Within-Mat Variability in Anatoxin-a and Homoanatoxin-a Production among Benthic Phormidium (Cyanobacteria) Strains
Abstract
:1. Introduction
2. Results
| Strain | Location | anaF PCR | LC-MS data (mg kg−1) | ||||
|---|---|---|---|---|---|---|---|
| ATX | dhATX | HTX | dhHTX | TOTAL | |||
| CYN103 | WR | + | 5.92 | 205.92 | ND | ND | 211.83 |
| CYN104 | WR | + | 5.79 | 156.07 | ND | ND | 161.86 |
| CYN105 | WR | + | 0.28 | 40.80 | ND | ND | 41.08 |
| CYN106 | WR | + | 0.47 | 56.39 | ND | ND | 56.86 |
| CYN107 | WR | + | 0.93 | 9.13 | ND | ND | 10.06 |
| CYN108 | WR | - | ND | ND | ND | ND | ND |
| CYN109 | WR | + | 3.25 | 128.38 | ND | ND | 131.63 |
| CYN110 | WR | + | 6.40 | 171.80 | ND | ND | 178.20 |
| CYN111 | WR | + | 1.08 | 71.90 | ND | ND | 72.98 |
| CYN112 | WR | + | 2.38 | 165.75 | 1.00 | ND | 169.13 |
| CYN113 | HR-1-1 | + | 0.04 | 66.91 | ND | ND | 66.95 |
| CYN115 | HR-1-1 | + | 0.25 | 28.57 | ND | ND | 28.83 |
| CYN116 | HR-1-1 | + | 0.04 | 31.29 | ND | ND | 31.33 |
| CYN117 | HR-1-1 | + | 0.12 | 152.16 | ND | ND | 152.28 |
| CYN118 | HR-2 | + | 0.02 | 2.19 | ND | ND | 2.21 |
| CYN119 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN120 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN121 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN122 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN123 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN124 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN125 | HR-2 | - | ND | ND | ND | ND | ND |
| CYN126 | HR-1-2 | + | 0.19 | 115.23 | ND | ND | 115.42 |
| CYN127 | HR-1-2 | + | 0.63 | 79.92 | ND | ND | 80.55 |
| CYN128 | HR-1-2 | + | 0.12 | 32.58 | ND | ND | 32.70 |
| CYN129 | HR-1-2 | - | ND | ND | ND | ND | ND |
| CYN130 | HR-1-2 | - | ND | ND | ND | ND | ND |
| CYN131 | HR-1-2 | + | ND | 4.75 | ND | ND | 4.75 |
| CYN132 | HR-1-2 | + | 0.25 | 74.32 | ND | ND | 74.58 |
| CYN133 | HR-1-2 | + | 0.06 | 73.56 | ND | ND | 73.62 |
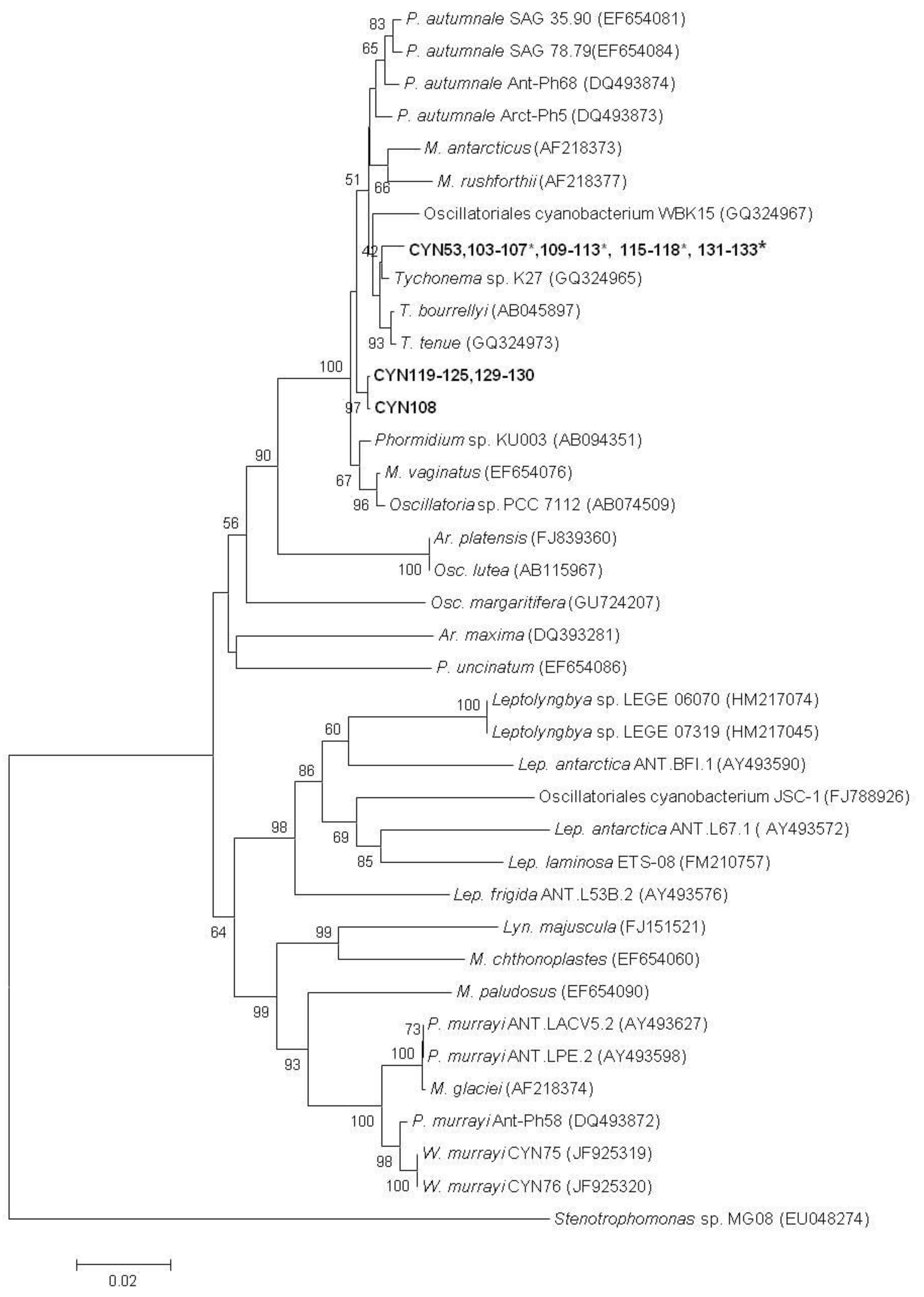
3. Discussion
4. Methods
4.1. Sample Sites and Collection
4.2. Cyanobacterial Culture Isolation
4.3. Anatoxin-a and Homoanatoxin-a Extraction and Detection
4.4. Morphological and Molecular Analysis
5. Conclusions
Acknowledgements
Conflict of Interest
References
- Gugger, M.; Lenoir, S.; Berger, C.; Ledreux, A.; Druart, J.C.; Humbert, J.F.; Guette, C.; Bernard, C. First report in a river in France of the benthic cyanobacterium Phormidium favosum producing anatoxin-a associated with dog neurotoxicosis. Toxicon 2005, 45, 919–928. [Google Scholar] [CrossRef]
- Heath, M.; Wood, S.A.; Ryan, K. Polyphasic assessment of fresh-water benthic mat forming cyanobacteria isolated from New Zealand. FEMS Ecol. Microbiol. 2010, 73, 95–109. [Google Scholar]
