Predatory and Defensive Strategies in Cone Snails
Abstract
1. Introduction
1.1. Envenomation Strategies in Cone Snails
1.2. Reality Check on the Concept of Cabals
1.3. Defensive Strategies
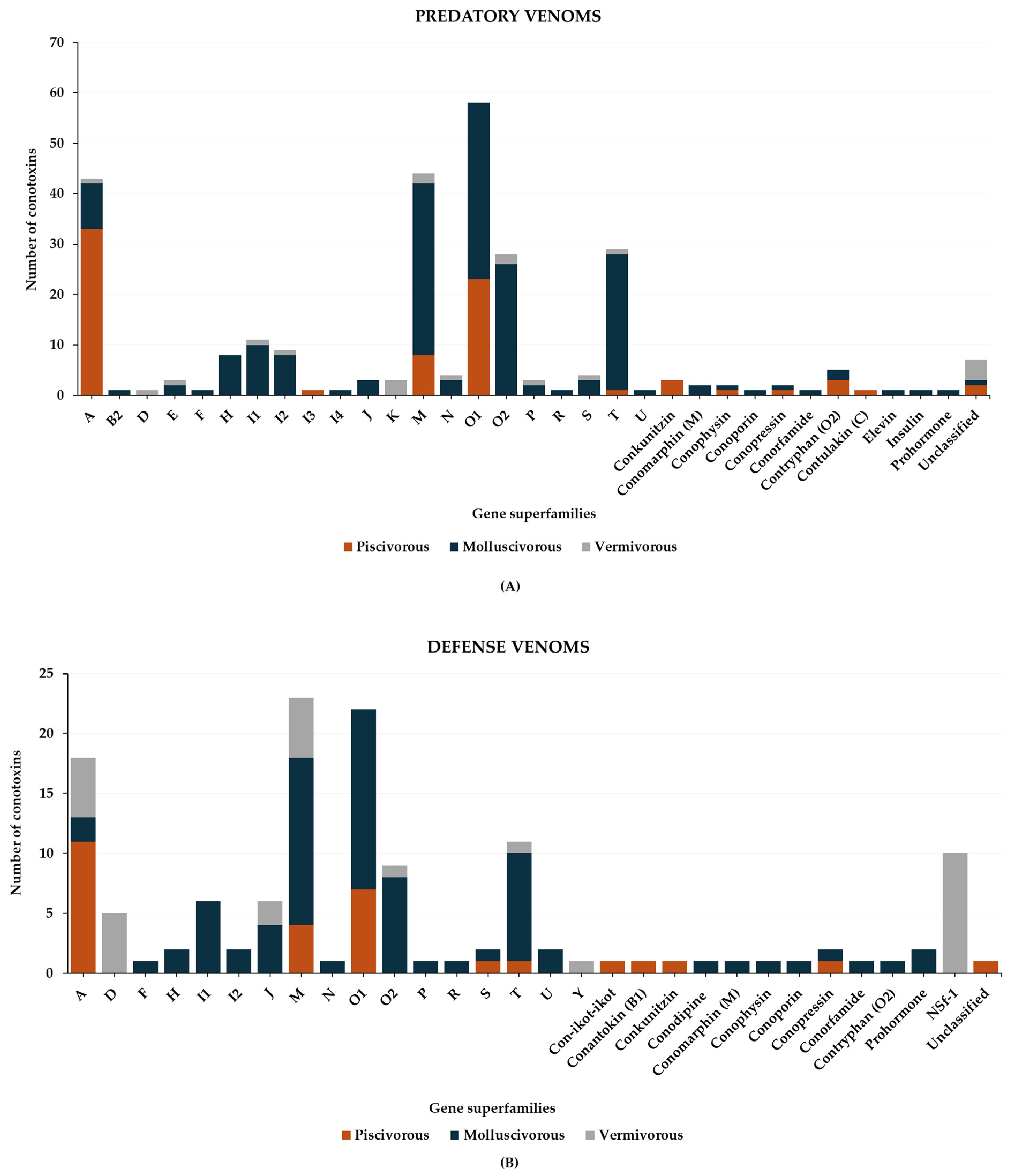
2. Piscivorous Cone Snails
2.1. Predatory Venom
2.2. Defensive Venom
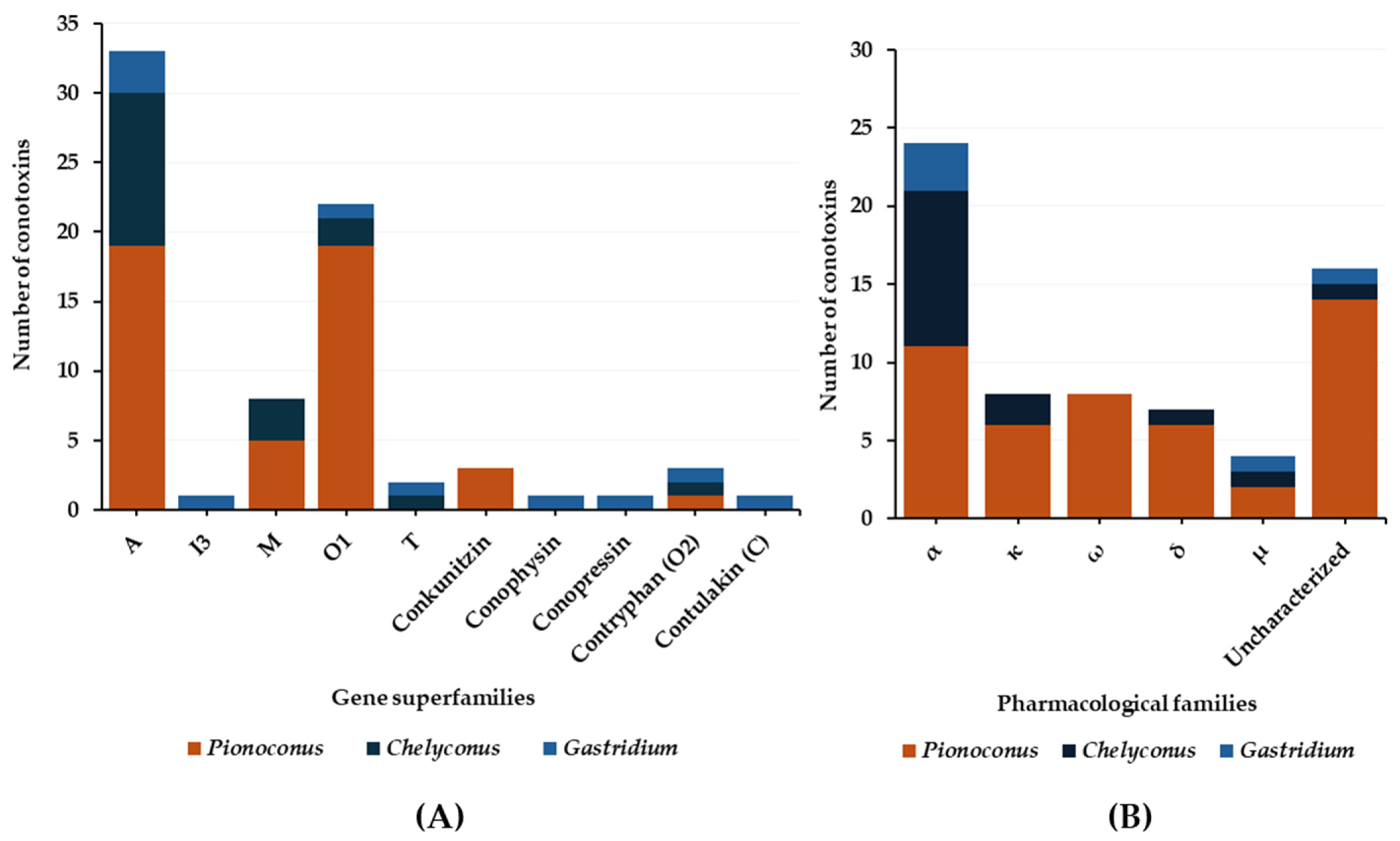
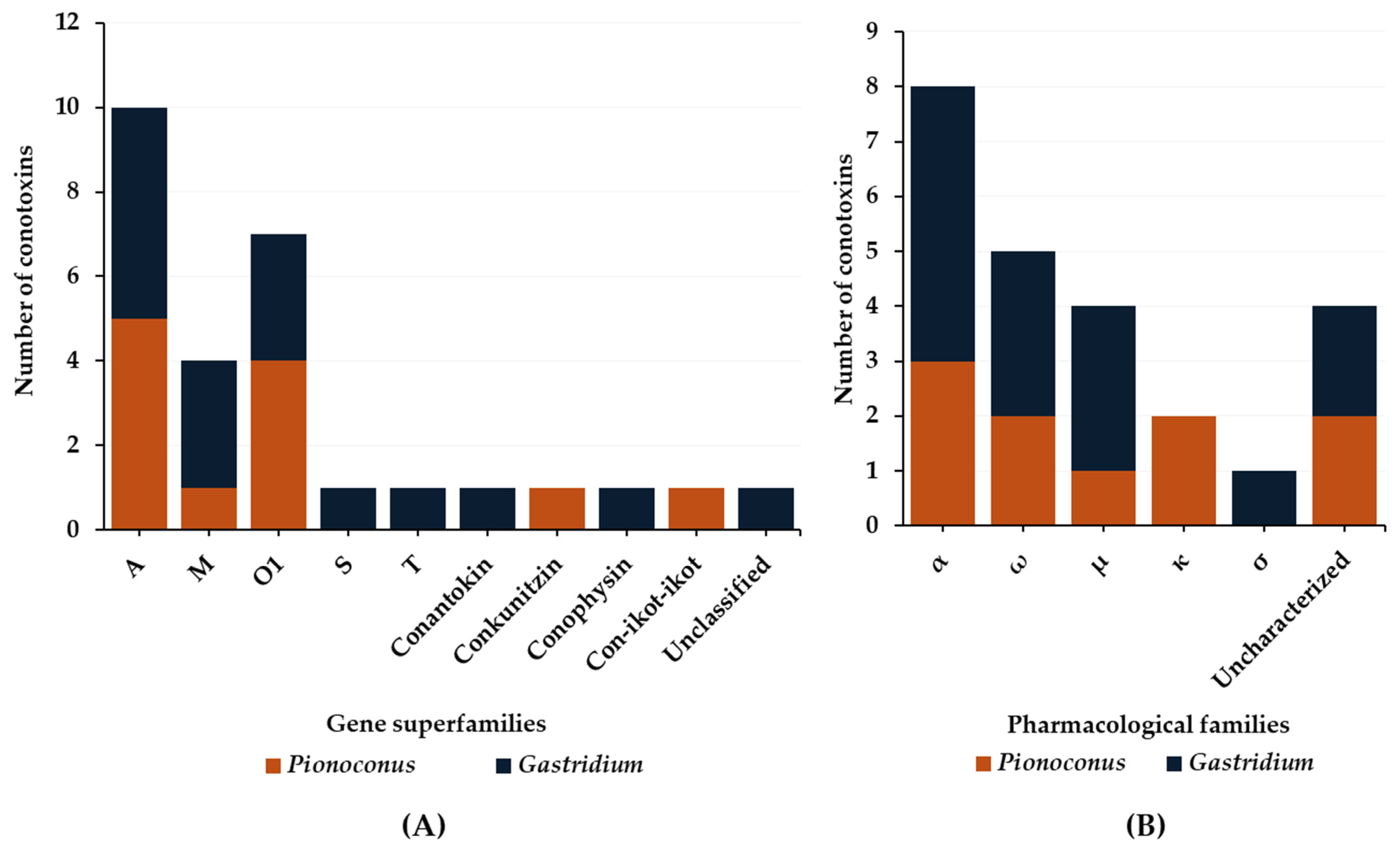
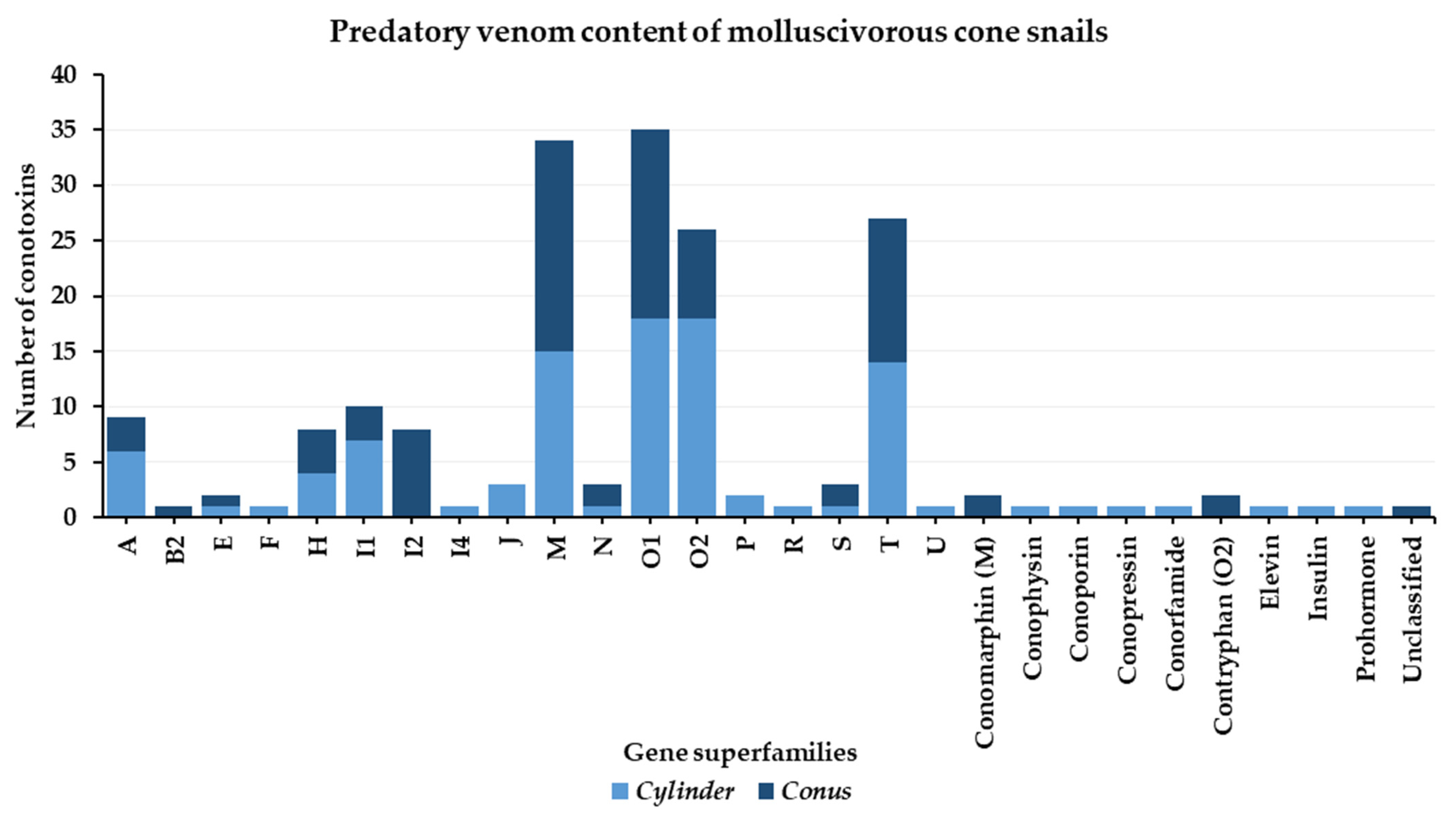
3. Molluscivorous Cone Snails
3.1. Predatory Venom
3.2. Defense Venom
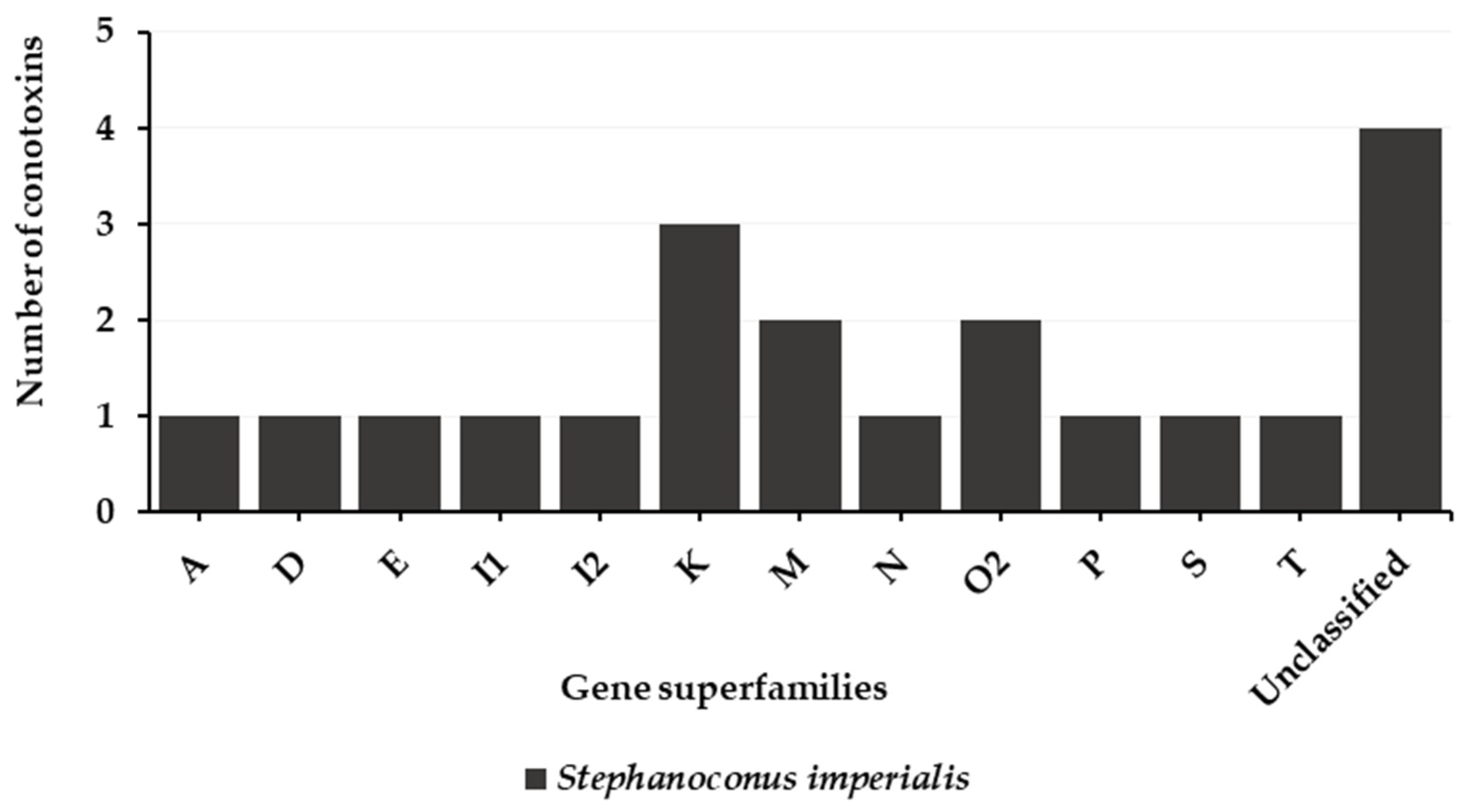
4. Vermivorous Cone Snails
4.1. Predatory Venom
4.2. Defense Venom
5. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kohn, A.J. Microhabitats, Abundance and Food of Conus on Atoll Reefs in the Maldive and Chagos Islands. Ecology 1968, 49, 1046–1062. [Google Scholar] [CrossRef]
- Puillandre, N.; Duda, T.F.; Meyer, C.; Olivera, B.M.; Bouchet, P. One, Four or 100 Genera? A New Classification of the Cone Snails. J. Molluscan Stud. 2015, 81, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.; Zhang, J.; Shi, Q. Cone Snails: A Big Store of Conotoxins for Novel Drug Discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef]
- Olivera, B.M. “Conus” Venom Peptides: Reflections from the Biology of Clades and Species. Annu. Rev. Ecol. Syst. 2002, 33, 25–47. [Google Scholar] [CrossRef]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, Synthesis, and Structure–Activity Relationships of Conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Puillandre, N.; Koua, D.; Favreau, P.; Olivera, B.M.; Stöcklin, R. Molecular Phylogeny, Classification and Evolution of Conopeptides. J. Mol. Evol. 2012, 74, 297–309. [Google Scholar] [CrossRef] [PubMed]
- Lebbe, E.K.M.; Tytgat, J. In the Picture: Disulfide-Poor Conopeptides, a Class of Pharmacologically Interesting Compounds. J. Venom. Anim. Toxins Trop. Dis. 2016, 22, 30. [Google Scholar] [CrossRef]
- Kaas, Q.; Westermann, J.-C.; Craik, D.J. Conopeptide Characterization and Classifications: An Analysis Using ConoServer. Toxicon 2010, 55, 1491–1509. [Google Scholar] [CrossRef]
- Jin, A.-H.; Dutertre, S.; Dutt, M.; Lavergne, V.; Jones, A.; Lewis, R.; Alewood, P. Transcriptomic-Proteomic Correlation in the Predation-Evoked Venom of the Cone Snail, Conus Imperialis. Mar. Drugs 2019, 17, 177. [Google Scholar] [CrossRef]
- Jin, A.-H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef]
- Kohn, A.J.; Nishi, M.; Pernet, B. Snail Spears and Scimitars: A Character Analysis of Conus Radular Teeth. J. Molluscan Stud. 1999, 65, 461–481. [Google Scholar] [CrossRef]
