Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update
Abstract
:1. Introduction
| Gene | Predicted Size (aa) | Putative Gene Product Function | ST Gene Ortholog | Identity aa (%) | AF Gene Ortholog | Identity aa (%) |
|---|---|---|---|---|---|---|
| dotA | 263 | Versicolorin reductase | stcU | 79.1 | aflM | 80.2 |
| dotB | 414 | Oxidase | stcC | 24.0 | ||
| dotC | 602 | MFS transporter | aflT | 31.2 | ||
| dotD | 322 | Thioesterase | stcA | 37.9 | aflC | 34.8 |
| pksA | 2399 | Polyketide synthase | stcA | 57.0 | aflC | 54.8 |
| cypA | 511 | Averufin monooxygenase | stcB | 59.8 | aflV | 59.3 |
| avfA | 301 | Oxidase | stcO | 43.7 | aflI | 47.8 |
| epoA | 420 | Epoxide hydrolase | ||||
| moxA | 626 | Monooxygenase | stcW | 59.0 | aflW | 55.1 |
| DS31 | 231 | Translation elongation factor | stcT | 41.1 | ||
| hexA | (321) | Fatty acid synthase (partial) | stcJ | 41.3 | aflA | 48.8 |
| hypC | 262 | Anthrone oxidase | stcM | 47.9 | hypC | 35.2 |
| vbsA | 643 | Versicolorin B synthase | stcN | 69.1 | aflK | 72.0 |
| norB | 392 | Norsolorinic acid reductase | stcV | 43.4 | aflF | 60.7 |
2. The Dothistromin Toxin
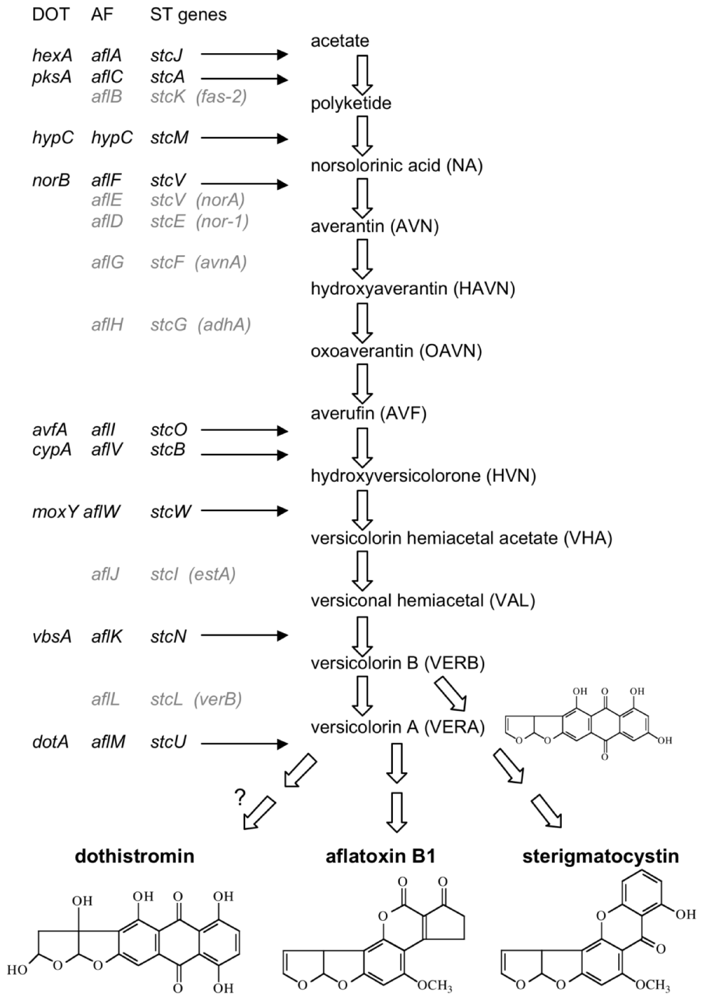
3. The Genetic Basis of Dothistromin Biosynthesis
3.1. AF/ST-like Genes and Gene Organization
| Gene Product | M. graminicola | M. fijiensis | C. heterostrophus | |||
|---|---|---|---|---|---|---|
| e-Value(% aa ID) | Protein ID (chromosome) | e-Value(% aa ID) | Protein ID (scaffold) | e-Value(% aa ID) | Protein ID (scaffold) | |
| DsDotA | 0 (71) | 87994 (11) | 0 (72) | 145765 (10) | 0 (65) | 25675 (10) |
| DsDotC | 0 (57) | 100062 (4) | 0 (57) | 58758 (8) | 0 (53) | 82192 (3) |
| DsPksA | 0 (33) | 96592 (11) | 0 (34) | 216850 (10) | 0 (34) | 30478 (10) |
| DsCypA | 1e-45 (27) | 32226 (1) | 0 (34) | 190367 (8) | 0 (38) | 63323 (5) |
| DsAvfA | [2e-09] | no hits | [3e-19] | |||
| DsMoxA | 0 (71) | 60715 (7) | 0 (69) | 56721 (7) | 0 (66) | 13307 (9) |
| DS31 | 0 (46) | 55916 (2) | 0 (48) | 70922 (2) | 0 (54) | 117471 (8) |
| DsHexA | 0 (40) | 108403 (3) | 0 (40) | 212667 (12) | 0 (40) | 102681 (5) |
| DsHypC | [9e-08] | 9e-23 (35) | 34418 (8) | [1e-13] | ||
| DsVbsA | 0 (42) | 71382 (4) | 0 (44) | 86877 (7) | 0 (42) | 113329 (25) |
| DsNorB | 0 (59) | 105021 (7) | 0 (61) | 36962 (9) | 0 (58) | 109470 (15) |
| ApAflP | 7e-37 (25) | 25671 (2) | 2e-25 (30) | 99112 (8) | 2e-30 (26) | 62952 (5) |
