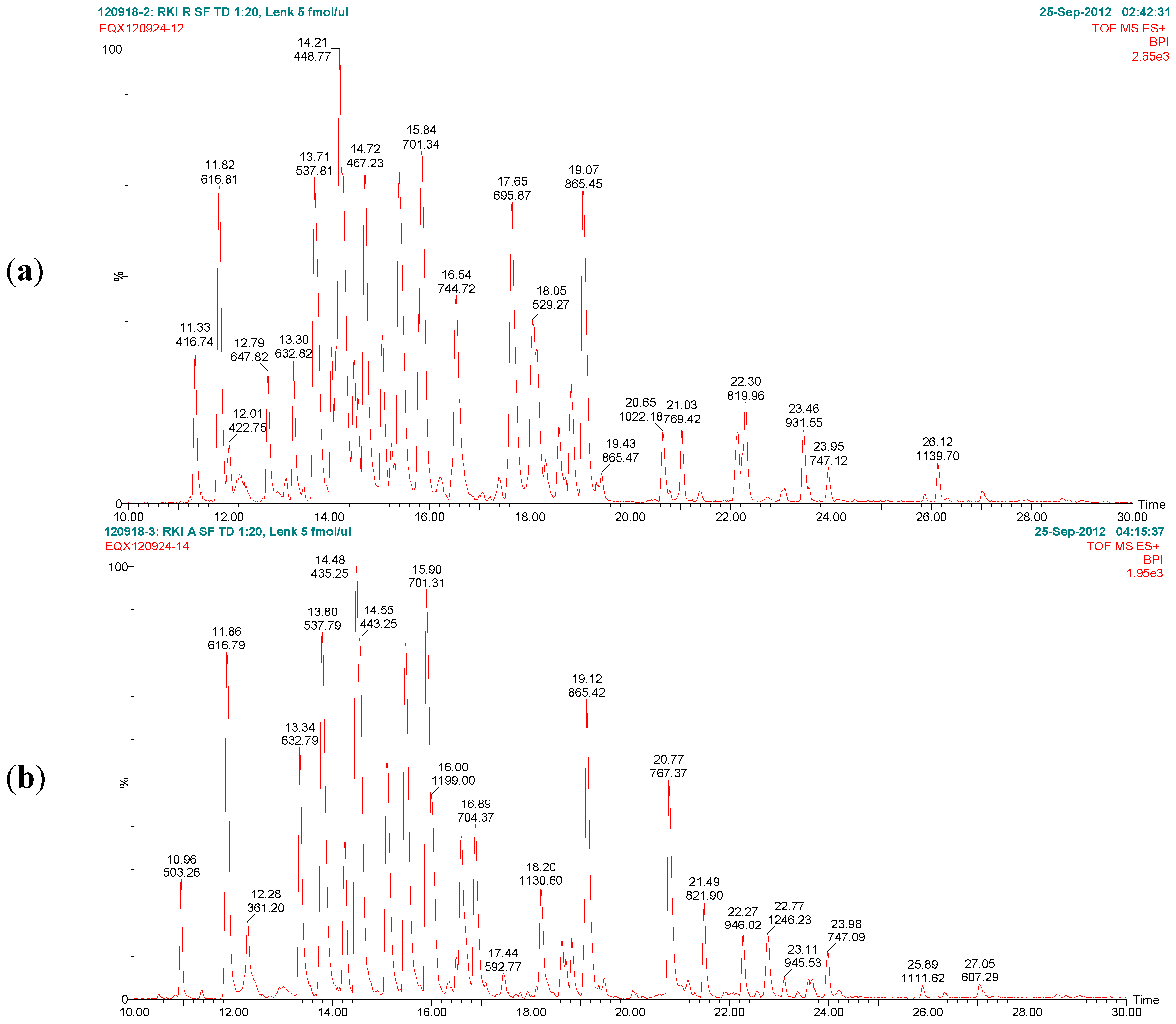
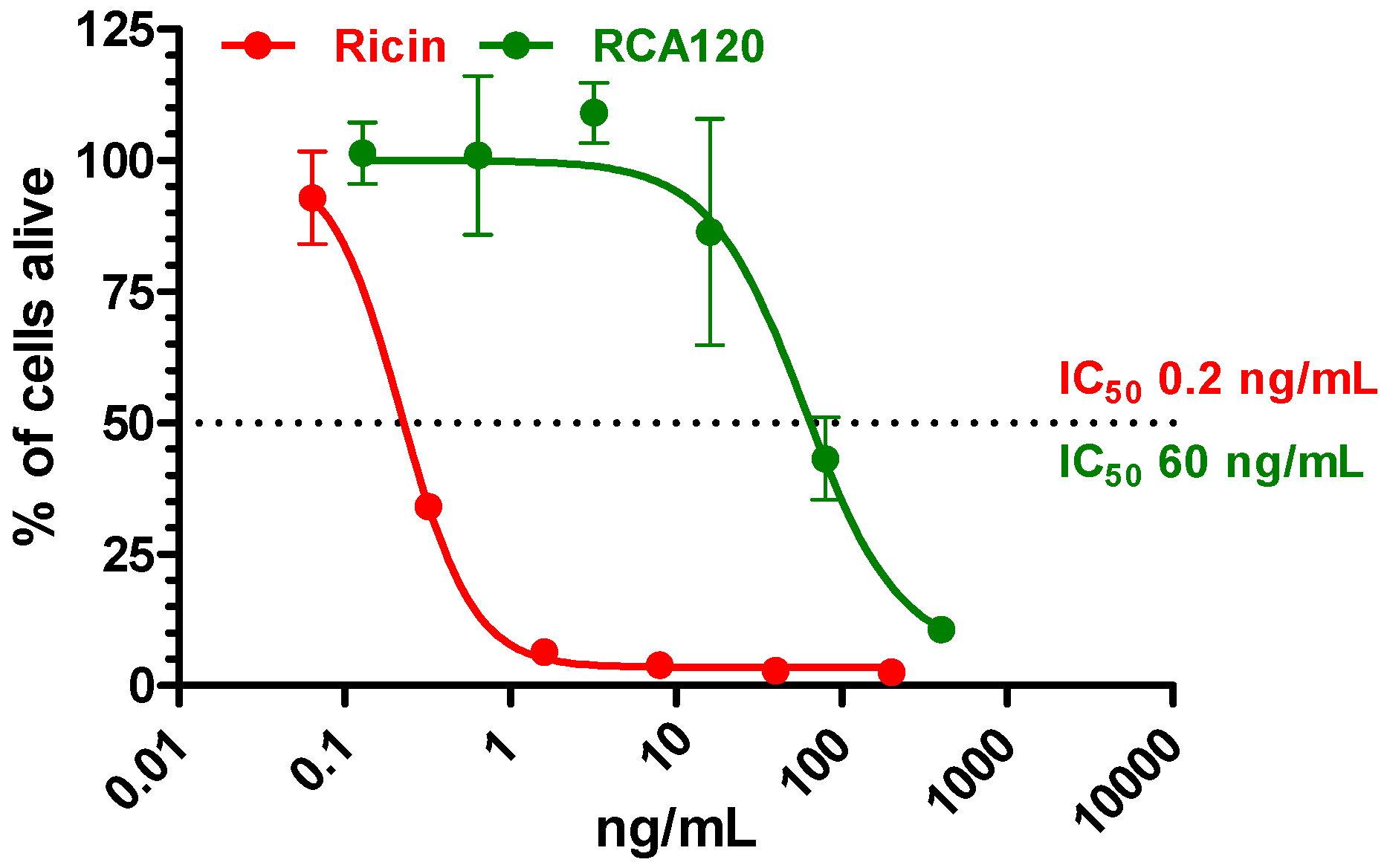
Figure 7 exemplarily shows the base peak chromatograms of the trypsin digests of ricin and RCA120 preparations obtained by LC-ESI MS/MS analysis. The peptides from ricin D observed in the ricin preparation covered 95% of the sequence of the A chain and 74% of the B chain. In the RCA120 preparation, peptides representing 90% of the A chain and 79% of the B chain of RCA120 were detected. The full sequences of ricin D, ricin E, and RCA120 are displayed in
Table 1. Sequences displayed in red show peptides which were experimentally detected by combining LC-ESI MS/MS and MALDI-TOF MS/MS data (this Section and
Section 2.2.2).