- Kohn, A.J. Piscivorous Gastropods of the Genus Conus. Proc. Natl. Acad. Sci. USA 1956, 42, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Olivera, B.M.; Seger, J.; Horvath, M.P.; Fedosov, A.E. Prey-Capture Strategies of Fish-Hunting Cone Snails: Behavior, Neurobiology and Evolution. Brain Behav. Evol. 2015, 86, 58–74. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M. Biology of Conus on Shores of the Dampier Archipelago, Northwestern Australia; University of Washington Seattle: Seattle, WA, USA, 2003. [Google Scholar]
- Salisbury, S.M.; Martin, G.G.; Kier, W.M.; Schulz, J.R. Venom Kinematics during Prey Capture in Conus: The Biomechanics of a Rapid Injection System. J. Exp. Biol. 2010, 213, 673–682. [Google Scholar] [CrossRef] [PubMed]
- Prator, C.A.; Murayama, K.M.; Schulz, J.R. Venom Variation during Prey Capture by the Cone Snail, Conus textile. PLoS ONE 2014, 9, e98991. [Google Scholar] [CrossRef] [PubMed]
- Himaya, S.W.A.; Marí, F.; Lewis, R.J. Accelerated Proteomic Visualization of Individual Predatory Venoms of Conus Purpurascens Reveals Separately Evolved Predation-Evoked Venom Cabals. Sci. Rep. 2018, 8, 330. [Google Scholar] [CrossRef] [PubMed]
- Abalde, S.; Tenorio, M.J.; Afonso, C.M.L.; Zardoya, R. Conotoxin Diversity in Chelyconus ermineus (Born, 1778) and the Convergent Origin of Piscivory in the Atlantic and Indo-Pacific Cones. Genome Biol. Evol. 2018, 10, 2643–2662. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Bandyopadhyay, P.K.; Olivera, B.M.; Yandell, M. Elucidation of the Molecular Envenomation Strategy of the Cone Snail Conus Geographus through Transcriptome Sequencing of Its Venom Duct. BMC Genom. 2012, 13, 284. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Gajewiak, J.; Karanth, S.; Robinson, S.D.; Ueberheide, B.; Douglass, A.D.; Schlegel, A.; Imperial, J.S.; Watkins, M.; Bandyopadhyay, P.K.; et al. Specialized Insulin Is Used for Chemical Warfare by Fish-Hunting Cone Snails. Proc. Natl. Acad. Sci. USA 2015, 112, 1743–1748. [Google Scholar] [CrossRef]
- Robinson, S.D.; Li, Q.; Bandyopadhyay, P.K.; Gajewiak, J.; Yandell, M.; Papenfuss, A.T.; Purcell, A.W.; Norton, R.S.; Safavi-Hemami, H. Hormone-like Peptides in the Venoms of Marine Cone Snails. Gen. Comp. Endocrinol. 2017, 244, 11–18. [Google Scholar] [CrossRef]
- Hopkins, C.; Grilley, M.; Miller, C.; Shon, K.-J.; Cruz, L.J.; Gray, W.R.; Dykert, J.; Rivier, J.; Yoshikami, D.; Olivera, B.M. A New Family of Conus Peptides Targeted to the Nicotinic Acetylcholine Receptor (*). J. Biol. Chem. 1995, 270, 22361–22367. [Google Scholar] [CrossRef]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V.; Gilly, W.F.; Schulz, J.R. Intraspecific Variation of Venom Injected by Fish-Hunting Conus Snails. J. Exp. Biol. 2005, 208, 2873–2883. [Google Scholar] [CrossRef]
