Genetics of Dothistromin Biosynthesis in the Peanut Pathogen Passalora arachidicola
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Culture Conditions and Densitometric Quantification of Dothistromin
2.2. Identification of Dothistromin Gene Fragments by Degenerate PCR
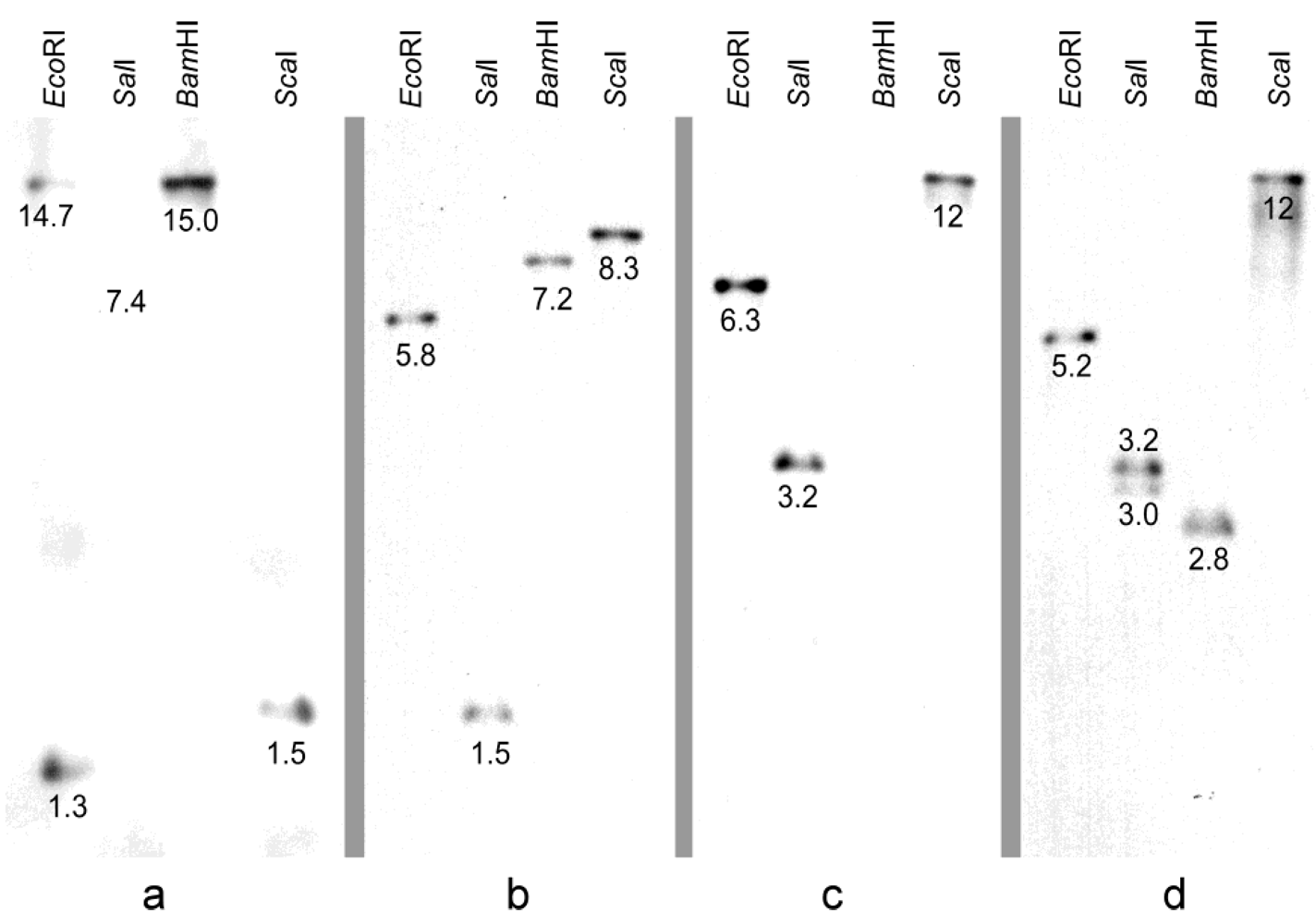
2.3. Isolation and Identification of Clustered Dothistromin Genes
2.4. RNA Isolation and RT-PCR
2.5. Preparation of NZ46 Pa-dotA and Pa-vbsA Gene Complementation Constructs
2.6. Functional Identification of P. Arachidicola Dothistromin Genes by Complementation of D. Septosporum Mutants
2.7. Bioinformatics
3. Results and Discussion
3.1. Isolation of P. arachidicola Dothistromin Genes cypA, dotA, pksA and vbsA and Functional Identification by Complementation
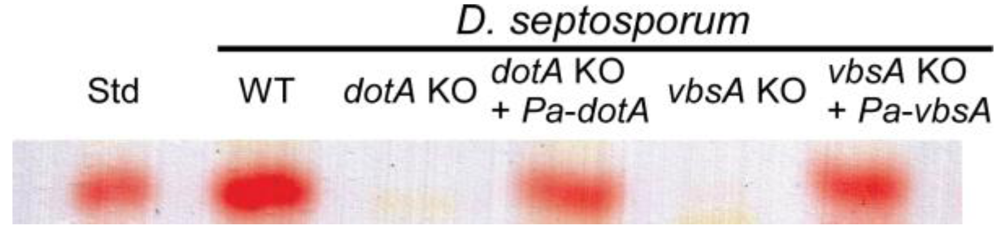
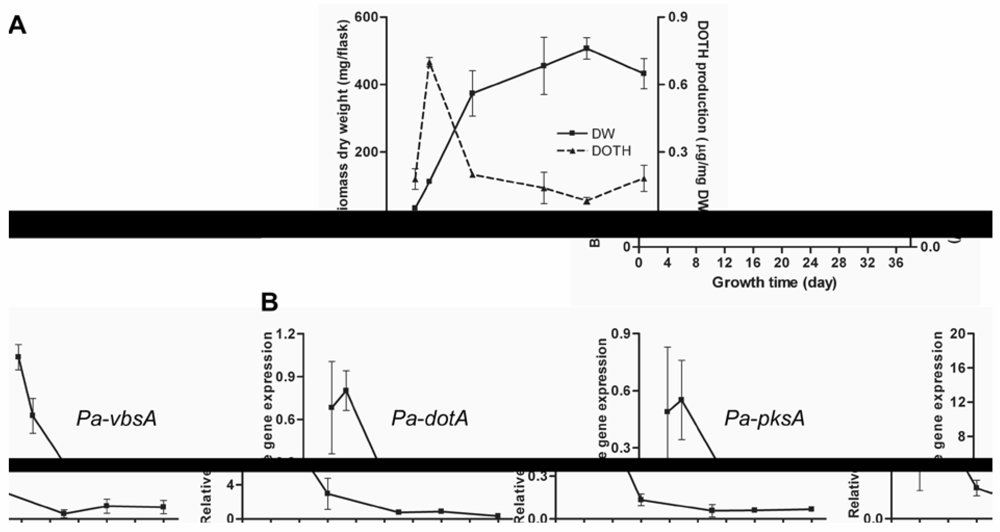
3.2. Regulation of Dothistromin Biosynthesis in P. Arachidicola
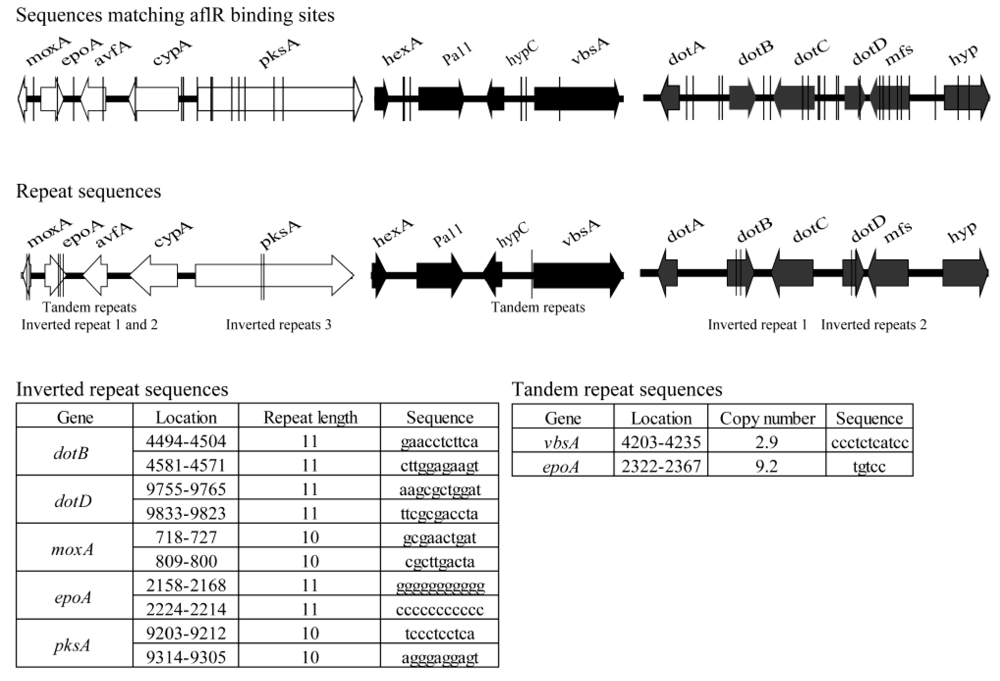
3.3. Identification and Characterization of Three Groups of Dothistromin Genes
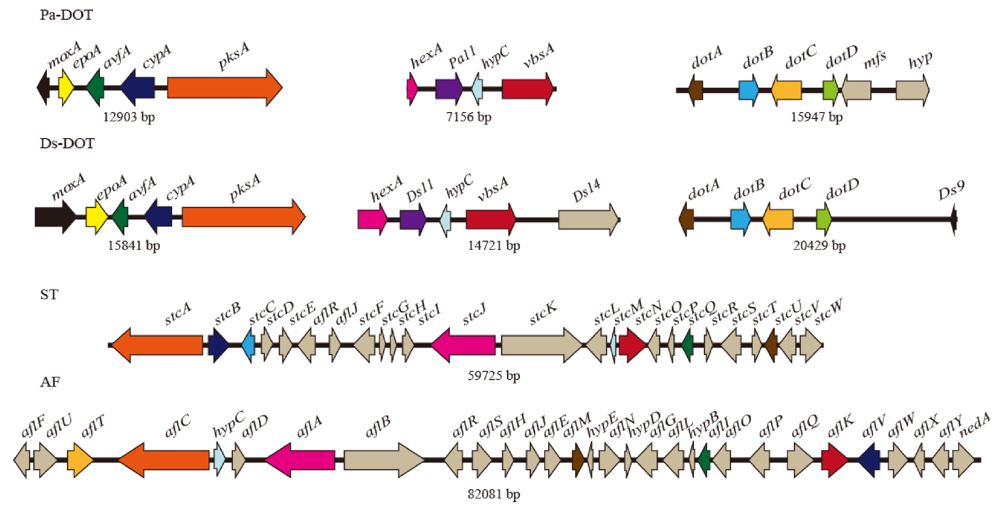
| doth gene | Putative function | % nt ID | % aa ID | ST/AF ortholog | % aa ID | Intron number | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pa/Ds | Pa/Ds | An | Ap | Pa/An | Ds/An | Pa/Ap | Ds/Ap | Pa | Ds | An | Ap | ||
| dotA | ketoreductase | 81.8 | 95.8 | stcU | aflM | 78.8 | 79.1 | 79.1 | 80.2 | 2 | 2 | 2 | 2 |
| dotB | oxidase | 77.9 | 84.3 | stcC | - | 15.6 | 24.0 | - | - | 0 | 0 | 0 | - |
| dotC | MFS transporter | 76.1 | 82.3 | - | aflT | - | - | 25.7 | 31.2 | 2 | 3 | - | 5 |
| dotD | thioesterase | 74.3 | 75.5 | stcA | aflC | 28.1 | 37.9 | 31.6 | 34.8 | 0 | 0 | 2 | 5 |