Figure 7.
Base peak chromatograms of the trypsin digests of (a) purified ricin and (b) purified RCA120. The relative intensity is plotted against the retention time. Peaks are labeled with retention time and the m/z value of the base peak.
Figure 7.
Base peak chromatograms of the trypsin digests of (a) purified ricin and (b) purified RCA120. The relative intensity is plotted against the retention time. Peaks are labeled with retention time and the m/z value of the base peak.
Table 2 and
Table 3 list the theoretical trypsin digest peptides obtained from chains A and B of ricin D (Uniprot [
56] ID: P02879), and those from chain B of ricin E (NCBI Genpept [
57] ID: GI:225419 ) are found in
Table 4.
The corresponding trypsin digest peptides from RCA120 (Uniprot [
56] ID: P06750) and for chain B (NCBI Genpept [
57] ID: GI:225114) are shown in
Table 5 and
Table 6. In each of the tables, the molecular ions of the experimentally observed peptides are indicated by bold typeface. Also, the peptides that can be used to distinguish ricin D from ricin E and RCA120 from ricin D are indicated in bold typeface.
Table 1.
The amino acid sequences of ricin D, ricin E, and RCA120 identified in purified ricin and purified RCA120. The experimentally detected sequences using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI MS/MS) and matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry (MALDI-TOF MS/MS) as trypsin digest peptides are indicated in red.
Table 1.
The amino acid sequences of ricin D, ricin E, and RCA120 identified in purified ricin and purified RCA120. The experimentally detected sequences using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI MS/MS) and matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry (MALDI-TOF MS/MS) as trypsin digest peptides are indicated in red.
| Sequence coverage of ricin preparation: |
|---|
| Ricin D chain A (Uniprot P02879, NCBI GI:132567) |
| 1 | QYPIIN FTTAGATVQS YTNFIRAVRLTTGADVRH EIPVLPNRVG LPINQRFILV |
| 61 | ELSNHAELSV TLALDVTNAY VVGYRAGNSA YFFHPDNQED AEAITHLFTD VQNRYTFAFG |
| 121 | GNYDRLEQLA GNLRENIELG NGPLEEAISA LYYYSTGGTQ LPTLARSFII CIQMISEAAR |
| 181 | FQYIEGEMRIRYNRRSAP DPSVITLENS WGRLSTAIQE SNQGAFASPI QLQRF |
| 241 | SVYDVSILIP IIALMVYRCA PPPSSQF |
| Ricin D chain B (Uniprot P02879, NCBI GI:132567) |
| 1 | ADVCMDPEPI VRIVGRNGLC VDVRFHN GNAIQLWPCK SNTDANQLWT LKRDNTIRSN |
| 61 | GKCLTTYGYS PGVYVMIYDC NTAATDATRW QIWDNGTIIN PR |
| 121 | AE QQWALYADGS |
| 181 | IRPQQNRDNC LTSDSNIRET VVKILSCGP ASSGQRWMFKN DGTILNLYSG LVLDVRASDP |
| 241 | SLKQIILYPL HGDPNQIWLP LF |
| Ricin E chain B (NCBI GI:225419) |
| 1 | ADVCMDPEPI VRIVGRNGLC VDVRFHN GNAIQLWPCK SNTDANQLWT LKRDNTIRSN |
| 61 | GKCLTTYGYP SGVYVMIYDC NTAATDATRW QIWDNGTIIN PR |
| 121 | VW LEDCTSEKAE QQWALYADGS |
| 181 | IRPQQNRDNC LTTDANIKILSCGPA SSGQRWMFKN DGTILNLYNG LVLDVR |
| 241 | QIIVHPV HGNLNQIWLP LF |
| Sequence coverage of RCA120 preparation: |
| RCA120 chain A (Uniprot P06750, NCBI GI:113504) |
| 1 | QYPIIN FTTADATVES YTNFIRH EIPVLPNRVG LPISQRFILV |
| 61 | ELSNHAELSV TLALDVTNAY VVGCRAGNSA YFFHPDNQED AEAITHLFTD VQNSFTFAFG |
| 121 | GNYDRLEQLG GLRENIELGT GPLEDAISAL YYYSTCGTQI PTLARSFMVC IQMISEAARF |
| 181 | QYIEGEMRYNRRSAPD PSVITLENSW GRLSTAIQES NQGAFASPIQ LQRFN |
| 241 | VYDVSILIPI IALMVYRCAP PPSSQF |
| RCA120 chain B (NCBI GI:225114) |
| 1 | ADVCMDPEPI VRIVGRNGLC VDVGEEFD GNPIQLWPCK SNTDWNQLWT LRK |
| 61 | CLTISKSS PGQQVVIYNC STATVGATRW QIWDNRTIIN PTSGLVLAAT SGNSGTK |
| 121 | VW LEDCTSEKAE QQWALYADGS |
| 181 | IRPQQNRDNC LTTDANIKILSCGPA SSGQRWMFKN DGTILNLYNG LVLDVRRSDP |
| 241 | SLKQIIVHPV HGNLNQIWLP LF |
Table 2.
Ricin D and E chain A trypsin digest peptides (Uniprot [
56] ID: P02879, NCBI Genpept [
57] ID: GI:132567), reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
Table 2.
Ricin D and E chain A trypsin digest peptides (Uniprot [56] ID: P02879, NCBI Genpept [57] ID: GI:132567), reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
| T# (a) | Amino Acid Sequence | (M+H)+ | (M+2H)2+ | (M+3H)3+ | (M+4H)4+ |
|---|
| T1a | IFPK | 504.3180 | - | - | - |
| T2a-glyc | QYPIINFTTAGATVQSYTNFIR | 3675.6946
* | 1838.3509
* | 1225.9031 * | 919.6791
* |
| T3a | AVR | 345.2245 | - | - | - |
| T4a | GR | 232.1404 | - | - | - |
| T5a | LTTGADVR | 832.4523 | 416.7298 | - | - |
| T6a | HEIPVLPNR | 1074.6054 | 537.8063 | - | - |
| T7a | VGLPINQR | 896.5312 | 448.7692 | - | - |
| T8a | FILVELSNHAELSVTLALDVTNAYVVGYR | 3206.7095 | 1603.8584 | 1069.5747 | 802.4328 |
| T9a | AGNSAYFFHPDNQEDAEAITHLFTDVQNR | 3307.5038 | 1654.2555 | 1103.1728 | 827.6314 |
| T10a | YTFAFGGNYDR | 1310.5800 | 655.7936 | - | - |
| T11a | LEQLAGNLR | 1013.5738 | 507.2905 | - | - |
| T12a | ENIELGNGPLEEAISALYYYSTGGTQLPTLAR | 3440.7219 | 1720.8646 | 1147.5788 | 860.9359 |
| T13a | SFIICIQMISEAAR | 1638.8342 | 819.9207 | - | - |
| T14a | FQYIEGEMR | 1172.5404 | 586.7738 | - | - |
| T15a | TR | 276.1666 | - | - | - |
| T16a | IR | 288.2030 | - | - | - |
| T17a | YNR | 452.2252 | - | - | - |
| T18a | R | 175.1189 | - | - | - |
| T19a | SAPDPSVITLENSWGR | 1728.8551 | 864.9312 | - | - |
| T20a | LSTAIQESNQGAFASPIQLQR | 2259.1727 | 1130.0900 | 753.7291 | - |
| T21a | R | 175.1189 | - | - | - |
| T22a | NGSK | 405.2092 | - | - | - |
| T23a | FSVYDVSILIPIIALMVYR | 2212.2450 | 1106.6261 | 738.0865 | - |
| T24a | CAPPPSSQF | 990.4349 | 495.7211 | - | - |
Table 3.