- Dutertre, S.; Biass, D.; Stöcklin, R.; Favreau, P. Dramatic Intraspecimen Variations within the Injected Venom of Conus consors: An Unsuspected Contribution to Venom Diversity. Toxicon 2010, 55, 1453–1462. [Google Scholar] [CrossRef]
- Himaya, S.W.A.; Jin, A.-H.; Dutertre, S.; Giacomotto, J.; Mohialdeen, H.; Vetter, I.; Alewood, P.F.; Lewis, R.J. Comparative Venomics Reveals the Complex Prey Capture Strategy of the Piscivorous Cone Snail Conus catus. J. Proteome Res. 2015, 14, 4372–4381. [Google Scholar] [CrossRef]
- Kelley, W.P.; Schulz, J.R.; Jakubowski, J.A.; Gilly, W.F.; Sweedler, J.V. Two Toxins from Conus striatus That Individually Induce Tetanic Paralysis. Biochemistry 2006, 45, 14212–14222. [Google Scholar] [CrossRef]
- Le Gall, F.; Favreau, P.; Benoit, E.; Mattei, C.; Bouet, F.; Menou, J.-L.; Ménez, A.; Letourneux, Y.; Molgó, J. A New Conotoxin Isolated from Conus Consors Venom Acting Selectively on Axons and Motor Nerve Terminals through a Na+-Dependent Mechanism. Eur. J. Neurosci. 1999, 11, 3134–3142. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.-H.; Vetter, I.; Hamilton, B.; Sunagar, K.; Lavergne, V.; Dutertre, V.; Fry, B.G.; Antunes, A.; Venter, D.J.; et al. Evolution of Separate Predation- and Defence-Evoked Venoms in Carnivorous Cone Snails. Nat. Commun. 2014, 5, 3521. [Google Scholar] [CrossRef]
- Kohn, A.J. Conus Envenomation of Humans: In Fact and Fiction. Toxins 2019, 11, 10. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wüster, W.; Vonk, F.J.; Harrison, R.A.; Fry, B.G. Complex Cocktails: The Evolutionary Novelty of Venoms. Trends Ecol. Evol. 2013, 28, 219–229. [Google Scholar] [CrossRef]
- Zhao, Y.; Antunes, A. Biomedical Potential of the Neglected Molluscivorous and Vermivorous Conus Species. Mar. Drugs 2022, 20, 105. [Google Scholar] [CrossRef]
- Himaya, S.W.A.; Jin, A.-H.; Hamilton, B.; Rai, S.K.; Alewood, P.; Lewis, R.J. Venom Duct Origins of Prey Capture and Defensive Conotoxins in Piscivorous Conus striatus. Sci. Rep. 2021, 11, 13282. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.-H.; Alewood, P.F.; Lewis, R.J. Intraspecific Variations in Conus geographus Defence-Evoked Venom and Estimation of the Human Lethal Dose. Toxicon 2014, 91, 135–144. [Google Scholar] [CrossRef]
- Shells For Sale|The Largest Official Seashells Website|Conchology. Available online: https://www.conchology.be/ (accessed on 18 July 2023).