- Wood, S.A.; Kuhajek, J.; de Winton, M.; Phillips, N.R. Species composition and cyanotoxin production in periphyton mats from three lakes of varying trophic status. FEMS Microbiol. Ecol. 2012, 79, 312–326. [Google Scholar] [CrossRef]
- Wood, S.A.; Selwood, A.I.; Rueckert, A.; Holland, P.T.; Milne, J.R.; Smith, K.F.; Smits, B.; Watts, L.F.; Cary, C.S. First report of homoanatoxin-a and associated dog neurotoxicosis in New Zealand. Toxicon 2007, 50, 292–301. [Google Scholar] [CrossRef]
- Carmichael, W.W. The toxins of cyanobacteria. Sci. Am. 1994, 270, 78–86. [Google Scholar] [CrossRef]
- Faassen, E.J.; Harkema, L.; Begeman, L.; Lurling, M. First report of (homo)anatoxin-a and dog neurotoxicosis after ingestion of benthic cyanobacteria in The Netherlands. Toxicon 2012, 60, 378–384. [Google Scholar] [CrossRef]
- Puschner, B.; Pratt, C.; Tor, E.R. Treatment and diagnosis of a dog with fulminant neurological deterioration due to anatoxin-a intoxication. J. Vet. Emerg. Crit. Care 2010, 20, 518–522. [Google Scholar] [CrossRef]
- Wood, S.A.; Young, R.G. Review of Benthic Cyanobacteria Monitoring Programme 2012; Cawthron Report No. 2217; Horizon Regional Council: Palmerston North, New Zealand, 2012; p. 30. [Google Scholar]
- Wood, S.A.; Young, R. Benthic Cyanobacteria and Toxin Production in the Manawatu-Wanganui Region; Cawthron Report No. 1959; Horizon Regional Council: Palmerston North, New Zealand, 2011; p. 34. [Google Scholar]
- Wood, S.A.; Heath, M.W.; Kuhajek, J.; Ryan, K.G. Fine-scale spatial variability in anatoxin-a and homoanatoxin-a concentrations in benthic cyanobacterial mats: Implication for monitoring and management. J. Appl. Microbiol. 2010, 109, 2011–2018. [Google Scholar] [CrossRef]
- Kotak, B.G.; Lam, A.K.Y.; Prepas, E.E.; Kenefick, S.L.; Hrudey, S.E. Variability of the hepatotoxin microcystin-LR in hypereutrophic drinking water lakes. J. Phycol. 1995, 31, 248–263. [Google Scholar]
- Vezie, C.; Brient, L.; Sivonen, K.; Bertru, G.; Lefeuvre, J.C.; Salkinoja-Salonen, M. Variation of microcystin content of cyanobacterial blooms and isolated strains in Grand-Lieu lake (France). Microbial. Ecol. 1998, 35, 126–135. [Google Scholar] [CrossRef]
- Ballot, A.; Fastner, J.; Lentz, M.; Wiedner, C. First report of anatoxin-a-producing cyanobacterium Aphanizomenon issatschenkoi in northeastern Germany. Toxicon 2010, 56, 964–971. [Google Scholar] [CrossRef]
- Velzeboer, R.M.A.; Baker, P.D.; Rositano, J.; Heresztyn, T.; Codd, G.A.; Raggett, S.L. Geographical patterns of occurrence and composition of saxitoxins in the cyanobacterial genus Anabaena (Nostocales, Cyanophyta) in Australia. Phycologia 2000, 39, 395–407. [Google Scholar] [CrossRef]
- Vaitomaa, J.; Rantala, A.; Halinen, K.; Rouhiainen, L.; Tallberg, P.; Mokelke, L.; Sivonen, K. Quantitative real-time PCR for determination of microcystin synthetase E copy number for Microcystis and Anabaena in lakes. Appl. Environ. Microbiol. 2003, 69, 7289–7297. [Google Scholar] [CrossRef]