Ricin D chain B trypsin digest peptides (Uniprot [
56] ID: P02879, NCBI Genpept [
57] ID: GI:132567), reduced and carbamidomethylated. Ricin/RCA120 distinguishing peptides and observed molecular ions are given in bold.
Table 3.
Ricin D chain B trypsin digest peptides (Uniprot [56] ID: P02879, NCBI Genpept [57] ID: GI:132567), reduced and carbamidomethylated. Ricin/RCA120 distinguishing peptides and observed molecular ions are given in bold.
| T# (a) | Amino Acid Sequence | (M+H)+ | (M+2H)2+ | (M+3H)3+ | (M+4H)4+ |
|---|
| T1b | ADVCMDPEPIVR | 1401.6501 | 701.3287 | - | - |
| T2b | IVGR | 444.2929 | - | - | - |
| T3b | NGLCVDVR | 932.4618 | 466.7345 | - | - |
| T4b | DGR | 347.1673 | - | - | - |
| T5b | FHNGNAIQLWPCK | 1584.7740 | 792.8906 | - | - |
| T6b | SNTDANQLWTLK | 1390.6961 | 695.8517 | - | - |
| T7b | R | 175.1189 | - | - | - |
| T8b | DNTIR | 618.3205 | - | - | - |
| T9b | SNGK | 405.2092 | - | - | - |
| T10b | CLTTYGYSPGVYVMIYDCNTAATDATR | 3063.3532 | 1532.1802 | 1021.7893 | 766.5938 |
| T11b-glyc | WQIWDNGTIINPR | 2991.2987
* | 1496.1530
* | 997.7711 * | 748.5801
* |
| T12b-glyc | SSLVLAATSGNSGTTLTVQTNIYAVSQGWLPTNNTQPFVTTIVGLYGLCLQANSGQVWIEDCSSEK | [7047.4666] | - | - | - |
| T13b | AEQQWALYADGSIRPQQNR | 2231.0952 | 1116.0512 | 744.3699 | - |
| T14b | DNCLTSDSNIR | 1294.5692 | 647.7882 | - | - |
| T15b | ETVVK | 575.3399 | - | - | - |
| T16b | ILSCGPASSGQR | 1232.6052 | 616.8062 | - | - |
| T17b | WMFK | 611.3010 | 306.1541 | - | - |
| T18b | NDGTILNLYSGLVLDVR | 1862.0018 | 931.5045 | - | - |
| T19b | ASDPSLK | 717.3777 | 359.1925 | - | - |
| T20b | QIILYPLHGDPNQIWLPLF | 2277.2430 | 1139.1251 | 759.7525 | - |
Table 4.
Ricin E chain B trypsin digest peptides (NCBI Genpept [
57] ID: GI:225419), reduced and carbamidomethylated. Ricin D/E-distinguishing sequence and observed molecular ions are given in bold.
Table 4.
Ricin E chain B trypsin digest peptides (NCBI Genpept [57] ID: GI:225419), reduced and carbamidomethylated. Ricin D/E-distinguishing sequence and observed molecular ions are given in bold.
| T# (a) | Amino Acid Sequence | (M+H)+ | (M+2H)2+ | (M+3H)3+ | (M+4H)4+ |
|---|
| T1b | ADVCMDPEPIVR | 1401.6501 | 701.3287 | - | - |
| T2b | IVGR | 444.2929 | - | - | - |
| T3b | NGLCVDVR | 932.4618 | 466.7345 | - | - |
| T4b | DGR | 347.1673 | - | - | - |
| T5b | FHNGNAIQLWPCK | 1584.7740 | 792.8906 | - | - |
| T6b | SNTDANQLWTLK | 1390.6961 | 695.8517 | - | - |
| T7b | R | 175.1189 | - | - | - |
| T8b | DNTIR | 618.3205 | - | - | - |
| T9b | SNGK | 405.2092 | - | - | - |
| T10b | CLTTYGYPSGVYVMIYDCNTAATDATR | 3063.3532 | 1532.1802 | 1021.7893 | 766.5938 |
| T11b glyc | WQIWDNGTIINPR | - | 1496.1530 * | 997.7711 * | 748.5801 * |
| T12b glyc | SSLVLAATSGNSGTTLTVQTNIYAVSQGWLPTNNTQPFVTTIVGLYGMCLQANSGK | [5831.9258] | - | - | - |
| T13b | VWLEDCTSEK | 1266.5670 | 633.7871 | - | - |
| T14b | AEQQWALYADGSIRPQQNR | 2231.0952 | 1116.0512 | 744.3699 | - |
| T15b | DNCLTTDANIK | 1264.5838 | 632.7955 | - | - |
| T16b | GTVVK | 503.3187 | - | - | - |
| T17b | ILSCGPASSGQR | 1232.6052 | 616.8062 | - | - |
| T18b | WMFK | 611.3010 | 306.1541 | - | - |
| T19b | NDGTILNLYNGLVLDVR | 1889.0127 | 945.0100 | - | - |
| T20b | R | 175.1189 | - | - | - |
| T21b | SDPSLK | 646.3406 | 323.6739 | - | - |
| T22b | QIIVHPVHGNLNQIWLPLF | 2238.2545 | 1119.6309 | 746.7564 | - |
Table 5.
RCA120 chain A (Uniprot [
56] ID: P06750, NCBI Genpept [
57] ID: GI:113504) trypsin digest peptides, reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
Table 5.
RCA120 chain A (Uniprot [56] ID: P06750, NCBI Genpept [57] ID: GI:113504) trypsin digest peptides, reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
| T# (a) | Amino Acid Sequence | (M+H)+ | (M+2H)2+ | (M+3H)3+ | (M+4H)4+ |
|---|
| T1a | IFPK | 504.3180 | - | - | - |
| T2a glyc | QYPIINFTTADATVESYTNFIR | 3734.6841
* | 1867.8457
* | 1245.5662 * | 934.4265
* |
| T3a | AVR | 345.2245 | - | - | - |
| T4a | SHLTTGADVR | 1056.5432 | 528.7752 | - | - |
| T5a | HEIPVLPNR | 1074.6054 | 537.8063 | - | - |
| T6a | VGLPISQR | 869.5203 | 435.2638 | - | - |
| T7a | FILVELSNHAELSVTLALDVTNAYVVGCR | 3203.6768 | 1602.3420 | 1068.5638 | 801.6747 |
| T8a | AGNSAYFFHPDNQEDAEAITLFTDVQNSFTFAFGGNYDR | 4514.0020 | 2257.5046 | 1505.3389 | 1129.2560 |
| T9a | LEQLGGLR | 885.5152 | 443.2612 | - | - |
| T10a | ENIELGTGPLEDAISALYYYSTCGTQIPTLAR | 3516.7202 | 1758.8637 | 1172.9116 | 879.9355 |
| T11a | SFMVCIQMISEAAR | 1642.7750 | 821.8911 | - | - |
| T12a | FQYIEGEMR | 1172.5404 | 586.7738 | - | - |
| T13a | TR | 276.1666 | - | - | - |
| T14a | IR | 288.2030 | - | - | - |
| T15a | YNR | 452.2252 | - | - | - |
| T16a | R | 175.1189 | - | - | - |
| T17a | SAPDPSVITLENSWGR | 1728.8551 | 864.9312 | - | - |
| T18a | LSTAIQESNQGAFASPIQLQ R | 2259.1727 | 1130.0900 | 753.7291 | - |
| T19a | R | 175.1189 | - | - | - |
| T20a | NGSK | 405.2092 | - | - | - |
| T21a | FNVYDVSILIPIIALMVYR | 2239.2559 | 1120.1316 | 747.0902 | - |
| T22a | CAPPPSSQF | 990.4349 | 495.7211 | - | - |
Table 6.