- Gajewiak, J.; Azam, L.; Imperial, J.; Walewska, A.; Green, B.R.; Bandyopadhyay, P.K.; Raghuraman, S.; Ueberheide, B.; Bern, M.; Zhou, H.M.; et al. A Disulfide Tether Stabilizes the Block of Sodium Channels by the Conotoxin μO§-GVIIJ. Proc. Natl. Acad. Sci. USA 2014, 111, 2758–2763. [Google Scholar] [CrossRef]
- Teichert, R.W.; Jacobsen, R.; Terlau, H.; Yoshikami, D.; Olivera, B.M. Discovery and Characterization of the Short κA-Conotoxins: A Novel Subfamily of Excitatory Conotoxins. Toxicon 2007, 49, 318–328. [Google Scholar] [CrossRef]
- Biass, D.; Dutertre, S.; Gerbault, A.; Menou, J.-L.; Offord, R.; Favreau, P.; Stöcklin, R. Comparative Proteomic Study of the Venom of the Piscivorous Cone Snail Conus consors. J. Proteom. 2009, 72, 210–218. [Google Scholar] [CrossRef]
- Violette, A.; Biass, D.; Dutertre, S.; Koua, D.; Piquemal, D.; Pierrat, F.; Stöcklin, R.; Favreau, P. Large-Scale Discovery of Conopeptides and Conoproteins in the Injectable Venom of a Fish-Hunting Cone Snail Using a Combined Proteomic and Transcriptomic Approach. J. Proteom. 2012, 75, 5215–5225. [Google Scholar] [CrossRef]
- Kapono, C.A.; Thapa, P.; Cabalteja, C.C.; Guendisch, D.; Collier, A.C.; Bingham, J.-P. Conotoxin Truncation as a Post-Translational Modification to Increase the Pharmacological Diversity within the Milked Venom of Conus magus. Toxicon 2013, 70, 170–178. [Google Scholar] [CrossRef]
- Saintmont, F.; Cazals, G.; Bich, C.; Dutertre, S. Proteomic Analysis of the Predatory Venom of Conus striatus Reveals Novel and Population-Specific κA-Conotoxin SIVC. Toxins 2022, 14, 799. [Google Scholar] [CrossRef]
- Teichert, R.W.; Rivier, J.; Dykert, J.; Cervini, L.; Gulyas, J.; Bulaj, G.; Ellison, M.; Olivera, B.M. αA-Conotoxin OIVA Defines a New αA-Conotoxin Subfamily of Nicotinic Acetylcholine Receptor Inhibitors. Toxicon 2004, 44, 207–214. [Google Scholar] [CrossRef]
- Ramilo, C.A.; Zafaralla, G.C.; Nadasdi, L.; Hammerland, L.G.; Yoshikami, D.; Gray, W.R.; Kristipati, R.; Ramachandran, J.; Miljanich, G. Novel Alpha- and Omega-Conotoxins and Conus striatus Venom. Biochemistry 1992, 31, 9919–9926. [Google Scholar] [CrossRef]
- Biass, D.; Violette, A.; Hulo, N.; Lisacek, F.; Favreau, P.; Stöcklin, R. Uncovering Intense Protein Diversification in a Cone Snail Venom Gland Using an Integrative Venomics Approach. J. Proteome Res. 2015, 14, 628–638. [Google Scholar] [CrossRef] [PubMed]
- Quinton, L.; Servent, D.; Girard, E.; Molgó, J.; Le Caer, J.-P.; Malosse, C.; Haidar, E.A.; Lecoq, A.; Gilles, N.; Chamot-Rooke, J. Identification and Functional Characterization of a Novel α-Conotoxin (EIIA) from Conus ermineus. Anal. Bioanal. Chem. 2013, 405, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Terlau, H.; Olivera, B.M. Conus Venoms: A Rich Source of Novel Ion Channel-Targeted Peptides. Physiol. Rev. 2004, 84, 41–68. [Google Scholar] [CrossRef] [PubMed]
- López-Vera, E.; Jacobsen, R.B.; Ellison, M.; Olivera, B.M.; Teichert, R.W. A Novel Alpha Conotoxin (α-PIB) Isolated from C. purpurascens Is Selective for Skeletal Muscle Nicotinic Acetylcholine Receptors. Toxicon 2007, 49, 1193–1199. [Google Scholar] [CrossRef] [PubMed]
- Teichert, R.W.; Rivier, J.; Torres, J.; Dykert, J.; Miller, C.; Olivera, B.M. A Uniquely Selective Inhibitor of the Mammalian Fetal Neuromuscular Nicotinic Acetylcholine Receptor. J. Neurosci. 2005, 25, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Hoggard, M.F.; Rodriguez, A.M.; Cano, H.; Clark, E.; Tae, H.-S.; Adams, D.J.; Godenschwege, T.A.; Marí, F. In Vivo and In Vitro Testing of Native α-Conotoxins from the Injected Venom of Conus purpurascens. Neuropharmacology 2017, 127, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Van Wagoner, R.M.; Jacobsen, R.B.; Olivera, B.M.; Ireland, C.M. Characterization and Three-Dimensional Structure Determination of ψ-Conotoxin P IIIF, a Novel Noncompetitive Antagonist of Nicotinic Acetylcholine Receptors. Biochemistry 2003, 42, 6353–6362. [Google Scholar] [CrossRef]
- Jacobsen, R.; Yoshikami, D.; Ellison, M.; Martinez, J.; Gray, W.R.; Cartier, G.E.; Shon, K.-J.; Groebe, D.R.; Abramson, S.N.; Olivera, B.M.; et al. Differential Targeting of Nicotinic Acetylcholine Receptors by Novel αA-Conotoxins. J. Biol. Chem. 1997, 272, 22531–22537. [Google Scholar] [CrossRef]