| ApAflX | 0 (47) | 96127 (10) | no hits | 0 (36) | 95438 (27) | |
| ApAflR | [2e-07] | 7e-31 (11) | 198930 (8) | [5e-16] | ||
3.2. Dothistromin Genes
3.2.1. Functionally Analysed Dothistromin Genes
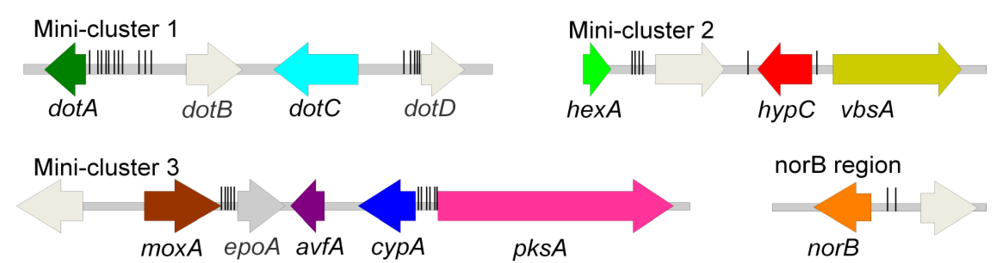
3.2.2. Other Putative Dothistromin Genes
3.3. Regulation of Dothistromin Biosynthesis
4. A Hypothesis for a Biological Function of Dothistromin
5. Outlook for Dothistromin Research with the D. septosporum Genome

6. Conclusions
Acknowledgements
References
- Barnes, I.; Crous, P.W.; Wingfield, B.D.; Wingfield, M.J. Multigene phylogenies reveal that red band needle blight of Pinus is caused by two distinct species of Dothistroma, D.septosporum and D. pini. Stud. Mycol. 2004, 50, 551–565. [Google Scholar]
- Watt, M.S.; Kriticos, D.J.; Alcaraz, S.; Brown, A.V.; Leriche, A. The host spectrum and potential geographic range of Dothistroma needle blight. Forest Ecol. Manag. 2009, 257, 1505–1519. [Google Scholar]
- Barnes, I.; Kirisits, T.; Akulov, A.; Chhetri, D.B.; Wingfield, B.D.; Bulgakov, T.S.; Wingfield, M.J. New host and country records of the Dothistroma needle blight pathogens from Europe and Asia. Forest Pathol. 2008, 38, 178–195. [Google Scholar]
- Barnes, I. Taxonomy, phylogeny and population biology of the red band needle blight pathogen and related species. 2009. [Google Scholar]
- Ioos, R.; Fabre, B.; Saurat, C.; Fourrier, C.; Frey, P.; Marçais, B. Development, comparison, and validation of real-time and conventional PCR tools for the detection of the fungal pathogens causing brown spot and red band needle blights of pines. Phytopathology 2010, 100, 105–114. [Google Scholar]
- Bradshaw, R.E. Dothistroma (red-band) needle blight of pines and the dothistromin toxin: A review. Forest Pathol. 2004, 34, 163–185. [Google Scholar]
- Drenkhan, R.; Hanso, M. Recent invasion of foliage fungi of pines (Pinus spp.) to the Northern Baltics. Forest. Stud. 2009, 51, 49–64. [Google Scholar]
- Welsh, C.; Lewis, K.; Woods, A. The outbreak history of Dothistroma needle blight: An emerging forest disease in northwestern British Columbia, Canada. Can. J. Forest Res. 2009, 39, 2505–2519. [Google Scholar]
- Woods, A.J.; Coates, D.K.; Hamann, A. Is an unprecedented Dothistroma needle blight epidemic related to climate change? BioScience 2005, 55, 761–769. [Google Scholar]
- EPPO. Mycosphaerella dearnessii and Mycosphaerella pini. EPPO Bull. 38, 349–362.