- Via-Ordorika, L.; Fastner, J.; Kurmayer, R.; Hisbergues, M.; Dittmann, E.; Komárek, J.; Erhard, M.; Chorus, I. Distribution of microcystin-producing and non-microcystin-producing Microcystis sp. in European freshwater bodies: Detection of microcystins and microcystin genes in individual colonies. Syst. Appl. Microbiol. 2004, 27, 582–602. [Google Scholar]
- Kurmayer, R.; Christiansen, G.; Fastner, J.; Börner, T. Abundance of active and inactive microcystin genotypes in populations of the toxic cyanobacterium Planktothrix spp. Environ. Microbiol. 2004, 6, 831–841. [Google Scholar] [CrossRef]
- Saker, M.L.; Fastner, J.; Dittmann, E.; Christiansen, G.; Vasconcelos, V.M. Variation between strains of the cyanobacterium Microcystis aeruginosa isolated from a Portuguese river. J. Appl. Microbiol. 2005, 99, 749–757. [Google Scholar] [CrossRef]
- Izaguirre, G.; Jungblut, A.D.; Neilan, B.A. Benthic cyanobacteria (Oscillatoriaceae) that produce microcystin-LR, isolated from four reservoirs in southern California. Water Res. 2007, 41, 492–498. [Google Scholar] [CrossRef]
- Seifert, M.; McGregor, G.; Eaglesham, G.; Wickramasinghe, W.; Shaw, G. First evidence for the production of cylindrospermopsin and deoxy-cylindrospermopsin by the freshwater benthic cyanobacterium, Lyngbya wollei (Farlow ex Gomont) Speziale and Dyck. Harmful Algae 2007, 6, 73–80. [Google Scholar] [CrossRef]
- Fiore, M.F.; Genuário, D.B.; da Silva, C.S.P.; Shishido, T.K.; Moraes, L.A.B.; Neto, R.C.; Silva-Stenico, M.E. Microcystin production by a freshwater spring cyanobacterium of the genus Fischerella. Toxicon 2009, 53, 754–761. [Google Scholar] [CrossRef]
- Wood, S.A.; Heath, M.; McGregor, G.; Holland, P.T.; Munday, R.; Ryan, K. Identification of a benthic microcystin producing filamentous cyanobacterium (Oscillatoriales) associated with a dog poisoning in New Zealand. Toxicon 2010, 55, 987–903. [Google Scholar]
- Smith, F.; Wood, S.A.; Ginkel, R.V.; Broady, P.; Gaw, S. First report of saxitoxin production by a species of the freshwater benthic cyanobacterium, Scytonema Agardh. Toxicon 2011, 57, 566–573. [Google Scholar] [CrossRef]
- Cadel-Six, S.; Peyraud-Thomas, C.; Brient, L.; de Marsac, N.T.; Rippka, R.; Méjean, A. Different genotypes of anatoxin-producing cyanobacteria coexist in the Tarn River, France. Appl. Environ. Microbiol. 2007, 73, 7605–7614. [Google Scholar] [CrossRef]
- Rinta-Kanto, J.M.; Ouellette, A.J.; Boyer, G.L.; Twiss, M.R.; Bridgeman, T.B.; Wilhelm, S.W. Quantification of toxic Microcystis spp. during the 2003 and 2004 blooms in western Lake Erie using quantitative real-time PCR. Environ. Sci. Technol. 2005, 39, 4198–4205. [Google Scholar] [CrossRef]