- Rivera-Ortiz, J.A.; Cano, H.; Marí, F. Intraspecies Variability and Conopeptide Profiling of the Injected Venom of Conus ermineus. Peptides 2011, 32, 306–316. [Google Scholar] [CrossRef]
- Echterbille, J.; Gilles, N.; Araóz, R.; Mourier, G.; Amar, M.; Servent, D.; De Pauw, E.; Quinton, L. Discovery and Characterization of EIIB, a New α-Conotoxin from Conus Ermineus Venom by nAChRs Affinity Capture Monitored by MALDI-TOF/TOF Mass Spectrometry. Toxicon 2017, 130, 1–10. [Google Scholar] [CrossRef]
- Martinez, J.S.; Olivera, B.M.; Gray, W.R.; Craig, A.G.; Groebe, D.R.; Abramson, S.N.; McIntosh, J.M. Alpha-Conotoxin EI, A New Nicotinic Acetylcholine Receptor Antagonist with Novel Selectivity. Biochemistry 1995, 34, 14519–14526. [Google Scholar] [CrossRef]
- Robinson, S.D.; Norton, R.S. Conotoxin Gene Superfamilies. Mar. Drugs 2014, 12, 6058–6101. [Google Scholar] [CrossRef]
- Shon, K.-J.; Grilley, M.M.; Marsh, M.; Yoshikami, D.; Hall, A.R.; Kurz, B.; Gray, W.R.; Imperial, J.S.; Hillyard, D.R.; Olivera, B.M. Purification, Characterization, Synthesis, and Cloning of the Lockjaw Peptide from Conus Purpurascens Venom. Biochemistry 1995, 34, 4913–4918. [Google Scholar] [CrossRef]
- Himaya, S.; Lewis, R. Venomics-Accelerated Cone Snail Venom Peptide Discovery. Int. J. Mol. Sci. 2018, 19, 788. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, Y.; Abe, T.; Satake, M.; Odani, S.; Suzuki, J.; Ishikawa, K. A Novel Sodium Channel Inhibitor from Conus geographus: Purification, Structure, and Pharmacological Properties. Biochemistry 1988, 27, 6256–6262. [Google Scholar] [CrossRef] [PubMed]
- Del Río-Sancho, S.; Cros, C.; Coutaz, B.; Cuendet, M.; Kalia, Y.N. Cutaneous Iontophoresis of μ-Conotoxin CnIIIC-A Potent NaV1.4 Antagonist with Analgesic, Anaesthetic and Myorelaxant Properties. Int. J. Pharm. 2017, 518, 59–65. [Google Scholar] [CrossRef]
- Zhang, M.-M.; Fiedler, B.; Green, B.R.; Catlin, P.; Watkins, M.; Garrett, J.E.; Smith, B.J.; Yoshikami, D.; Olivera, B.M.; Bulaj, G. Structural and Functional Diversities among μ-Conotoxins Targeting TTX-Resistant Sodium Channels. Biochemistry 2006, 45, 3723–3732. [Google Scholar] [CrossRef]
- Olivera, B.M.; McIntosh, J.M.; Curz, L.J.; Luque, F.A.; Gray, W.R. Purification and Sequence of a Presynaptic Peptide Toxin from Conus geographus Venom. Biochemistry 1984, 23, 5087–5090. [Google Scholar] [CrossRef]
- Lewis, R.J.; Nielsen, K.J.; Craik, D.J.; Loughnan, M.L.; Adams, D.A.; Sharpe, I.A.; Luchian, T.; Adams, D.J.; Bond, T.; Thomas, L.; et al. Novel ω-Conotoxins from Conus Catus Discriminate among Neuronal Calcium Channel Subtypes. J. Biol. Chem. 2000, 275, 35335–35344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Luo, W.; Liu, T.; Ni, Y.; Qin, Z. Neurotoxins Acting at Synaptic Sites: A Brief Review on Mechanisms and Clinical Applications. Toxins 2023, 15, 18. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Brogan, S.E.; Olivera, B.M. Pain Therapeutics from Cone Snail Venoms: From Ziconotide to Novel Non-Opioid Pathways. J. Proteom. 2019, 190, 12–20. [Google Scholar] [CrossRef]
- Zafaralla, G.C.; Ramilo, C.; Gray, W.R.; Karlstrom, R.; Olivera, B.M.; Cruz, L.J. Phylogenetic Specificity of Cholinergic Ligands:.Alpha.-Conotoxin SI. Biochemistry 1988, 27, 7102–7105. [Google Scholar] [CrossRef]
- Dutertre, S.; Croker, D.; Daly, N.L.; Andersson, Å.; Muttenthaler, M.; Lumsden, N.G.; Craik, D.J.; Alewood, P.F.; Guillon, G.; Lewis, R.J. Conopressin-T from Conus tulipa Reveals an Antagonist Switch in Vasopressin-like Peptides. J. Biol. Chem. 2008, 283, 7100–7108. [Google Scholar] [CrossRef]
- Haack, J.A.; Rivier, J.; Parks, T.N.; Mena, E.E.; Cruz, L.J.; Olivera, B.M. Conantokin-T. A Gamma-Carboxyglutamate Containing Peptide with N-Methyl-d-Aspartate Antagonist Activity. J. Biol. Chem. 1990, 265, 6025–6029. [Google Scholar] [CrossRef]
- Craig, A.G.; Norberg, T.; Griffin, D.; Hoeger, C.; Akhtar, M.; Schmidt, K.; Low, W.; Dykert, J.; Richelson, E.; Navarro, V.; et al. Contulakin-G, an O-Glycosylated Invertebrate Neurotensin. J. Biol. Chem. 1999, 274, 13752–13759. [Google Scholar] [CrossRef] [PubMed]