- Bassett, C.; Buchanan, M.; Gallagher, R.T.; Hodges, R.L. A toxic difuroanthraquinone from Dothistroma pini. Chem. Ind. 1970, 52, 1659–1660. [Google Scholar]
- Bradshaw, R.E.; Ganley, R.J.; Jones, W.T.; Dyer, P.S. High levels of dothistromin toxin produced by the forest pathogen Dothistroma pini. Mycol. Res. 2000, 104, 325–332. [Google Scholar]
- Shain, L.; Franich, R.A. Induction of Dothistroma blight symptoms with dothistromin. Physiol. Plant Pathol. 1981, 19, 49–55. [Google Scholar]
- Schwelm, A.; Barron, N.J.; Baker, J.; Dick, M.; Long, P.G.; Zhang, S.; Bradshaw, R.E. Dothistromin toxin is not required for dothistroma needle blight in Pinus radiata. Plant Pathol. 2009, 58, 293–304. [Google Scholar]
- Stoessl, A.; Abramowski, Z.; Lester, H.H.; Rock, G.L.; Towers, G.H.N. Further toxic properties of the fungal metabolite dothistromin. Mycopathologia 1990, 112, 179–186. [Google Scholar]
- Bradshaw, R.E.; Bhatnagar, D.; Ganley, R.J.; Gillman, C.J.; Monahan, B.J.; Seconi, J.M. Dothistroma pini, a forest pathogen, contains homologs of aflatoxin biosynthetic pathway genes. Appl. Environ. Microbiol. 2002, 68, 2885–2892. [Google Scholar]
- Bradshaw, R.E.; Jin, H.; Morgan, B.; Schwelm, A.; Teddy, O.; Young, C.; Zhang, S. A polyketide synthase gene required for biosynthesis of the aflatoxin-like toxin, dothistromin. Mycopathologia 2006, 161, 283–294. [Google Scholar]
- Zhang, S.; Schwelm, A.; Jin, H.; Collins, L.J.; Bradshaw, R.E. A fragmented aflatoxin-like gene cluster in the forest pathogen Dothistroma septosporum. Fungal Genet. Biol. 2007, 44, 1342–1354. [Google Scholar]
- Brown, D.W.; Yu, J.-H.; Kelkar, H.S.; Fernandes, M.; Nesbitt, T.C.; Keller, N.P.; Adams, T.H.; Leonard, T.J. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 1996, 93, 1418–1422. [Google Scholar]
- Yu, J.; Bhatnagar, D.; Cleveland, T.E. Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 2004, 564, 126–130. [Google Scholar]
- Stoessl, A. Dothistromin as a metabolite of Cercospora arachidicola. Mycopathologia. 1984, 86, 165–168. [Google Scholar]
- Assante, G.; Camarda, L.; Merlini, L.; Nasini, G. Dothistromin and 2-Epidothistromin from Cercospora smilacis. Phytochemistry 1977, 16, 125–126. [Google Scholar]
- Assante, G.; Locci, R.; Camarada, L.; Merlini, L.; Nasini, G. Screening of the genus Cercospora for secondary metabolites. Phytochemistry 1977, 16, 243–247. [Google Scholar]
- Assante, G. Isolation and characterization of new dothstromins from Mycosphaerella laricina hart. Phytopathol. Med. 1985, 24, 271–273. [Google Scholar]
- Gallagher, R.T.; Hodges, R. The chemistry of dothistromin, a difuroanthraquinone from Dothistroma pini. Aust. J. Chem. 1972, 25, 2399–2407. [Google Scholar]
- Varga, J.; Frisvad, J.C.; Samson, R.A. A reappraisal of fungi producing aflatoxins. World Mycotoxin J. 2009, 2, 263–277. [Google Scholar]
- Schmidt-Heydt, M.; Häckel, S.; Rüfer, C.E.; Geisen, R. A strain of Fusarium kyushuense is able to produce aflatoxin B1 and G1. Mycotoxin Res. 2009, 25, 141–147. [Google Scholar]
- Franich, R.A.; Carson, M.J.; Carson, S.D. Synthesis and accumulation of benzoic acid in Pinus radiata needles in response to tissue injury with dothistromin, and correlation with resistance of P. radiata families to Dothistroma pini. Physiol. Mol. Plant Pathol. 1986, 23, 268–286. [Google Scholar]