| mfs | MFS transporter | - | - | - | aflT | - | - | 14.3 | 7 | - | - | - | |
| pksA | polyketide synthase | 77.9 | 88.7 | stcA | aflC | 52.5 | 57.0 | 54.5 | 54.8 | (2) | 2 | 2 | 5 |
| cypA | monooxygenase | 82.3 | 92.8 | stcB | aflV | 55.5 | 59.8 | 60.0 | 59.3 | 2 | 2 | 3 | 2 |
| avfA | oxidase | 66.4 | 73.3 | stcO | aflI | 47.0 | 43.7 | 46.9 | 47.8 | 0 | 1 | 0 | 0 |
| epoA | epoxide hydrolase | 30 | 22.5 | - | - | - | - | - | - | 1 | 1 | - | - |
| moxA | monooxygenase | 71.9 | 88.9 | stcW | aflW | 52.5 | 59.0 | 57.3 | 55.1 | (3) | 5 | 3 | 0 |
| hexA | fatty acid synthase | 69.3 | 73.8 | stcJ | aflA | 40.8 | 41.3 | 47.0 | 48.8 | (0) | 3 | 1 | 2 |
| Pa11 | unknown | 81.1 | 85.2 | - | - | - | - | - | - | 3 | 2 | - | - |
| hypC | anthrone oxidase | 77.1 | 86.7 | stcM | hypC | 46.9 | 47.9 | 37.8 | 35.2 | 0 | 0 | 0 | 0 |
| vbsA | ver. B synthase | 85.8 | 94.8 | stcN | aflK | 70.9 | 69.1 | 71.2 | 72.0 | 1 | 1 | 2 | 1 |
4. Conclusions
Acknowledgements
Appendix
| Target gene | Primer name (in-house number) | Sequence (5’ - 3’) | Size of PCR product (bp) |
|---|---|---|---|
| Primers used for initial amplification of dothistromin genes by degenerate PCR | |||
| Pa-dotA | Keto-ver5’ (#356) | GGIATHGGIGCIGCIATHGC | 708 |
| Keto-ver3’ (#355) | TCRTCIACYTGYTCRTCIGTRAA | ||
| Pa-cypA | JSdmo1r (#132) | CGTCAGTGATGATGTCCG | 949 |
| dmoCA (#119) | ATTATGGACGCCACTGCTGG | ||
| Pa-vbsA | vbs-1aF (#362) | TNTTYGACTGGTAYCARTAYAC | 1351 |
| vbs-4R (#366) | TAGTTBAGDATYTCBTCRTCSGTCTG | ||
| Pa-pksA | pksCB2 (#152) | TGAAGAAGTATGTCGCCG | 1306 |
| OTpks KO/2 | CTGTCTAGAGTGTTGGTCTCCATCC | ||