RCA120 chain B (NCBI Genpept [
57] ID: GI:225114) trypsin digest peptides, reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
Table 6.
RCA120 chain B (NCBI Genpept [57] ID: GI:225114) trypsin digest peptides, reduced and carbamidomethylated. Ricin/RCA120-distinguishing peptides and observed molecular ions are given in bold.
| T# (a) | Amino Acid Sequence | (M+H)+ | (M+2H)2+ | (M+3H)3+ | (M+4H)4+ |
|---|
| T1b | ADVCMDPEPIVR | 1401.6501 | 701.3287 | - | - |
| T2b | IVGR | 444.2929 | - | - | - |
| T3b | NGLCVDVFGEEFTDGNPIQLWPCK | 2795.2803 | 1398.1438 | 932.4316 | 699.5755 |
| T4b | SNTDWNQLWTLR | 1533.7444 | 767.3758 | - | - |
| T5b | K | 147.1128 | - | - | - |
| T6b | DSTIR | 591.3096 | - | - | - |
| T7b | SDGK | 406.1932 | - | - | - |
| T8b | CLTISK | 721.3913 | 361.1993 | - | - |
| T9b glyc | SSPGQQVVIYNCSTATVGATR | 3366.4960
* | 1683.7516
* | 1122.8369 * | 842.3795
* |
| T10b-glyc | WQIWDNR | 2395.9657
* | 1198.4865 * | 799.3268
* | - |
| T11b | TIINPTSGLVLAATSGNSGTK | 2002.0815 | 1001.5444 | 668.0320 | - |
| T12b glyc | LTVQTNIYAVSQGWLPTNNTQPFVTTIVGLYGMCLQANSGK | [4485.2581] | - | - | - |
| T13b | VWLEDCTSEK | 1266.5670 | 633.7871 | - | |
| T14b | AEQQWALYADGSIRPQQNR | 2231.0952 | 1116.0512 | 744.3699 | - |
| T15b | DNCLTTDANIK | 1264.5838 | 632.7955 | - | - |
| T16b | GTVVK | 503.3187 | - | - | - |
| T17b | ILSCGPASSGQR | 1232.6052 | 616.8062 | - | - |
| T18b | WMFK | 611.3010 | 306.1541 | - | - |
| T19b | NDGTILNLYNGLVLDVR | 1889.0127 | 945.0100 | - | - |
| T20b | R | 175.1189 | - | - | - |
| T21b | SDPSLK | 646.3406 | 323.6739 | - | - |
| T22b | QIIVHPVHGNLNQIWLPLF | 2238.2545 | 1119.6309 | 746.7564 | - |
In the E isoform of ricin, the amino acid sequence of chain A is identical to that of ricin D. The differences between D and E are located at the B chain, where approximately half the chain at the carboxyl terminal end has the same amino acid sequence as RCA120. In the
R. communis cultivar used here, both ricin isoforms were found. Several peptides detected in the purified ricin preparation were attributed to ricin E, e.g., T13b, T15b, T19b, and T22b (
Table 4).
Besides the sequence GI:225419 [
17], two other sequences for ricin E, GI:2169612 and GI:225896 [
18], are published in the NCBI Genpept database (The National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD, USA) [
57]. The peptide molecular ions at 616
2+ and 746
3+ matched T17b and T22b of GI:225419 and distinguished it from the other ricin E sequences (GI:225896, GI:2169612).
The ricin E sequence GI:225419 has a Pro-Ser at position 70–71 [
17] identical to that published for ricin D by the same group [
58,
59]. Pro70-Ser71 in the T10b peptide from this ricin preparation could not be confirmed (see
Supplementary Information, Figure S1). Instead, data supported Ser-Pro at position 70–71, which corresponds to other sequences published for ricin E (GI:225896 [
18], GI:2169612) and ricin D (GI:132567 [
60]). The discrepancy could be due to an actual difference between isoforms or flaws in the older data using sequencing methods available at that time. A comparison to X-ray structures could not be performed, since no structure for the B chain of ricin E is available so far.
The NCBI database contains a number of sequence variants for the B chain of RCA120, where sequence GI:225114 differs from other RCA120 sequences, e.g. Uniprot P06750 (NCBI Genpept GI:113504; X-ray structure: 1RZO). In the purified RCA120 preparation, the two peptides identified as T9 glycopeptide and T11 of the RCA120 chain B sequence GI:225114 distinguished this isoform from other RCA120 sequences, e.g., GI:113504 (
Table 6;
Supplementary Information Figures S2 and S3). The product ion spectrum of the T3 peptide of chain B, however, was consistent with the amino acid sequence of GI:113504 and not GI:225114 (
Supplementary Information Figure S4). Again, this inconsistency could be due to lower reliability of older sequencing methods.
Twenty-eight peptides from ricin and 27 from RCA120 were detected by the nano-LC-MS method used in this investigation. Nineteen of the ricin peptides were proteotypic, i.e., not found in other proteins including RCA120. For further in-depth characterization, increased sequence coverage could be obtained on sections resistant to trypsin by using proteases with alternative cleavage specificity, e.g., chymotrypsin or pepsin.
For identification and quantification, sequence coverage is less important, and instrument capacity and sensitivity requirements might restrict the number of monitored peptides. Peptides suitable for identification and quantification of ricin,
i.e., proteotypic and detected at high intensity, are consistent with those proposed earlier [
44,
61,
62] with the exception of T19b, which is polar and was not retained by the trapping column in the nano-LC system used here. Diagnostic peptides for ricin with high identification potential observed here were T5, T7, T9, T10, and T11 from the A chain and T6, T11, T18, and T20 from the B chain. When the sample was reduced and alkylated before digestion, both T13 from chain A and T14 from chain B (ricin D) also exhibited good electrospray response and excellent MS/MS sequence information. B chain peptides T3 and T5 were both prone to deamidation, which made them less suitable as target peptides for quantification and unequivocal identification.
In RCA120 T6 and T9 from chain A together with T4 from chain B were conserved in all isoforms and should be selected as primary target peptides for unequivocal identification of RCA120 as well as T11a when the sample is reduced and alkylated. Ricin E could be distinguished from ricin D by the B chain peptides highlighted in
Table 4, primarily T13, T15, T19, and T22.
Peptides common to ricin and RCA120 suitable for detection and identification of both proteins when combined with proteotypic peptides were T6, T14, T19, and T20 from chain A and T13 and T16 from chain B of ricin. The disulfide linked peptide connecting the A and B chains in ricin and RCA120 had a relatively low electrospray response, but in samples at toxicologically relevant concentrations it was useful as an indication of potentially active toxin.
When matched to the NCBIdatabase, the majority of the diagnostic ricin peptides were unique, except for T5a which was found in a
Pomacea flagellata hemagglutinin (a freshwater snail) and B chain T3 which appeared in related ribosome-inactivating proteins [
44]. The glycopeptide T11b also matched
Viscum album lectin 1 (viscumin) and T19b three other proteins. As the databases continue to grow, overlaps with both functionally related and purely coincidental new proteins could occur.
Depending on the scenario, the number of peptides required for identification and their sequence coverage need to be considered. In contrast to recommendations for proteomic identification of unknown proteins in complex mixtures in the life sciences [
63,
64,
65], the identification of target proteins in public health and forensic investigations involves the comparison of experimental data with authentic reference standards. Criteria for chromatographic retention time and ion ratios as well as the mass of digestion products and MS/MS sequence information will need to be specified [
66].
The World Anti-Doping Agency (WADA) has published detailed identification criteria for proteins. Reporting of a unique amino acid sequence and minimum sequence coverage of 10% was specified [
67]. Identification criteria for ricin in the context of verification of the Chemical Weapons Convention was proposed by a working group under the OPCW Scientific Advisory Board [
68]. The MS criteria recommended reporting a minimum of two peptides from each of the chains with detailed experimental data and, furthermore, that the uniqueness of the peptides needs to be specified. Supporting data from immunological techniques, ricin functionality assay, molecular weight determination or PCR was additionally required for unambiguous identification.