- Bayrhuber, M.; Vijayan, V.; Ferber, M.; Graf, R.; Korukottu, J.; Imperial, J.; Garrett, J.E.; Olivera, B.M.; Terlau, H.; Zweckstetter, M.; et al. Conkunitzin-S1 Is the First Member of a New Kunitz-Type Neurotoxin Family. J. Biol. Chem. 2005, 280, 23766–23770. [Google Scholar] [CrossRef] [PubMed]
- Möller, C.; Marí, F. 9.3 KDa Components of the Injected Venom of Conus purpurascens Define a New Five-Disulfide Conotoxin Framework. Pept. Sci. 2011, 96, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Walker, C.S.; Jensen, S.; Ellison, M.; Matta, J.A.; Lee, W.Y.; Imperial, J.S.; Duclos, N.; Brockie, P.J.; Madsen, D.M.; Isaac, J.T.R.; et al. A Novel Conus Snail Polypeptide Causes Excitotoxicity by Blocking Desensitization of AMPA Receptors. Curr. Biol. 2009, 19, 900–908. [Google Scholar] [CrossRef]
- Möller, C.; Clark, E.; Safavi-Hemami, H.; DeCaprio, A.; Marí, F. Isolation and Characterization of Conohyal-P1, a Hyaluronidase from the Injected Venom of Conus purpurascens. J. Proteom. 2017, 164, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Möller, C.; Davis, W.C.; Clark, E.; DeCaprio, A.; Marí, F. Conodipine-P1-3, the First Phospholipases A2 Characterized from Injected Cone Snail Venom. Mol. Cell. Proteom. MCP 2019, 18, 876–891. [Google Scholar] [CrossRef] [PubMed]
- Möller, C.; Vanderweit, N.; Bubis, J.; Marí, F. Comparative Analysis of Proteases in the Injected and Dissected Venom of Cone Snail Species. Toxicon 2013, 65, 59–67. [Google Scholar] [CrossRef]
- Safavi-Hemami, H.; Möller, C.; Marí, F.; Purcell, A.W. High Molecular Weight Components of the Injected Venom of Fish-Hunting Cone Snails Target the Vascular System. J. Proteom. 2013, 91, 97–105. [Google Scholar] [CrossRef]
- Nicke, A.; Loughnan, M.L.; Millard, E.L.; Alewood, P.F.; Adams, D.J.; Daly, N.L.; Craik, D.J.; Lewis, R.J. Isolation, Structure, and Activity of GID, a Novel A4/7-Conotoxin with an Extended N-Terminal Sequence. J. Biol. Chem. 2003, 278, 3137–3144. [Google Scholar] [CrossRef] [PubMed]
- England, L.J.; Imperial, J.; Jacobsen, R.; Craig, A.G.; Gulyas, J.; Akhtar, M.; Rivier, J.; Julius, D.; Olivera, B.M. Inactivation of a Serotonin-Gated Ion Channel by a Polypeptide Toxin from Marine Snails. Science 1998, 281, 575–578. [Google Scholar] [CrossRef]
- Diversity of Conus Neuropeptides. Available online: https://www.science.org/doi/10.1126/science.2165278 (accessed on 14 December 2023).
- Jakubowski, J.A.; Sweedler, J.V. Sequencing and Mass Profiling Highly Modified Conotoxins Using Global Reduction/Alkylation Followed by Mass Spectrometry. Anal. Chem. 2004, 76, 6541–6547. [Google Scholar] [CrossRef] [PubMed]
- Jakubowski, J.A.; Kelley, W.P.; Sweedler, J.V. Screening for Post-Translational Modifications in Conotoxins Using Liquid Chromatography/Mass Spectrometry: An Important Component of Conotoxin Discovery. Toxicon 2006, 47, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Abalde, S.; Dutertre, S.; Zardoya, R. A Combined Transcriptomics and Proteomics Approach Reveals the Differences in the Predatory and Defensive Venoms of the Molluscivorous Cone Snail Cylinder ammiralis (Caenogastropoda: Conidae). Toxins 2021, 13, 642. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, S.; Jin, A.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep Venomics Reveals the Mechanism for Expanded Peptide Diversity in Cone Snail Venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [PubMed]
- Corpuz, G.P.; Jacobsen, R.B.; Jimenez, E.C.; Watkins, M.; Walker, C.; Colledge, C.; Garrett, J.E.; McDougal, O.; Li, W.; Gray, W.R.; et al. Definition of the M-Conotoxin Superfamily: Characterization of Novel Peptides from Molluscivorous Conus Venoms. Biochemistry 2005, 44, 8176–8186. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shao, X.; Li, M.; Wang, S.; Chi, C.; Wang, C. Mr1e, a Conotoxin from Conus Marmoreus with a Novel Disulfide Pattern. Acta Biochim. Biophys. Sin. 2008, 40, 391–396. [Google Scholar] [CrossRef]
- McIntosh, J.M.; Hasson, A.; Spira, M.E.; Gray, W.R.; Li, W.; Marsh, M.; Hillyard, D.R.; Olivera, B.M. A New Family of Conotoxins That Blocks Voltage-Gated Sodium Channels. J. Biol. Chem. 1995, 270, 16796–16802. [Google Scholar] [CrossRef]