| Primers used for dothistromin gene expression by RT-PCR (Figure 2) (* indicates primer flanks an intron to prevent gDNA amplification) | |||
| Pa-tub | SZDP-146 (#865)* | CGGCTCCGGCGTGTACAATG | 214 |
| SZDP-147 (#866) | GGCCCAGTTGTTTCCAGCAC | ||
| Pa-dotA | SZDP-143 (#862)* | ACAGGTTGCGAAGATGATGG | 474 |
| SZDP-145(#864)* | AGGCCAACACGTTCTAGAGG | ||
| Pa-pksA | MGPA65 (#803) | GTCTCCAGAATTGCCTTGAC | 665 |
| SZDP-142 (#861)* | TCACATTATGTGGTTCGGGC | ||
| Pa-vbsA | MGPA31 (#768) | GGGCTCGATGCTCAATACTC | 366 |
| SZDP-140 (#859)* | CGATTGTGGAGACCCTCTTG | ||
| Primers used for construction of the complementation construct pR280 | |||
| Pa-dotA orf and 3’UTR | SZDP-137 (#724) | CACACCACCTCATCTCCCATAATGTCTGCCGACAACTTCC | 1276 |
| SZDP-130 (#599) | GCGGCCGCTCCCTCTTATCGTTTCATAG | ||
| Ds-dotA promoter | SZDP-135 (#722) | ATAGGGCGAATTGGGTACCG | 962 |
| SZDP-136 (#723) | GGAAGTTGTCGGCAGACATTATGGGAGATGAGGTGGTGTG | ||
| Primers used for inverse PCR | |||
| Pa-dotA end | SZDP-82 (#538) | GCAACGAAGAATTGACCACG | 1000 |
| SZDP-83 (#539) | GATATGTACGCTGCTGTTGC | ||
| Pa-moxA end | MGPA66 (#804) | TGAGCCTGTGGCTCCTGTTG | 2100 |
| MGPA69 (#807) | GTACTGTCTGCGCACAACTC | ||
| Pa-pksA end | MGPA22 (#757) | GATCTCCCTTCGAGTCTTTG | 900 |
| MGPA35 (#772) | TCGACAGCTTGAGCTCTATG | ||
| Pa-hexA and Pa-vbsA ends | MGPA46 (#783) | TGGTGCCGTGGTCATCCTAC | 2200 |
| MGPA79 (#886) | ATCGCCGAACACTTCCACTC | ||
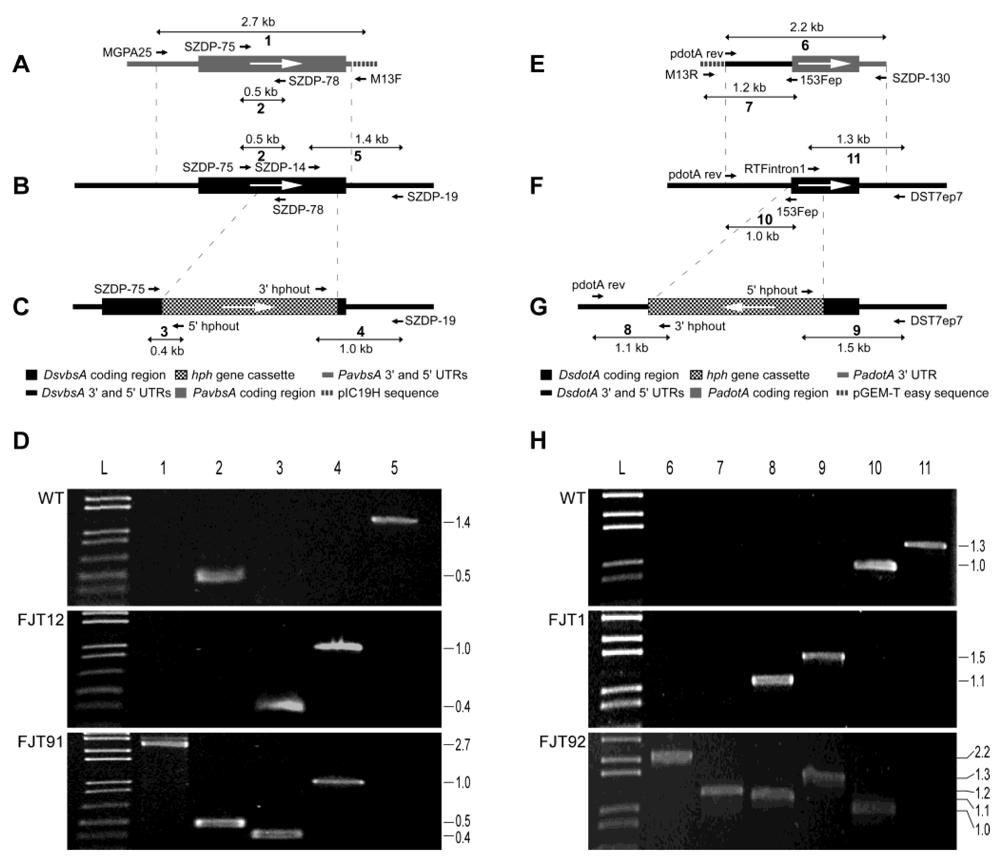
| Primers used for identification of gene knockout and complementation (Figure S2) | |||
| Reaction | Primer name | Sequence (5’ - 3’) | Product (bp) |
| 1 | MGPA25 (#762) | GGGACATGTTGCTGGTTGTG | 2691 |
| M13 forward | GTAAAACGACGGCCAG | ||