Generally, no peptides from other
R. communis components were detected in the trypsin digests of purified ricin and purified RCA120, e.g., 2S-albumin or the peptide biomarkers RCB-1 to -3 [
69]. The next step was to analyze potential cross-contaminations of RCA120 in the purified ricin preparation and,
vice versa, ricin in the purified RCA120 preparation. The concentration of the impurities in the preparations was calculated using the protein concentrations obtained from UV
280 measurement and the responses of peptides specific for ricin and RCA120.
The amount of RCA120 in the purified ricin sample was calculated from the peak areas determined from the extracted ion chromatograms, using the RCA120 preparation as the analytical standard. Similarly, the amount of ricin in purified RCA120 was determined using the ricin preparation as the analytical standard. Based on this procedure, the amount of RCA120 in the ricin preparation was 1.4% ± 0.4% relative to ricin based on the average of chain A peptides, and 4.5% ± 1.5% based on chain B peptides. Therefore, the purity of ricin was specified to ≥97% based on the average of chain A and B peptides.
For the R. communis agglutinin preparation, the amount of ricin found was 0.5% ± 0.2% calculated for chain A peptides and 1.4% ± 0.7% for chain B peptides, resulting in an average purity of >99%. For both protein preparations, these results were in good agreement with the purity estimated from CGE analysis.
Overall, the purity of the ricin and the RCA120 preparation characterized in this work was very good. While for a commercial preparation of ricin a similar purity of 96% was reported, the corresponding RCA120 preparation showed a purity of only 68% [
43]. Additionally, our toxin preparations were derived from a defined
R. communis cultivar, while the cultivar used for purification concerning the commercial products is not known.
As described above, the concentration of RCA120 in the purified ricin sample was consistently higher when calculated using chain B peptides than peptides from chain A. The same observation was made for ricin in the purified RCA120 sample. This was unexpected, considering the nature of the sample and the purification scheme used. The purification process consisted of an acid extraction at pH 4 and ammonium sulfate precipitation of proteins. Affinity separation using a lactosyl-Sepharose-4B column was performed to isolate ricin and RCA120 from other constituents in the precipitate. Finally, size exclusion chromatography was used to separate ricin from RCA120. A possible explanation for the excess of chain B peptides over chain A peptides measured as contamination in both preparations could be a decomposition process or a reduction of the A–B interchain disulfide bond during the extraction and precipitation process. Chain B of both ricin and RCA120 would be isolated together with the intact proteins in the affinity step and could end up in the ricin size exclusion chromatography fraction to an extent depending on the size exclusion separation efficiency.
The amount of ricin E was determined in the ricin preparation using the chain B peptides that distinguish the E isoform from ricin D (
Table 4). However, since they are shared with RCA120, the result had to be corrected by the content of RCA120 which can be determined from the intensity of the relevant RCA120 peptides, e.g., T6, T9, and T11 from chain A and T3 and T4 from chain B. Based on this procedure, the ratio of ricin D to ricin E was estimated to be close to one for the ricin preparation purified from
R. communis carmencita in this work (data not shown).
2.2.2. Analysis of Trace Contaminants by MALDI-TOF MS/MS Analysis
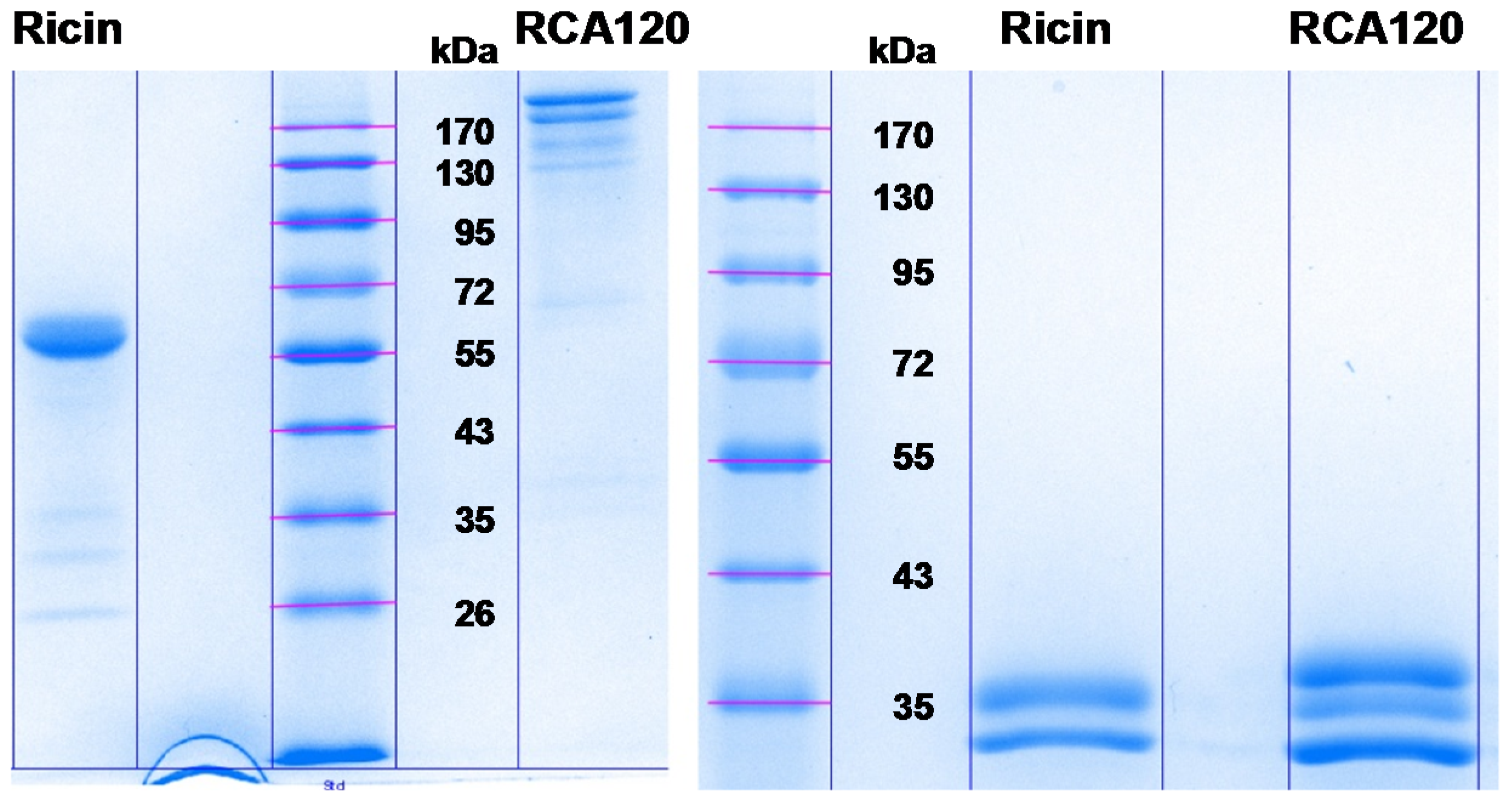
The aim of the following experiments was to identify any impurities which might be present in both the ricin and the RCA120 preparation. As described in
Section 2.1.1, the separation of larger amounts of both purified proteins by SDS-PAGE resulted in a number of faint lower-molecular-weight protein bands of unknown identity (
Figure 2). It has remained unclear from the experiments conducted so far whether these protein bands were fragmented ricin or RCA120 proteins or irrelevant contaminants co-purified from the seeds. Therefore, both purified preparations were separated on an SDS-PAGE, and Coomassie-stained bands were cut out, and proteins were reduced, alkylated, and digested with trypsin, followed by a MALDI-TOF MS/MS analysis.