- Grant, M.A.; Hansson, K.; Furie, B.C.; Furie, B.; Stenflo, J.; Rigby, A.C. The Metal-Free and Calcium-Bound Structures of a γ-Carboxyglutamic Acid-Containing Contryphan from Conus Marmoreus, Glacontryphan-M. J. Biol. Chem. 2004, 279, 32464–32473. [Google Scholar] [CrossRef]
- Prashanth, J.R.; Dutertre, S.; Jin, A.H.; Lavergne, V.; Hamilton, B.; Cardoso, F.C.; Griffin, J.; Venter, D.J.; Alewood, P.F.; Lewis, R.J. The Role of Defensive Ecological Interactions in the Evolution of Conotoxins. Mol. Ecol. 2016, 25, 598–615. [Google Scholar] [CrossRef]
- Jin, A.-H.; Vetter, I.; Himaya, S.W.A.; Alewood, P.F.; Lewis, R.J.; Dutertre, S. Transcriptome and Proteome of Conus Planorbis Identify the Nicotinic Receptors as Primary Target for the Defensive Venom. Proteomics 2015, 15, 4030–4040. [Google Scholar] [CrossRef]
- Casewell, N.R.; Jackson, T.N.W.; Laustsen, A.H.; Sunagar, K. Causes and Consequences of Snake Venom Variation. Trends Pharmacol. Sci. 2020, 41, 570–581. [Google Scholar] [CrossRef] [PubMed]
- Rogalski, A.; Himaya, S.W.A.; Lewis, R.J. Coordinated Adaptations Define the Ontogenetic Shift from Worm- to Fish-Hunting in a Venomous Cone Snail. Nat. Commun. 2023, 14, 3287. [Google Scholar] [CrossRef] [PubMed]
- Prunier, G.; Cherkaoui, M.; Lysiak, A.; Langella, O.; Blein-Nicolas, M.; Lollier, V.; Benoist, E.; Jean, G.; Fertin, G.; Rogniaux, H.; et al. Fast Alignment of Mass Spectra in Large Proteomics Datasets, Capturing Dissimilarities Arising from Multiple Complex Modifications of Peptides. BMC Bioinform. 2023, 24, 421. [Google Scholar] [CrossRef] [PubMed]
- Prashanth, J.R.; Dutertre, S.; Rai, S.K.; Lewis, R.J. Venomics Reveals a Non-Compartmentalised Venom Gland in the Early Diverged Vermivorous Conus Distans. Toxins 2022, 14, 226. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Torres, J.P.; Watkins, M.; Paguigan, N.; Niu, C.; Imperial, J.S.; Tun, J.; Safavi-Hemami, H.; Finol-Urdaneta, R.K.; Neves, J.L.B.; et al. Non-Peptidic Small Molecule Components from Cone Snail Venoms. Front. Pharmacol. 2021, 12, 655981. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.L.B.; Lin, Z.; Imperial, J.S.; Antunes, A.; Vasconcelos, V.; Olivera, B.M.; Schmidt, E.W. Small Molecules in the Cone Snail Arsenal. Org. Lett. 2015, 17, 4933–4935. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.P.; Lin, Z.; Watkins, M.; Salcedo, P.F.; Baskin, R.P.; Elhabian, S.; Safavi-Hemami, H.; Taylor, D.; Tun, J.; Concepcion, G.P.; et al. Small-Molecule Mimicry Hunting Strategy in the Imperial Cone Snail, Conus imperialis. Sci. Adv. 2021, 7, eabf2704. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.A.; Robinson, S.D.; Hamilton, B.F.; Undheim, E.A.B.; King, G.F. Deadly Proteomes: A Practical Guide to Proteotranscriptomics of Animal Venoms. Proteomics 2020, 20, 1900324. [Google Scholar] [CrossRef] [PubMed]

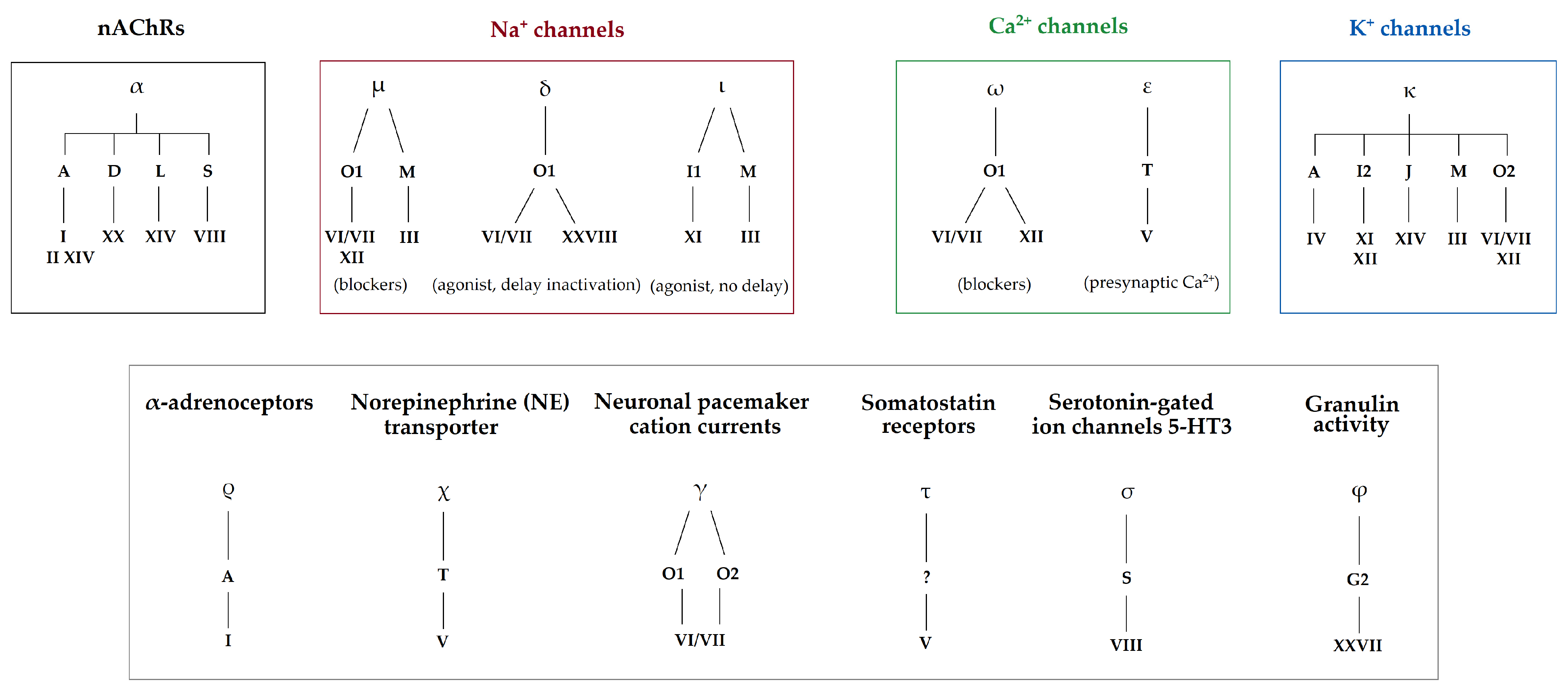

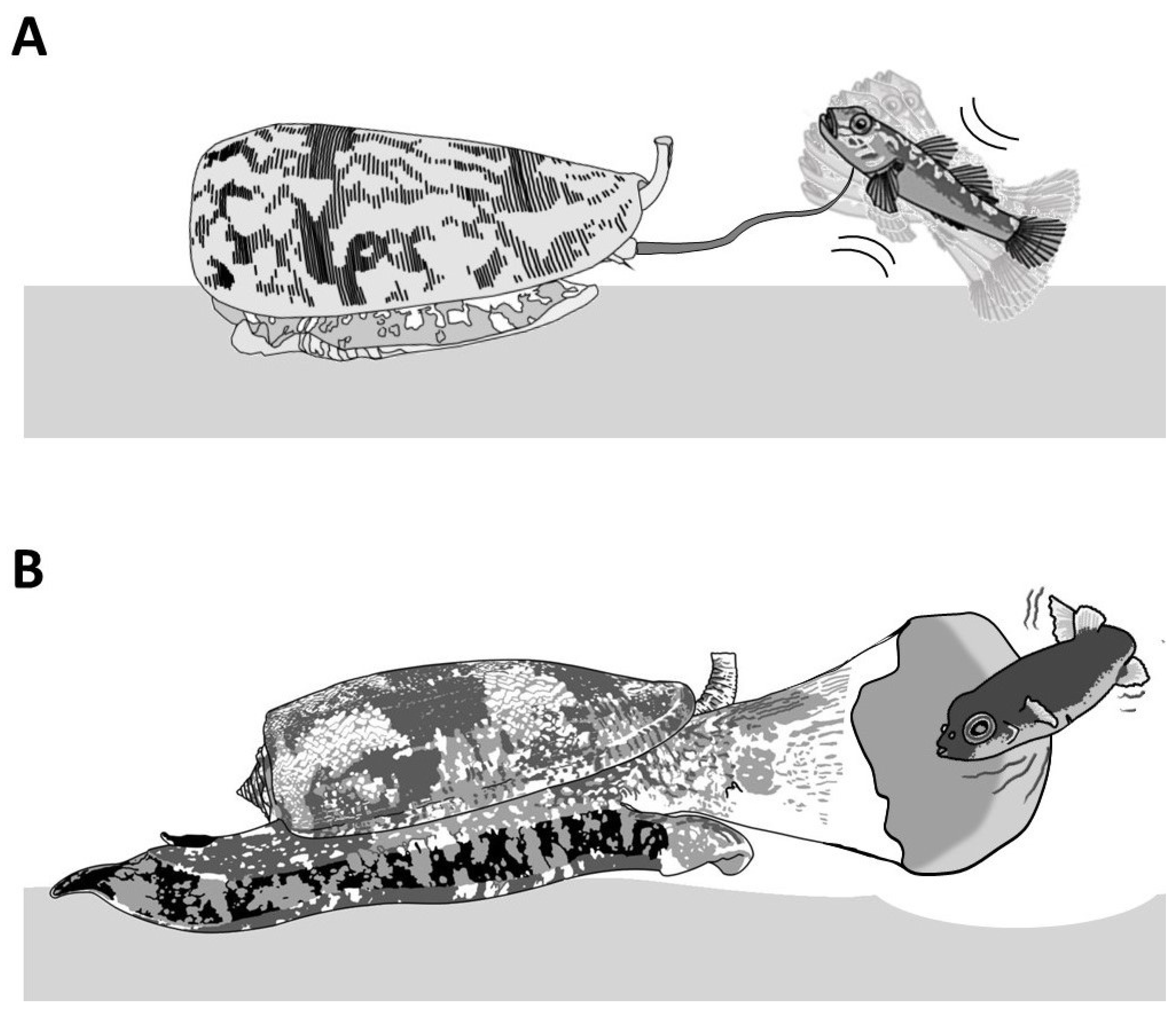
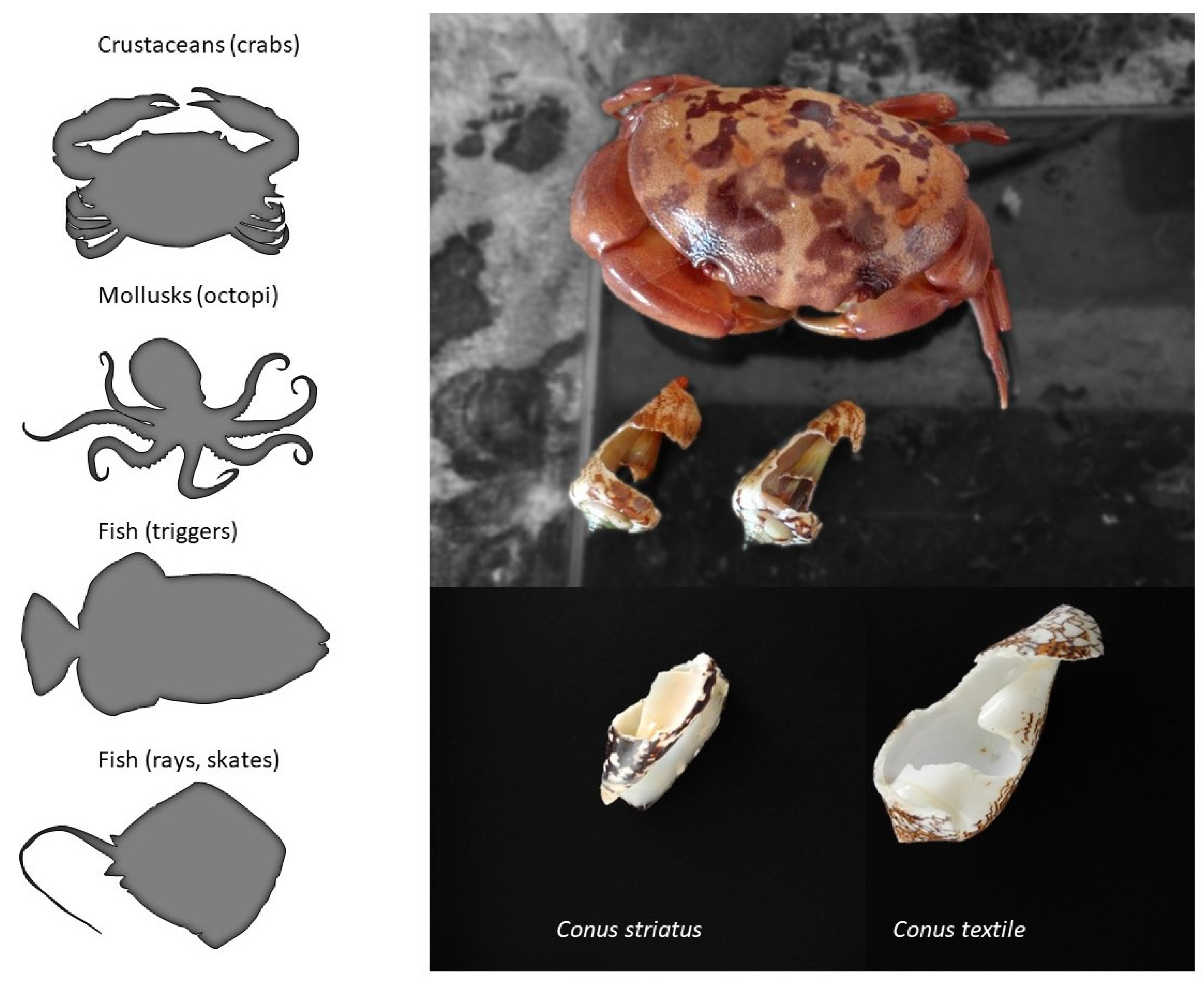
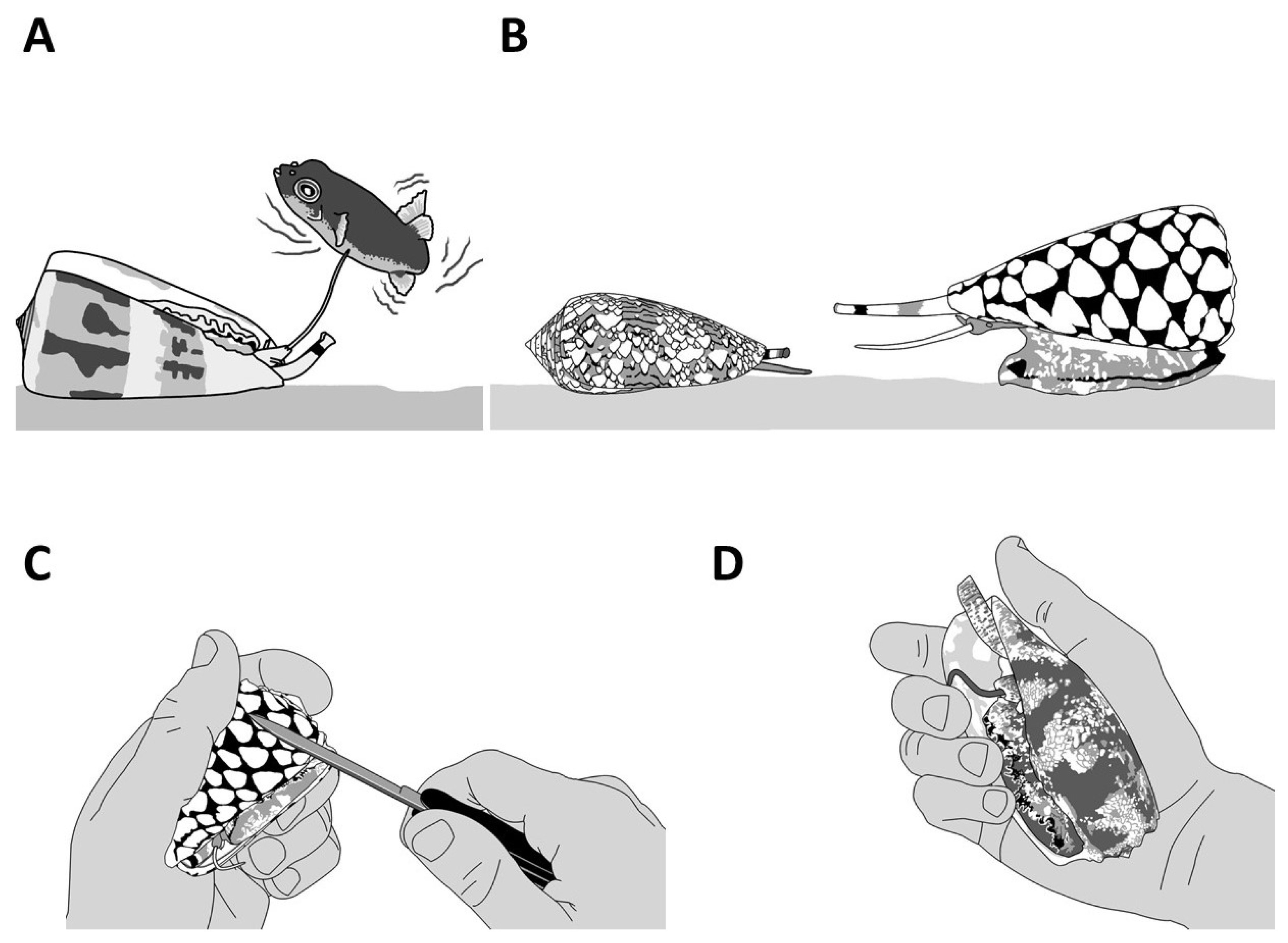
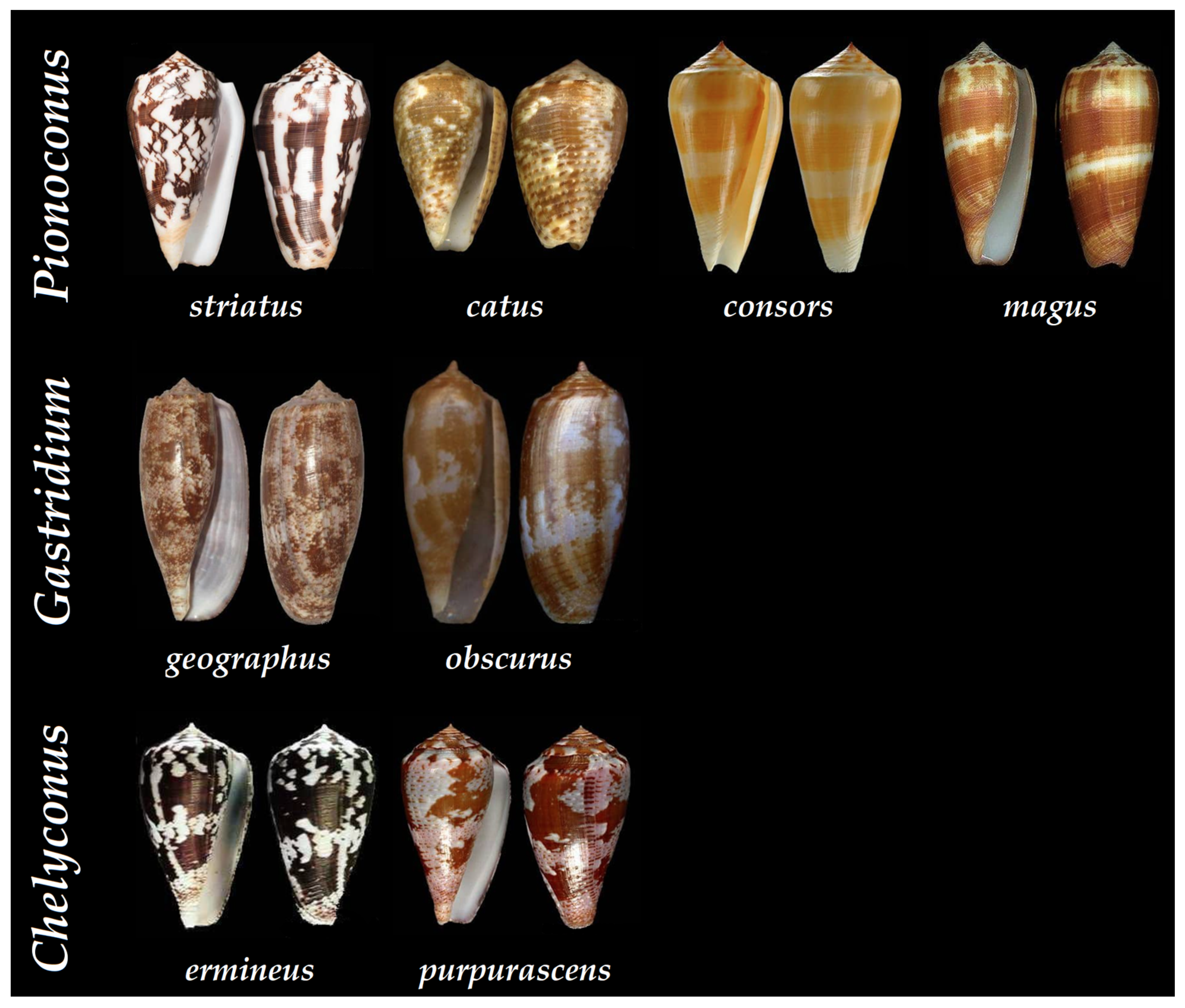




| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | striatus | κA-SIVA | ZKSLVP(gSr)VITTCCGYDOGTMCOOCRCTNSCX | A | IV | [32] |
| κA-SIVB | ZKELVP(gSr)VITTCCGYDOGTMCOOCRCTNSCOTKOKKOX | A | IV | [32] | ||
| κA-SIVC | AOAL(I)VVTATTNCCGYTGOACHOCL(I)CTQTC | IV | [36] | |||
| catus | C4.41 | QKELVPSTITTCCGHEPGTMCPKCMCDNTCPPQKEEKTRPQ | A | IV | [25] | |
| C1.5 | QKELVPSTITTCCGNGTGDNVDPKCMCDNTSSPKKKKRP | A | I | [25] | ||
| consors | κA-CcTx | AOWLVP(gSr)QITTCCGYNOGTMCOSCMCTNTC | A | IV | [24,37] | |
| Chelyconus | purpurascens | κA-PIVE | DCCGVKLEMCHPCLCDNSCKNYGKX | A | IV | [17,38] |
| κA-PIVF | DCCGVKLEMCHPCLCDNSCKKSGKX | A | IV | [17,38] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | striatus | α-SI | ICCNPACGPKYSCX | A | I | [23,32,39] |
| α-SIA | YCCHPACGKNFDCX | A | I | [23,32] | ||
| α-SII | GCCCNPACGPNYGCGTSCS | A | II | [23,32,39] | ||
| consors | α-CnIB | CCHPACGKYYSCX | A | I | [24,37] | |
| α-CnIA | GRCCHPACGKYYSCX | A | I | [24,37] | ||
| CnIG | CCHPACGKYFKCX | I | [37] | |||