| 2 | SZDP-75 (#531) | CAGTCCACGTTGCGTATCCA | 480 |
| SZDP-78 (#534) | GGCCGAGGATGATGGTGTCT | ||
| 3 | SZDP-75 (#531) | CAGTCCACGTTGCGTATCCA | 370 |
| 5’hphout (#35) | GAATCTCCGGTGTGGAAGA | ||
| 4 | 3’hphout (#36) | TCCTTGAACTCTCAAGCCTACAG | 953 |
| SZDP-19 (#475) | GTCGGATCGAAGCTCTTCAT | ||
| 5 | SZDP-14 (#470) | GTGTCGGCCAGAACATGCAA | 1389 |
| SZDP-19 (#475) | GTCGGATCGAAGCTCTTCAT | ||
| 6 | pdotA rev (#679) | GTCTCTTAAGAATCTTCGTCGCTCAGCT | 2234 |
| SZDP-130 (#599) | GCGGCCGCTCCCTCTTATCGTTTCATAG | ||
| 7 | M13 reverse | CAGGAAACAGCTATGAC | 1183 |
| 153Fep (#59) | GTCGACGGACATTATGGGAGATG | ||
| 8 | pdotA rev (#679) | GTCTCTTAAGAATCTTCGTCGCTCAGCT | 1077 |
| 3’hphout (#36) | TCCTTGAACTCTCAAGCCTACAG | ||
| 9 | 5’hphout (#35) | GAATCTCCGGTGTGGAAGA | 1516 |
| DsT7ep7 (#701) | ATTCGGCTACATGCCCTACAC | ||
| 10 | pdotA rev (#679) | GTCTCTTAAGAATCTTCGTCGCTCAGCT | 971 |
| 153Fep (#59) | GTCGACGGACATTATGGGAGATG | ||
| 11 | RTFintron1 (#73) | CTGGTGATGTACGTTGTAAAC | 1315 |
| DsT7ep7 (#701) | ATTCGGCTACATGCCCTACAC | ||
References
- Van der Does, H.C.; Rep, M. Virulence genes and the evolution of host specificity in plant-pathogenic fungi. Mol. Plant Microbe Interact. 2007, 20, 1175–1182. [Google Scholar]
- Friesen, T.L.; Faris, J.D.; Solomon, P.S.; Oliver, R.P. Host-specific toxins: Effectors of necrotrophic pathogenicity. Cell. Microbiol. 2008, 10, 1421–1428. [Google Scholar]
- Elliott, C.E.; Gardiner, D.M.; Thomas, G.; Cozijnsen, A.; De Wouw, A.V.; Howlett, B.J. Production of the toxin sirodesmin PL by Leptosphaeria maculans during infection of Brassica napus. Molec. Plant Pathol. 2007, 8, 791–802. [Google Scholar] [CrossRef]