- Galagan, J.E.; Calvo, S.E.; Cuomo, C.; Ma, L.J.; Wortman, J.R.; Batzoglou, S.; Lee, S.I.; Basturkmen, M.; Spevak, C.C.; Clutterbuck, J.; Kapitonov, V.; Jurka, J.; Scazzocchio, C.; Farman, M.; Butler, J.; Purcell, S.; Harris, S.; Braus, G.H.; Draht, O.; Busch, S.; D'Enfert, C.; Bouchier, C.; Goldman, G.H.; Bell-Pedersen, D.; Griffiths-Jones, S.; Doonan, J.H.; Yu, J.; Vienken, K.; Pain, A.; Freitag, M.; Selker, E.U.; Archer, D.B.; Penalva, M.A.; Oakley, B.R.; Momany, M.; Tanaka, T.; Kumagai, T.; Asai, K.; Machida, M.; Nierman, W.C.; Denning, D.W.; Caddick, M.; Hynes, M.; Paoletti, M.; Fischer, R.; Miller, B.; Dyer, P.; Sachs, M.S.; Osmani, S.A.; Birren, B.W. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 2005, 438, 1105–1115. [Google Scholar]
- Georgianna, D.R.; Payne, G.A. Genetic regulation of aflatoxin biosynthesis: From gene to genome. Fungal Genet. Biol. 2009, 46, 113–125. [Google Scholar]
- Yu, J.; Chang, P.-K.; Ehrlich, K.C.; Cary, J.W.; Bhatnagar, D.; Cleveland, T.E.; Payne, G.A.; Linz, J.E.; Woloshuk, C.P.; Bennett, J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004, 70, 1253–1262. [Google Scholar]
- McDonald, T.; Noordermeer, D.; Zhang, Y.-Q.; Hammond, T.M.; Keller, N.P. The ST cluster revisited: Lessons from a genetic model. In Aflatoxin and Food Safety; Abbas, H.K., Ed.; Taylor & Francis Group, LLC: Boca Raton, FL, USA, 2005; pp. 117–136. [Google Scholar]
- Cary, J.W.; Ehrlich, K.C. Aflatoxigenicity in Aspergillus: Molecular genetics, phylogenetic relationships and evolutionary implications. Mycopathologia 2006, 162, 167–177. [Google Scholar]
- Chang, P.-K.; Yu, J. Characterization of a partial duplication of the aflatoxin gene cluster in Aspergillus parasiticus ATCC 56775. Appl. Microbiol. Biotech. 2002, 58, 632–636. [Google Scholar]
- Tanaka, A.; Tapper, B.A.; Popay, A.; Parker, E.J.; Scott, B. A symbiosis expressed non-ribosomal peptide synthetase from a mutualistic fungal endophyte of perennial ryegrass confers protection to the symbiotum from insect herbivory. Molec. Microbiol. 2005, 57, 1036–1050. [Google Scholar]
- Young, C.A.; Felitti, S.; Shields, K.; Spangenberg, G.; Johnson, R.D.; Bryan, G.T.; Saikia, S.; Scott, B. A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte Neotyphodium lolii. Fungal Genet. Biol. 2006, 43, 679–693. [Google Scholar]
- Carbone, I.; Jakobek, J.L.; Ramirez-Prado, J.H.; Horn, B.W. Recombination, balancing selection and adaptive evolution in the aflatoxin gene cluster of Aspergillus parasiticus. Molec. Ecol. 2007, 16, 4401–4417. [Google Scholar]
- Carbone, I.; Ramirez-Prado, J.H.; Jakobek, J.L.; Horn, B.W. Gene duplication, modularity and adaptation in the evolution of the aflatoxin gene cluster. BMC Evol. Biol. 2007, 7, 111. [Google Scholar]
- Shaw, G.J.; Chick, M.; Hodges, R. A 13C NMR study of the biosynthesis of the anthraquinone dothistromin by Dothistroma pini. Phytochemistry 1978, 17, 1743–1745. [Google Scholar]
- Guo, Y. Identification and characterization of dothistromin biosynthetic genes in the peanut pathogen Passalora arachidicola. 2008. [Google Scholar]