- Rantala-Ylinen, A.; Känä, S.; Wang, H.; Rouhiainen, L.; Wahlsten, M.; Rizzi, E.; Berg, K.; Gugger, M.; Sivonen, K. Anatoxin-a synthetase gene cluster of the cyanobacterium Anabaena sp. strain 37 and molecular methods to detect potential producers. Appl. Environ. Microbiol. 2011, 77, 7271–7278. [Google Scholar] [CrossRef]
- Wood, S.A.; Rhodes, L.; Adams, S.; Adamson, J.; Smith, K.; Smith, J.; Tervit, R.; Cary, S.C. Maintenance of cyanotoxin production by cryopreserved New Zealand culture collection cyanobacteria. N.Z. J. Mar. Freshwat. Res. 2008, 42, 277–283. [Google Scholar] [CrossRef]
- Devlin, J.P.; Edwards, O.E.; Gorham, P.R.; Hunter, M.R.; Pike, R.K.; Stavric, B. Anatoxin-a, a toxic alkaloid from Anabaena flos-aquae NCR-44. Can. J. Chem. 1977, 55, 1367–1371. [Google Scholar] [CrossRef]
- Skulberg, O.M.; Carmichael, W.W.; Andersen, R.A.; Matsunaga, S.; Moore, R.E.; Skulberg, R. Investigations of a neurotoxic Oscillatorialean strain (Cyanophyceae) and its toxin isolation and characterization of homoanatoxin-a. Environ. Tox. Chem. 1992, 11, 321–329. [Google Scholar] [CrossRef]
- Bates, H.A.; Rapoport, H. Synthesis of anatoxin-a via intramolecular cyclization of iminium salts. J. Am. Chem. Soc. 1979, 101, 1259–1265. [Google Scholar] [CrossRef]
- Comte, K.; Sabacka, M.; Carre-Mlouka, A.; Elster, J.; Komarek, J. Relationships between the Arctic and the Antarctic cyanobacteria; three Phormidium like strains evaluated by a polyphasic approach. FEMS Microbiol. Ecol. 2007, 59, 366–376. [Google Scholar] [CrossRef]
- Palinska, K.A.; Marquardt, J. Genotypic and phenotypic analysis of strains assigned to the widespread cyanobacterial morphospecies Phormidium autumnale (Oscillatoriales). Arch. Microbiol. 2008, 189, 325–335. [Google Scholar] [CrossRef]
- Lyra, C.; Suomalainen, S.; Gugger, M.; Vezie, C.; Sundman, P.; Paulin, L.; Sivonen, K. Molecular characterization of planktic cyanobacteria of Anabaena, Aphanizomenon, Microcystis and Planktothrix genera. Int. J. Syst. Evol. Microbiol. 2001, 51, 513–526. [Google Scholar]
- Neilan, B.A.; Jacobs, D.; del Dot, T.; Blackall, L.L.; Hawkins, P.R.; Cox, P.T.; Goodman, A.E. rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Bacteriol. 1997, 47, 693–697. [Google Scholar] [CrossRef]
- Baker, J.; Entsch, B.; Neilan, B.; McKay, D. Monitoring changing toxigenicity of a cyanobacterial bloom by molecular methods. Appl. Environ. Microbiol. 2002, 68, 6070–6076. [Google Scholar] [CrossRef]
- Ouellette, A.J.; Wilhelm, S.W. Toxic cyanobacteria: The evolving molecular toolbox. Front. Ecol. Environ. 2003, 1, 359–366. [Google Scholar] [CrossRef]
- Komárek, J. Problems in Cyanobacterial Taxonomy: Implications for Most Common Toxin Producing Species. In Workshop the Blooms of Toxic Algae in Fresh Waters: Health Emergency and Control Measures; Melchiorre, S., Viaggiu, E., Bruno, M., Eds.; National Institute of Health: Rome, Italy, 2000; pp. 6–43. [Google Scholar]