- Stoessl, A.; Abramowski, Z.; Lester, H.H.; Rock, G.L.; Towers, G.H.N. Further toxic properties of the fungal metabolite dothistromin. Mycopathologia 1990, 112, 179–186. [Google Scholar]
- Stoessl, A. Dothistromin as a metabolite of Cercospora arachidicola. Mycopathologia 1984, 86, 165–168. [Google Scholar] [CrossRef] [PubMed]
- Schwelm, A.; Barron, N.; Baker, J.; Dick, M.; Long, P.G.; Zhang, S.; Bradshaw, R.E. Dothistromin toxin is not required for Dothistroma needle blight in Pinus radiata. Plant Pathol. 2009, 58, 293–304. [Google Scholar] [CrossRef]
- Shain, L.; Franich, R.A. Induction of Dothistroma blight symptoms with dothistromin. Physiol. Plant Pathol. 1981, 19, 49–55. [Google Scholar]
- Schwelm, A.; Barron, N.J.; Zhang, S.; Bradshaw, R.E. Early expression of aflatoxin-like dothistromin genes in the forest pathogen Dothistroma septosporum. Mycol. Res. 2008, 112, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Skory, C.D.; Chang, P.; Linz, J.E. Regulated expression of the nor-1 and ver-1 genes associated with aflatoxin biosynthesis. Appl. Environ. Microbiol. 1993, 59, 1642–1646. [Google Scholar] [PubMed]
- Zhang, S.; Schwelm, A.; Jin, H.P.; Collins, L.J.; Bradshaw, R.E. A fragmented aflatoxin-like gene cluster in the forest pathogen Dothistroma septosporum. Fungal Genet. Biol. 2007, 44, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Cary, J.W.; Ehrlich, K.C.; Beltz, S.B.; Harris-Coward, P.; Klich, M.A. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia 2009, 101, 352–362. [Google Scholar]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 2006, 43, 679–693. [Google Scholar] [CrossRef] [PubMed]