- Kodama, M.; Rose, M.S.; Yang, G.; Yun, S.H.; Yoder, O.C.; Turgeon, B.G. The translocation-associated Tox1 locus of Cochliobolus heterostrophus is two genetic elements on two different chromosomes. Genetics 1999, 151, 585–596. [Google Scholar]
- Kimura, M.; Tokai, T.; Takahashi-Ando, N.; Ohsato, S.; Fujimura, M. Molecular and genetic studies of Fusarium trichothecene biosynthesis: Pathways, genes, and evolution. Biosci. Biotech. Biochem. 2007, 71, 2105–2123. [Google Scholar]
- Nicholson, M.J.; Koulman, A.; Monahan, B.J.; Pritchard, B.L.; Payne, G.A.; Scott, B. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl. Environ. Microbiol. 2009, 75, 7469–7481. [Google Scholar]
- Cary, J.; Ehrlich, K.; Beltz, S.; Harris-Coward, P.; Klich, M. Characterization of the Aspergillus ochraceoroseus aflatoxin/sterigmatocystin biosynthetic gene cluster. Mycologia 2009, 101, 352–362. [Google Scholar]
- Nierman, W.C.; Pain, A.; Anderson, M.J.; Wortman, J.R.; Kim, H.S.; Arroyo, J.; Berriman, M.; Abe, K.; Archer, D.B.; Bermejo, C.; Bennett, J.; Bowyer, P.; Chen, D.; Collins, M.; Coulsen, R.; Davies, R.; Dyer, P.S.; Farman, M.; Fedorova, N.; Feldblyum, T.V.; Fischer, R.; Fosker, N.; Fraser, A.; Garcia, J.L.; Garcia, M.J.; Goble, A.; Goldman, G.H.; Gomi, K.; Griffith-Jones, S.; Gwilliam, R.; Haas, B.; Haas, H.; Harris, D.; Horiuchi, H.; Huang, J.; Humphray, S.; Jimenez, J.; Keller, N.; Khouri, H.; Kitamoto, K.; Kobayashi, T.; Konzack, S.; Kulkarni, R.; Kumagai, T.; Lafton, A.; Latge, J.P.; Li, W.X.; Lord, A.; Majoros, W.H.; May, G.S.; Miller, B.L.; Mohamoud, Y.; Molina, M.; Monod, M.; Mouyna, I.; Mulligan, S.; Murphy, L.; O'Neil, S.; Paulsen, I.; Penalva, M.A.; Pertea, M.; Price, C.; Pritchard, B.L.; Quail, M.A.; Rabbinowitsch, E.; Rawlins, N.; Rajandream, M.A.; Reichard, U.; Renauld, H.; Robson, G.D.; de Cordoba, S.R.; Rodriguez-Pena, J.M.; Ronning, C.M.; Rutter, S.; Salzberg, S.L.; Sanchez, M.; Sanchez-Ferrero, J.C.; Saunders, D.; Seeger, K.; Squares, R.; Squares, S.; Takeuchi, M.; Tekaia, F.; Turner, G.; de Aldana, C.R.V.; Weidman, J.; White, O.; Woodward, J.; Yu, J.H.; Fraser, C.; Galagan, J.E.; Asai, K.; Machida, M.; Hall, N.; Barrell, B.; Denning, D.W. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 2005, 438, 1151–1156. [Google Scholar]
- Graziani, S.; Vasnier, C.; Daboussi, M. Novel polyketide synthase from Nectria haematococca. Appl. Environ. Microbiol. 2004, 70, 2984–2988. [Google Scholar]
- Sakuno, E.; Kameyama, M.; Nakajima, H.; Yabe, K. Purification and gene cloning of a dehydrogenase from Lactobacillus brevis that catalyzes a reaction involved in aflatoxin biosynthesis. Biosci. Biotech. Biochem. 2008, 72, 724–734. [Google Scholar]
- Chiang, Y.-M.; Szewczyk, E.; Davidson, A.D.; Entwistle, R.; Keller, N.P.; Wang, C.C.C.; Oakley, B.R. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl. Environ. Microbiol. 2010, 76, 2067–2074. [Google Scholar]
- Kroken, S.; Glass, N.L.; Taylor, J.W.; Yoder, O.C.; Turgeon, B.G. Phylogenomic analysis of type 1 polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 2003, 100, 15670–15675. [Google Scholar]
- Ehrlich, K.C.; Montalbano, B.; Boue, S.M.; Bhatnagar, D. An aflatoxin biosynthesis cluster gene encodes a novel oxidase required for conversion of versicolorin A to sterigmatocystin. Appl. Environ. Microbiol. 2005, 71, 8963–8965. [Google Scholar]