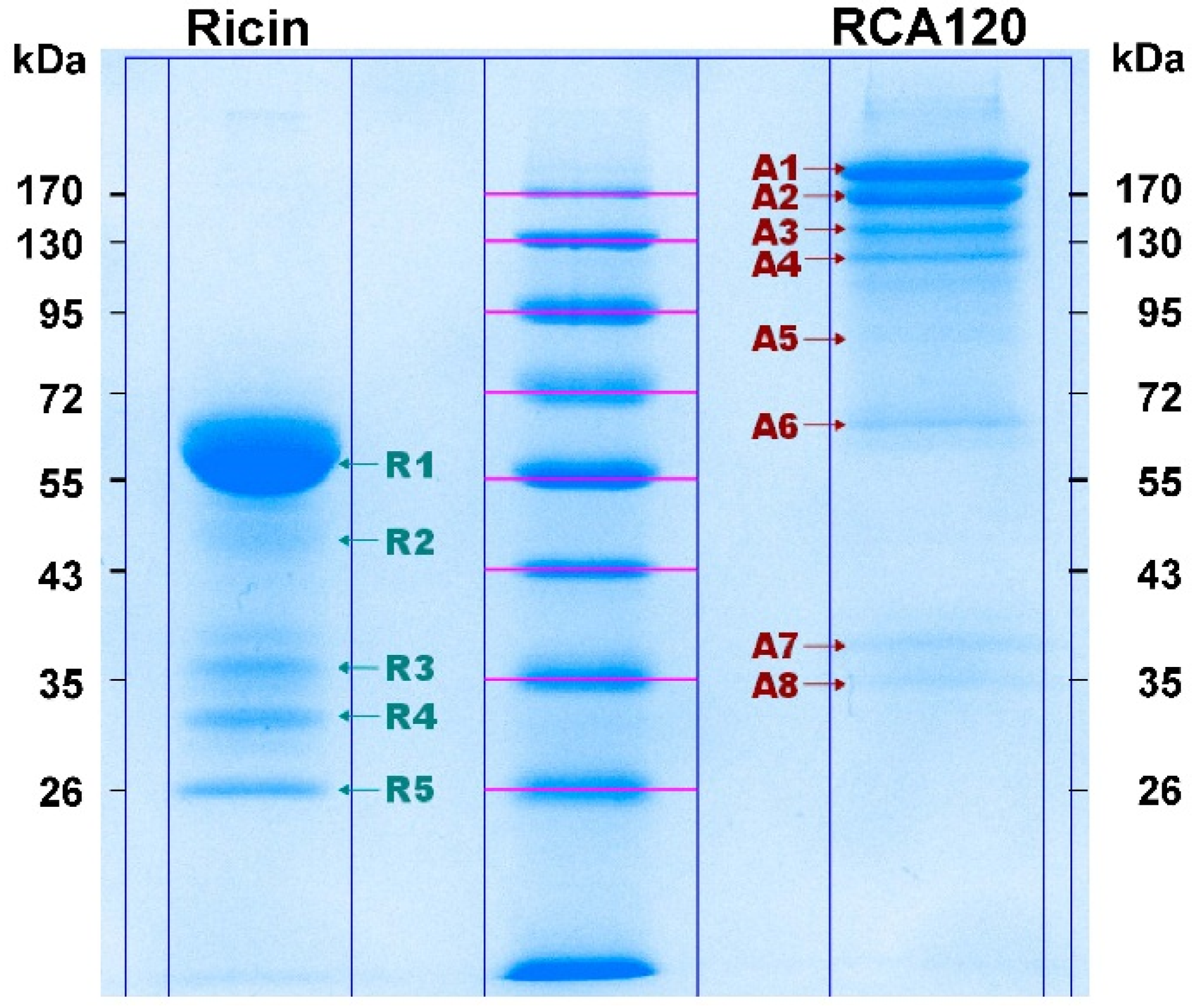
Figure 8 shows the Coomassie-stained gel which was used to cut out protein bands. The gel pieces corresponding to the individual protein bands were numbered sequentially R1 through R5 for the purified ricin preparation and A1 through A8 for the purified RCA120 preparation.
Figure 8.
Separation of purified ricin and RCA120 by gel electrophoresis for subsequent MALDI-TOF MS/MS analysis. Purified ricin (15.1 µg, left) or purified RCA120 (10.3 µg, right) were separated on a 10% SDS-PAGE. The indicated protein bands were cut out for further analysis: bands R1 through R5 for ricin and bands A1 through A8 for RCA120, respectively. For an in-gel digest, bands were cut out, reduced, and alkylated, used for tryptic digestion, and finally eluted from the gel, followed by MALDI-TOF MS/MS analysis.
Figure 8.