- Carbone, I.; Ramirez-Prado, J.H.; Jakobek, J.L.; Horn, B.W. Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster. BMC Evol. Biol. 2007, 7, 111. [Google Scholar]
- Moore, G.G.; Singh, R.; Horn, B.W.; Carbone, I. Recombination and lineage-specific gene loss in the aflatoxin gene cluster of Aspergillus flavus. Molec. Ecol. 2009, 18, 4870–4887. [Google Scholar] [CrossRef]
- Cary, J.W.; Ehrlich, K.C. Aflatoxigenicity in Aspergillus: Molecular genetics, phylogenetic relationships and evolutionary implications. Mycopathologia 2006, 162, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Klich, M.A. Aspergillus flavus: The major producer of aflatoxin. Molec. Plant Pathol. 2007, 8, 713–722. [Google Scholar] [CrossRef]
- Wadia, K.D.R.; Butler, D.R. Relationships between temperature and latent periods of rust and leaf-spot diseases of groundnut. Plant Pathol. 1994, 43, 121–129. [Google Scholar]
- Bradshaw, R.E.; Ganley, R.J.; Jones, W.T.; Dyer, P.S. High levels of dothistromin toxin produced by the forest pathogen Dothistroma pini. Mycol. Res. 2000, 104, 325–332. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 1987, 11–15. [Google Scholar]
- Ochman, H.; Gerber, A.S.; Hartl, D.L. Genetic applications of an inverse polymerase chain reaction. Genetics 1988, 120, 621–623. [Google Scholar]
- Bradshaw, R.E.; Bhatnagar, D.; Ganley, R.J.; Gillman, C.J.; Monahan, B.J.; Seconi, J.M. Dothistroma pini, a forest pathogen, contains homologs of aflatoxin. Appl. Environ. Microbiol. 2002, 68, 2885–2892. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, R.E.; Jin, H.; Morgan, B.S.; Schwelm, A.; Teddy, O.R.; Young, C.A.; Zhang, S. A polyketide synthase gene required for biosynthesis of the aflatoxin-like. Mycopathologia 2006, 161, 283–294. [Google Scholar]