- Henry, K.M.; Townsend, C.A. Ordering the reductive and cytochrome P450 oxidative steps in demethylsterigmatocystin formation yields general insights into the biosynthesis of aflatoxin and related fungal metabolites. J. Am. Chem. Soc. 2005, 127, 3724–3733. [Google Scholar]
- Danks, A.V.; Hodges, R. Polyhydroxyanthraquinones from Dothistroma pini. Australian J. Chem. 1974, 27, 1603–1606. [Google Scholar]
- Schwelm, A. Investigation of dothistromin gene expression in Dothistroma septosporum and the putative role of dothistromin toxin. 2007. [Google Scholar]
- Bradshaw, R.E.; Feng, Z.; Schwelm, A.; Yang, Y.; Zhang, S. Functional analysis of a putative dothistromin toxin MFS transporter gene. Toxins 2009, 1, 173–187. [Google Scholar]
- Martin, J.F.; Casqueiro, J.; Liras, P. Secretion systems for secondary metabolites: How producer cells send out messages of intercellular communication. Curr. Opin. Microbiol. 2005, 8, 282–293. [Google Scholar]
- Chang, P.-K.; Yu, J.; Yu, J.-H. aflT, a MFS transporter-encoding gene located in the aflatoxin gene cluster, does not have a significant role in aflatoxin secretion. Fungal Genet. Biol. 2004, 41, 911–920. [Google Scholar]
- Chanda, A.; Roze, L.V.; Kang, S.; Artymovich, K.A.; Hicks, G.R.; Raikhel, N.V.; Calvo, A.M.; Linz, J.E. A key role for vesicles in fungal secondary metabolism. Proc. Natl. Acad. Sci. USA 2009, 106, 19533–19538. [Google Scholar]
- Ehrlich, K.C.; Li, P.; Scharfenstein, L.; Chang, P.-K. HypC is the anthrone oxidase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2010, 76, 3374–3377. [Google Scholar]
- Feng, Z. Further studies of dothistromin toxin genes in the fungal forest pathogen Dothistroma septosporum. 2007. [Google Scholar]
- Cary, J.W.; Wright, M.; Bhatnagar, D.; Lee, R.; Chu, F.S. Molecular characterization of an Aspergillus parasiticus dehydrogenase gene, norA, located on the aflatoxin biosynthesis gene cluster. Appl. Environ. Microbiol. 1996, 62, 360–366. [Google Scholar]
- Ehrlich, K.C.; Scharfenstein, L.L., Jr.; Montalbano, B.G.; Chang, P.-K. Are the genes nadA and norB involved in formation of aflatoxin G1? Int.J. Molec. Sci. 2008, 9, 1717–1729. [Google Scholar]
- Feng, G.H.; Leonard, T.J. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 1998, 64, 2275–2277. [Google Scholar]
- Cary, J.W.; Calvo, A.M. Regulation of Aspergillus mycotoxin biosynthesis. Toxin Rev. 2008, 27, 347–370. [Google Scholar]
- Yu, J.-H.; Keller, N. Regulation of secondary metabolism in filamentous fungi. Ann. Rev. Phytopathol. 2005, 43, 437–458. [Google Scholar]
- Zhang, Y.; Keller, N.; Tsitsigiannis, D.; Wilkinson, H.H. Secondary metabolite gene clusters. In Handbook of Industrial Mycology; An, Z., Ed.; Marcel Dekker: New York, NY, USA, 2005; Volume 22, pp. 355–385. [Google Scholar]
- Bayram, O.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.-J.; Keller, N.P.; Yu, J.-H.; Braus, G.H. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science 2008, 320, 1504–1506. [Google Scholar]
- Schwelm, A.; Barron, N.J.; Zhang, S.; Bradshaw, R.E. Early expression of aflatoxin-like dothistromin genes in the forest pathogen Dothistroma septosporum. Mycol. Res. 2008, 112, 138–146. [Google Scholar]