Separation of purified ricin and RCA120 by gel electrophoresis for subsequent MALDI-TOF MS/MS analysis. Purified ricin (15.1 µg, left) or purified RCA120 (10.3 µg, right) were separated on a 10% SDS-PAGE. The indicated protein bands were cut out for further analysis: bands R1 through R5 for ricin and bands A1 through A8 for RCA120, respectively. For an in-gel digest, bands were cut out, reduced, and alkylated, used for tryptic digestion, and finally eluted from the gel, followed by MALDI-TOF MS/MS analysis.
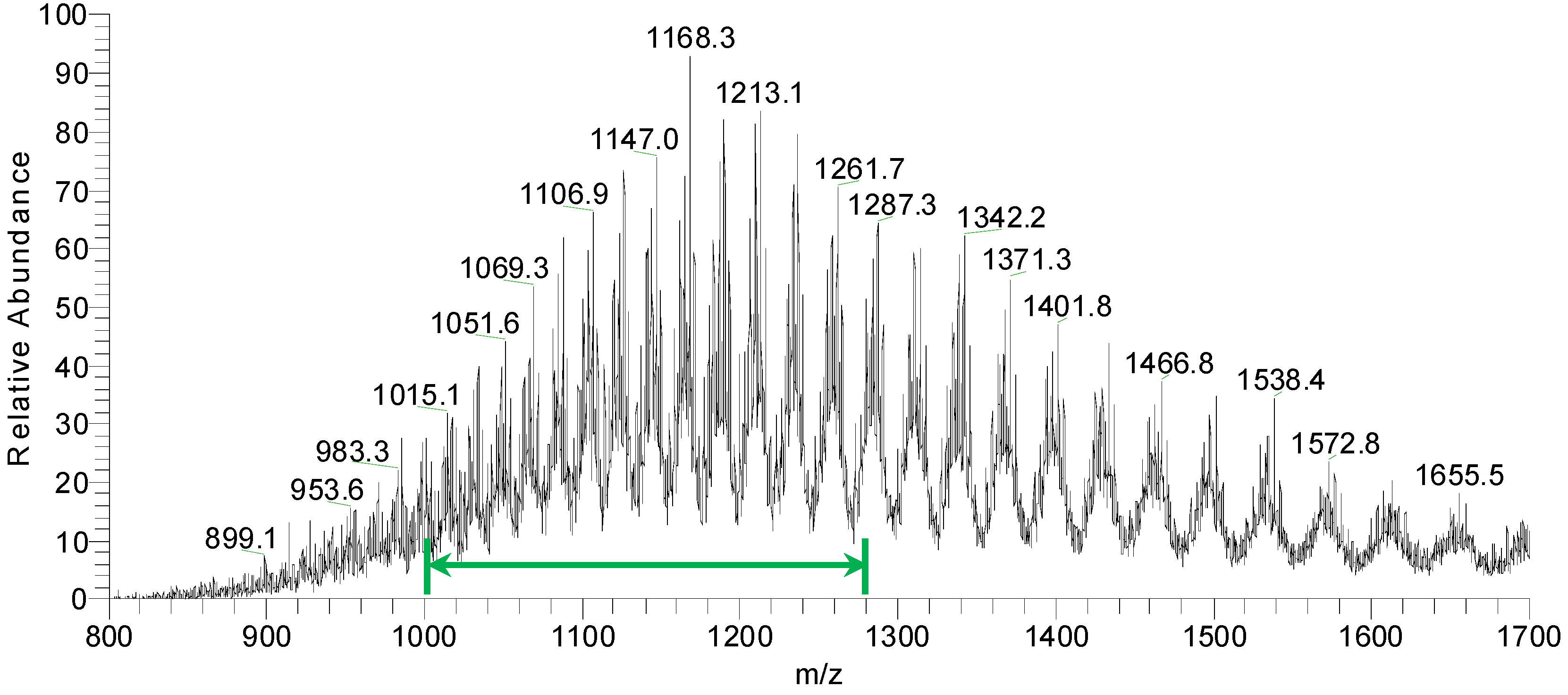
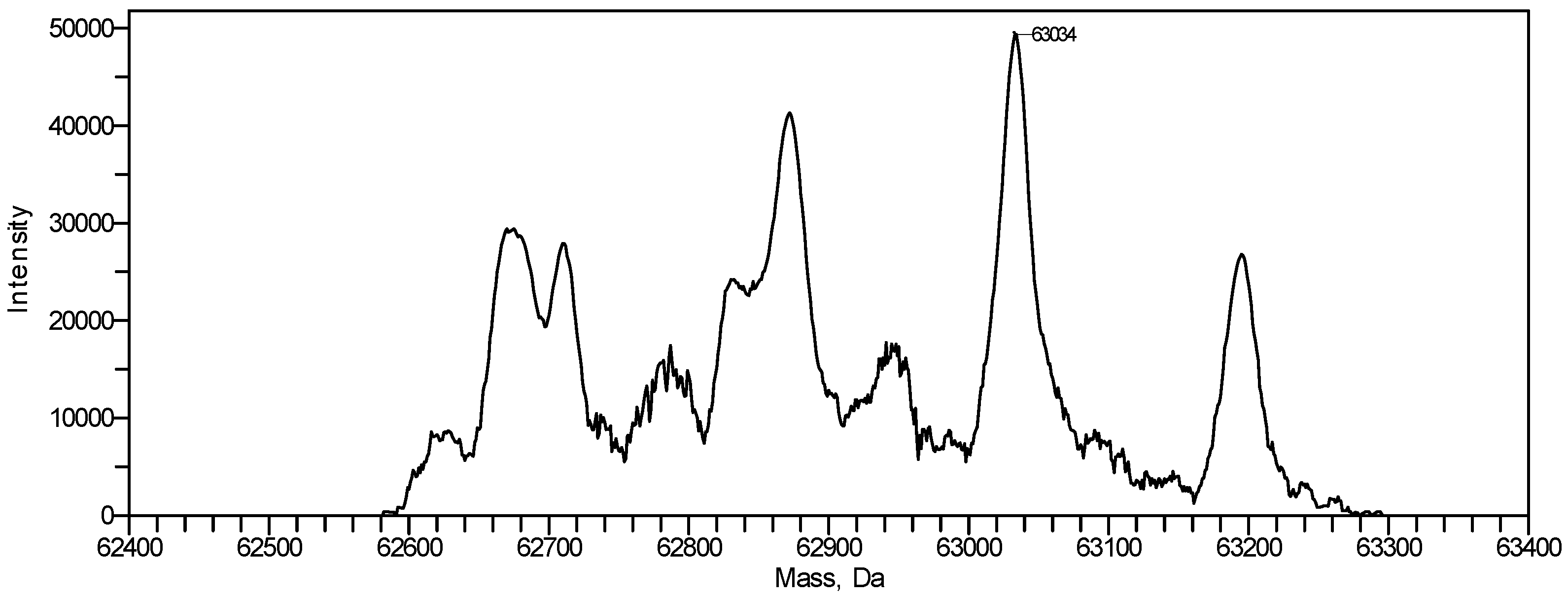
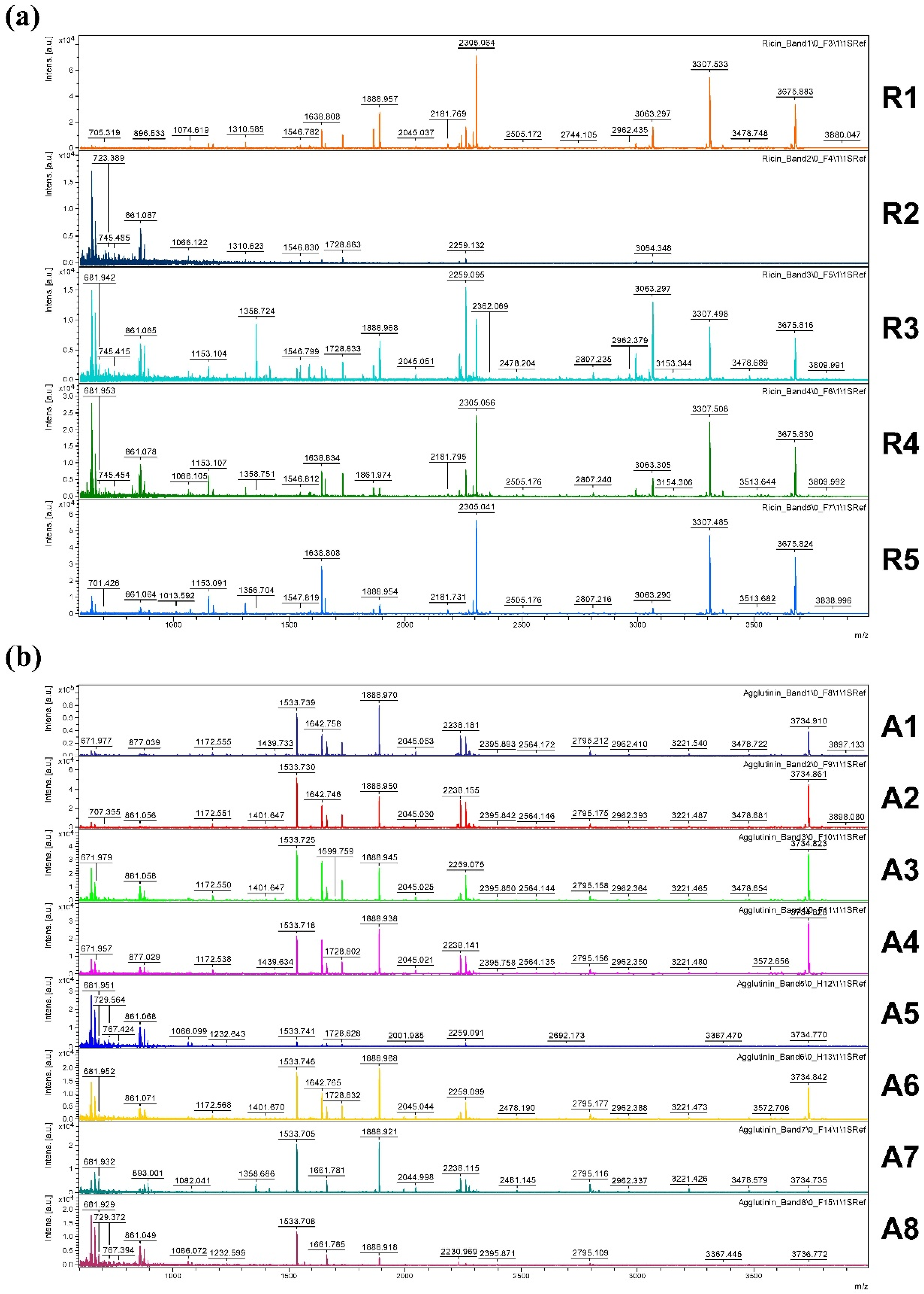
Figure 9 shows an overview of resulting mass spectra of trypsin-digested ricin and RCA120 bands. All ricin bands showed similar mass spectral patterns, except for band 2, presumably because of a lower protein amount. For the purified RCA120 preparation, all mass spectra were similar, except for bands 5 and 8, again presumably because of low protein amounts analyzed.