- Silar, P. Two new easy to use vectors for transformations. Fungal genet. Newsl 1995, 42, 73. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997, 25, 3389–3402. [Google Scholar]
- Crous, P.W.; Summerell, B.A.; Carnegie, A.J.; Wingfield, M.J.; Hunter, G.C.; Burgess, T.I.; Andjic, V.; Barber, P.A.; Groenewald, J.Z. Unravelling Mycosphaerella: Do you believe in genera? Persoonia 2009, 23, 99–118. [Google Scholar] [PubMed]
- Yu, J.; Chang, P.-K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, P.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar]
- Schwelm, A.; Bradshaw, R.E. Genetics of dothistromin biosynthesis of Dothistroma septosporum: An update. Toxins 2010, 2, 2680–2698. [Google Scholar]
© 2010 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhang, S.; Guo, Y.; Bradshaw, R.E. Genetics of Dothistromin Biosynthesis in the Peanut Pathogen Passalora arachidicola. Toxins 2010, 2, 2738-2753. https://doi.org/10.3390/toxins2122738
Zhang S, Guo Y, Bradshaw RE. Genetics of Dothistromin Biosynthesis in the Peanut Pathogen Passalora arachidicola. Toxins. 2010; 2(12):2738-2753. https://doi.org/10.3390/toxins2122738
Chicago/Turabian StyleZhang, Shuguang, Yanan Guo, and Rosie E. Bradshaw. 2010. "Genetics of Dothistromin Biosynthesis in the Peanut Pathogen Passalora arachidicola" Toxins 2, no. 12: 2738-2753. https://doi.org/10.3390/toxins2122738
APA StyleZhang, S., Guo, Y., & Bradshaw, R. E. (2010). Genetics of Dothistromin Biosynthesis in the Peanut Pathogen Passalora arachidicola. Toxins, 2(12), 2738-2753. https://doi.org/10.3390/toxins2122738