- Calvo, A.M.; Wilson, R.A.; Bok, J.W.; Keller, N.P. Relationship between secondary metabolism and fungal development. Microbiol. Molec. Biol. Rev. 2002, 66, 447–459. [Google Scholar]
- Wilkinson, H.H.; Ramaswamy, A.; Sim, S.C.; Keller, N.P. Increased conidiation associated with progression along the sterigmatocystin biosynthetic pathway. Mycologia 2004, 96, 1190–1198. [Google Scholar]
- Donzelli, B.G.G.; Krasnoff, S.B.; Churchill, A.C.L.; Vandenberg, J.D.; Gibson, D.M. Identification of a hybrid PKS–NRPS required for the biosynthesis of NG-391 in Metarhizium robertsii. Curr. Genet. 2010. [Google Scholar]
- Amaike, S.; Keller, N.P. Distinct roles for VeA and LaeA in development and pathogenesis of Aspergillus flavus. Euk. Cell 2009, 8, 1051–1060. [Google Scholar]
- Horowitz Brown, S.; Scott, J.B.; Bhaheetharan, J.; Sharpee, W.C.; Milde, L.; Wilson, R.A.; Keller, N.P. Oxygenase coordination is required for morphological transition and the host-fungus interaction of Aspergillus flavus. Mol. Plant Microbe Interact. 2009, 22, 882–894. [Google Scholar]
- Reverberi, M.; Ricelli, A.; Zjalic, S.; Fabbri, A.A.; Fanelli, C. Natural functions of mycotoxins and control of their biosynthesis in fungi. Appl. Microbiol. Biotechnol. 2010, 87, 899–911. [Google Scholar]
- Youngman, R.J.; Elstner, E.F. Photodynamic and reductive mechanism of oxygen activation by the fungal phytotoxins, cercosporin and dothistromin. In Oxygen Radicals in Chemistry and Biology; W. de Gruyter:: New York, NY, USA, , 1984; pp. 501–508. [Google Scholar]
- Ganley, R.J.; Newcombe, G. Fungal endophytes in seeds and needles of Pinus monticola. Mycol. Res. 2006, 110, 318–327. [Google Scholar]
- Zamora, P.; Martinez-Ruiz, C.; Diez, J.J. Fungi in needles and twigs of pine plantations in northern Spain. Fungal Diversity 2008, 30, 171–184. [Google Scholar]
- Deckert, R.J.; Melville, L.H.; Peterson, R.L. Structural features of a Lophodermium endophyte during the cryptic life-cycle phase in the foliage of Pinus strobus. Mycol. Res. 2001, 105, 991–997. [Google Scholar]
- Keon, J.; Antoniw, J.; Carzaniga, R.; Deller, S.; Ward, J.L.; Baker, J.M.; Beale, M.H.; Hammond-Kosack, K.; Rudd, J.J. Transcriptional adaptation of Mycosphaerella graminicola to programmed cell death (PCD) of its susceptible wheat host. Mol. Plant Microbe Interact. 2007, 20, 178–193. [Google Scholar]
© 2010 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Schwelm, A.; Bradshaw, R.E. Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update. Toxins 2010, 2, 2680-2698. https://doi.org/10.3390/toxins2112680
Schwelm A, Bradshaw RE. Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update. Toxins. 2010; 2(11):2680-2698. https://doi.org/10.3390/toxins2112680
Chicago/Turabian StyleSchwelm, Arne, and Rosie E. Bradshaw. 2010. "Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update" Toxins 2, no. 11: 2680-2698. https://doi.org/10.3390/toxins2112680
APA StyleSchwelm, A., & Bradshaw, R. E. (2010). Genetics of Dothistromin Biosynthesis of Dothistroma septosporum: An Update. Toxins, 2(11), 2680-2698. https://doi.org/10.3390/toxins2112680