- Bolch, C.J.S.; Blackburn, S.I. Isolation and purification of Australian isolates of the toxic cyanobacterium Microcystis aeruginosa Kütz. J. Appl. Phycol. 1996, 8, 5–13. [Google Scholar] [CrossRef]
- Komárek, J.; Anagnostidis, K. Cyanoprokaryota: 2. Teil: Oscillatoriales; Elsevier GmbH: München, Germany, 2005; Volume 19/2, pp. 1–759. [Google Scholar]
- McGregor, G.B. Freshwater Cyanoprokaryota of North-Eastern Australia 1: Oscillatoriales; Australian Biological Resources Study: Canberra, Australia, 2007; p. 123. [Google Scholar]
- Méjean, A.; Mann, S.; Maldiney, T.; Vassiliadis, G.; Lequin, O.; Ploux, O. Evidence that biosynthesis of the neurotoxic alkaloids anatoxin-a and homoanatoxin-a in the cyanobacterium Oscillatoria PCC 6506 occurs on a modular polyketide synthase initiated by l-proline. J. Am. Chem. Soc. 2009, 131, 7512–7513. [Google Scholar]
- Jungblut, A.; Neilan, B.A. Molecular identification and evolution of the cyclic peptide hepatotoxins, microcystin and nodularin, synthetase genes in three orders of cyanobacteria. Arch. Microbiol. 2006, 185, 107–114. [Google Scholar] [CrossRef]
- Frazão, B.; Martins, R.; Vasconcelos, V. Are known cyanotoxins involved in the toxicity of picoplanktonic and filamentous North Atlantic marine cyanobacteria? Mar. Drugs 2010, 8, 1908–1919. [Google Scholar] [CrossRef]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2008, 36, D25–D30. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Heath, M.; Wood, S.A.; Ryan, K. Spatial and temporal variability in Phormidium sp. abundance and toxin production in the Hutt River, New Zealand. Aquat. Environ. Microbiol. 2011, 64, 69–79. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wood, S.A.; Smith, F.M.J.; Heath, M.W.; Palfroy, T.; Gaw, S.; Young, R.G.; Ryan, K.G. Within-Mat Variability in Anatoxin-a and Homoanatoxin-a Production among Benthic Phormidium (Cyanobacteria) Strains. Toxins 2012, 4, 900-912. https://doi.org/10.3390/toxins4100900
Wood SA, Smith FMJ, Heath MW, Palfroy T, Gaw S, Young RG, Ryan KG. Within-Mat Variability in Anatoxin-a and Homoanatoxin-a Production among Benthic Phormidium (Cyanobacteria) Strains. Toxins. 2012; 4(10):900-912. https://doi.org/10.3390/toxins4100900
Chicago/Turabian StyleWood, Susanna A., Francine M. J. Smith, Mark W. Heath, Thomas Palfroy, Sally Gaw, Roger G. Young, and Ken G. Ryan. 2012. "Within-Mat Variability in Anatoxin-a and Homoanatoxin-a Production among Benthic Phormidium (Cyanobacteria) Strains" Toxins 4, no. 10: 900-912. https://doi.org/10.3390/toxins4100900
APA StyleWood, S. A., Smith, F. M. J., Heath, M. W., Palfroy, T., Gaw, S., Young, R. G., & Ryan, K. G. (2012). Within-Mat Variability in Anatoxin-a and Homoanatoxin-a Production among Benthic Phormidium (Cyanobacteria) Strains. Toxins, 4(10), 900-912. https://doi.org/10.3390/toxins4100900