Figure 9.
Overview spectra of trypsin-digested samples for gel lanes containing the purified ricin or RCA120 preparation. From top to bottom: analysis of (a) bands R1 through R5 of the ricin preparation and (b) bands A1 through A8 of the RCA120 preparation. Peaks are labeled with the corresponding m/z value of the base peak.
Figure 9.
Overview spectra of trypsin-digested samples for gel lanes containing the purified ricin or RCA120 preparation. From top to bottom: analysis of (a) bands R1 through R5 of the ricin preparation and (b) bands A1 through A8 of the RCA120 preparation. Peaks are labeled with the corresponding m/z value of the base peak.
Table 7,
Table 8 and
Table 9 list the specific peptides identified as products of trypsin digestion from chains A and B of ricin D (Uniprot [
56] ID: P02879) as well from chain B of ricin E (Uniprot [
56] ID: Q41143) which could be found in the mass spectra of the indicated gel bands.
Table 7.
Ricin D chain A-specific peptides obtained by trypsin digestion (Uniprot [
56] ID: P02879, NCBI Genpept [
57] ID: GI:132567) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
Table 7.
Ricin D chain A-specific peptides obtained by trypsin digestion (Uniprot [56] ID: P02879, NCBI Genpept [57] ID: GI:132567) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
| Position | Amino Acid Sequence | (M+H)+ | R1 * | R2 * | R3 * | R4 * | R5 * |
|---|
| 67–74 | VGLPINQR | 896.5312 | x | - | - | x | x |
| 161–169 | LEQLAGNLR | 1013.5738 | x | x | - | x | x |
| 150–160 | YTFAFGGNYDR | 1310.5800 | x | x | x | x | x |
| 190–203 | SFIICIQMISEAAR | 1638.8342 | x | x | x | x | x |
| 67–83 | LTTGADVRHEIPVLPNR | 1888.0399 | x | - | x | x | x |
| 150–169 | YTFAFGGNYDRLEQLAGNLR | 2305.1360 | x | - | x | x | x |
| 121–149 | AGNSAYFFHPDNQEDAEAIT HLFTDVQNR | 3307.5038 | x | - | x | x | x |
Table 8.
Ricin D chain B-specific peptides obtained by trypsin digestion (Uniprot [
56] ID: P02879, NCBI GenPept [
57] ID: GI:132567) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
Table 8.
Ricin D chain B-specific peptides obtained by trypsin digestion (Uniprot [56] ID: P02879, NCBI GenPept [57] ID: GI:132567) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
| Position | Amino Acid Sequence | (M+H)+ | R1 * | R2 * | R3 * | R4 * | R5 * |
|---|
| 355–366 | SNTDANQLWTLK | 1390.6961 | x | - | x | - | - |
| 355–367 | SNTDANQLWTLKR | 1546.7972 | x | x | x | x | x |
| 330–342 | FHNGNAIQLWPCK | 1584.7740 | x | - | x | x | - |
| 534–550 | NDGTILNLYSGLVLDVR | 1862.0018 | x | - | x | x | x |
| 365–391 | CLTTYGYSPGVYVMIYDCNTAATDATR | 3063.3532 | x | x | x | x | x |
Table 9.
Ricin E chain B-specific peptides obtained by trypsin digestion (Uniprot [
56] ID: Q41143) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
Table 9.
Ricin E chain B-specific peptides obtained by trypsin digestion (Uniprot [56] ID: Q41143) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
| Position | Amino Acid Sequence | (M+H)+ | R1 * | R2 * | R3 * | R4 * | R5 * |
|---|
| 220–236 | NDGTILNLYNGLVLDVR | 1889.0127 | x | - | x | x | x |
All ricin bands R1 to R5 contained ricin chains A and B. Ricin E chain B could be observed in the ricin bands 1, and 3 to 5. Cross-contamination of RCA120 in the ricin preparation was found in bands R1 and R3, but only in one RCA120 peptide fragment (m/z 1533, SNTDWNQLWTLR). Based on these data, cross-contamination of RCA120 in the ricin preparation was very low, corroborating results obtained by the LC-ESI MS/MS analysis.
The corresponding specific trypsin digest peptides from RCA120, Uniprot [
56] ID: P06750, are displayed in
Table 10 and
Table 11. In the RCA120 preparation, only RCA120 peptides (Uniprot [
56] ID: P06750) and no ricin peptides were detected by MALDI-TOF MS analysis. Measured peptides belonged to RCA120 chains A and B, while band 8 showed peptides for RCA120 chain B only.
In summary, one-dimensional gel electrophoresis showed that the ricin preparation can be separated into five distinct bands with one main fragment at about 63 kDa, whereas purified RCA120 can be separated into eight bands with two main fragments running at around 170 kDa. The identity of the ricin and RCA120 bands was verified by MALDI-TOF-MS/MS analysis after trypsin digestion. All ricin bands contained ricin chains A and B of ricin D (Uniprot [
56] ID: P02879). Ricin bands 1 and 3 to 5 also contained ricin E (Uniprot [
56] ID: Q41143), while bands R1 and R3 also contained one peptide fragment (
m/
z 1533, SNTDWNQLWTLR) of RCA120. The cross-contamination was negligible. In purified RCA120 only RCA120 (Uniprot [
56] ID: P06750) and no ricin was detected. Measured peptides belonged to RCA120 chains A and B, while band 8 only showed peptides for RCA120 chain B. Most importantly, the MALDI-TOF-MS/MS experiments gave no indication of the presence of irrelevant proteins co-purified from the
R. communis seeds.
Table 10.
RCA120 chain A-specific peptides obtained by trypsin digestion (Uniprot [
56] ID: P06750, NCBI Genpept [
57] ID: GI113504) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
Table 10.
RCA120 chain A-specific peptides obtained by trypsin digestion (Uniprot [56] ID: P06750, NCBI Genpept [57] ID: GI113504) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
| Position | Amino Acid Sequence | (M+H)+ | A1 * | A2 * | A3 * | A4 * | A5 * | A6 * | A7 * | A8 * |
|---|
| 73–80 | VGLPISQR | 869.5203 | x | x | - | - | - | - | - | - |
| 150–157 | LEQLGGLR | 885.5152 | x | x | x | x | - | x | - | - |
| 190–203 | SFMVCIQMISEAAR | 1642.7750 | x | x | x | x | x | x | - | - |
| 64–80 | HEIPVLPNRVGLPISQR | 1925.1079 | x | x | - | - | - | - | - | - |
| 263–281 | FNVYDVSILIPIIALMVYR | 2238.2545 | x | x | x | x | - | x | x | - |
| 158–189 | ENIELGTGPLEDAISALYYYSTCGTQIPTLAR | 3516.7202 | x | x | x | x | - | x | - | - |
Table 11.
RCA120 chain B-specific peptides obtained by trypsin digestion (Uniprot [
56] ID: P06750, NCBI Genpept [
57] ID: GI225114) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
Table 11.
RCA120 chain B-specific peptides obtained by trypsin digestion (Uniprot [56] ID: P06750, NCBI Genpept [57] ID: GI225114) with one missed cleavage, reduced and carbamidomethylated in the mass range of 600–4000, and obtained by mass spectrometry analysis of the respective gel bands.
| Position | Amino Acid Sequence | (M+H)+ | A1 * | A2 * | A3 * | A4 * | A5 * | A6 * | A7 * | A8 * |
|---|
| 343–354 | SNTDWNQLWTLR | 1533.7444 | x | x | x | x | x | x | x | x |
| 343–355 | SNTDWNQLWTLRK | 1661.8394 | x | x | x | x | x | x | x | x |
| 319–342 | NGLCVDVFGEEFTDGNPIQLWPCK | 2795.2803 | x | x | x | x | - | x | x | x |