| CnIJ | GRCCHPACGGKYFKCX | A | I | [37] | ||
| CnIH | NGRCCHPACGKHFSCX | A | I | [37] | ||
| CnIK | NGRCCHPACGKYYSCX | A | I | [37] | ||
| CnIL | DGRCCHPACGKYYSCX | A | I | [37] | ||
| catus | α-C4.3 | NGRCCHPACGKHFSC | A | I | [25] | |
| α-CIB | GCCSNPVCHLEHPNACX | A | I | [25] | ||
| α-C1.3 | GCCSNPVCHLEHSNLCX | A | I | [25] | ||
| magus | α-MI | GRCCHPACGKNYSCX | A | I | [40] | |
| α-MII | GCCSNPVCHLEHSNLCX | A | I | [40] | ||
| α-MIC | CCHPACGKNYSCX | A | I | [40] | ||
| Gastridium | geographus | α-GIC | GCCSHPACAGNNQHICX | A | I | [28,33] |
| α-GIA | ECCHPACGRHYSCGK | A | I | [28,33] | ||
| α-GII | ECCHPACGKHFSCX | A | I | [28,33] | ||
| α-GID | IRD(Gla)CCSNPACRVNNPHVC | A | I | [28,33] | ||
| obscurus | α-OIVA | CCGVONAACHOCVCKNTCX | A | IV | [28,41] | |
| α-OIVB | CCGVONAACPOCVCNKTCGX | A | IV | [28,42] | ||
| Chelyconus | purpurascens | α-PIB | ZSOGCCWNPACVKNRCX | A | I | [17,43] |
| α-PIC | SGCCKHOACGKNRC | A | I | [17,44] | ||
| α-PIVA | GCCGSYONAACHOCSCKDROSYCGQX | A | IV | [17] | ||
| α-PIIIE | HOOCCLYGKCRRYPGCSSASCCQRX | M | III | [17] | ||
| α-PIIIF | GOOCCLYGSCROFOGCYNALCCRKX | M | III | [17,45] | ||
| ermineus | α-EIVA | GCCGPYONAACHOCGCKVGROOYCDROSGGX | A | IV | [46,47] | |
| α-EIVB | GCCGKYONAACHOCGCTVGROOYCDROSGGX | A | IV | [46,47] | ||
| α-EIIA | ZTOGCCWNPACVKNRCX | A | I | [47,48] | ||
| α-EIIB | ZTOGCCWHPACGKNRCX | A | I | [47,48] | ||
| α-EI | RDOCCYHPTCNMSNPQICX | A | I | [47,49] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | consors | δ-CnVIA | YECYSTGTFCGINGGLCCSNLCLFFVCLTFS | O1 | VI/VII | [37] |
| δ-CnVIB | DECFSOGTFCGTKOGLCCSARCFSFFCISLEFX | O1 | VI/VII | [37] | ||
| δ-CnVIC | DECFSOGTFCGIKOGLCCSARCFSFFCISLEFX | O1 | VI/VII | [37] | ||
| striatus | δ-SVIE | DGCSSGGTFCGIHOGLCCSEFCFLWCITFID | O1 | VI/VII | [32] | |
| catus | δ-CVIE-2 | YGCSNAGAFCGIHOGLCCSELCLVWCT | O1 | VI/VII | [25] | |
| δ-C6.2 | DGCYNAGTFCGIROGLCCSEFCFLWCITFVDSX | O1 | VI/VII | [25] | ||
| Chelyconus | purpurascens | δ-PVIA | EACYAPGTFCGIKPGLCCSEFCLPGVCFGX | O1 | VI/VII | [55,56] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | consors | μ-CnIIIB | ZGCCGEPNLCFTRWCRNNARCCRQQ | M | III | [24,37] |
| μ-CnIIIC | ZGCCNGPKGCSSKWCRDHARCCX | M | III | [24,37] | ||
| striatus | μ-S3-G02 | QKCCGEGSSCPKYFKNNFICGCC | M | III | [32] | |
| Chelyconus | purpurascens | μ-PIIIA | ZRLCCGFOKSCRSRQCKOHRCC | M | III | [56] |
| Gastridium | geographus | μ-Conotoxin-GS | ACSGRGSRCOOQCCMGLRCGRGNPQKCIGAH(Gla)DV | O1 | VI/VII | [28] |
| μ-GIIIA | RDCCTOOKKCKDRQCKOQRCCAX | M | III | [28,33] | ||
| μ-GIIIB | RDCCTOORKCKDRRCKOMKCCAX | M | III | [33] | ||
| μ-GIIIC | RDCCTOOKKCKDRRCKOLKCCA | M | III | [28,33] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | striatus | ω-SVIA | CRSSGSOCGVTSICCGRCYRGKCTX | O1 | VI/VII | [39,64] |
| ω-SVIB | CKLKGQSCRKTSYDCCSGSCGRSGKCX | O1 | VI/VII | [39,64] | ||
| SO4 | ATDCIEAGNYCGPTVMKICCGFCSPYSKICMNYPKN | O1 | VI/VII | [23,32] | ||
| SO5 | STSCMEAGSYCGSTTRICCGYCAYFGKKCIDYPSN | O1 | VI/VII | [23,32] | ||
| consors | ω-CnVIIA | CKGKGAOCTRL(Mox)YDCCHGSCSSSKGRCX | O1 | VI/VII | [24,37,51] | |
| magus | ω-MVIIA | CKGKGAKCSRLMYDCCTGSCRSGKCX | O1 | VI/VII | [40] | |
| ω-MVIIB | CKGKGASCHRTSYDCCTGSCNRGKCX | O1 | VI/VII | [40] | ||
| catus | ω-Catus-C2 | CQGRGASCRKTMYNCCSGSCNRGRC | O1 | VI/VII | [25] | |
| ω-CVIA | CKSTGASCRRTSYDCCTGSCRSGRCX | O1 | VI/VII | [25,61] | ||
| ω-CVID | CKSKGAKCSKLMYDCCSGSCSGTVGRCX | O1 | VI/VII | [25,61] | ||
| Gastridium | geographus | ω-GVIA | CKSOGSSCSOTSYNCCRSCNOYTKRCY | O1 | VI/VII | [28,33] |
| ω-GVIIA | CKSOGTOCSRGMRDCCTSCLLYSNKCRRY | O1 | VI/VII | [28,33] | ||
| ω-GVIIB | CKSOGTOCSRGMRDCCTSCLSYSNKCRRY | O1 | VI/VII | [28,33] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Pionoconus | striatus | Conkunitzin S1 | KDRPSLCDLPADSGSGTKAEKRIYYNSARKQCLRFDYTGQGGNENNFRRTYDCQRTCLYT | Conkunitzin | XIV | [32] |
| Str54 | PSYCNLPADSGSGTKPEQRIYYNSAKKQCVTFTYNGKGGNGNNFSRTNDCRQTCQYPLYACISGCRCET | Conkunitzin | [32] | |||
| Con-ikot-ikot S | SGPADCCRMKECCTDRVNECLQRYSGREDKFVSFCYQEATVTCGSFNEIVGCCYGYQMCMIRVVKPNSLSGAHEACKTVSCGNPCA | Con-ikot-ikot | [32] | |||
| Conkunitzin S2 | ARPKDRPSYCNLPADSGSGTKPEQRIYYNSAKKQCVTFTYNGKGGNGNNFSRTNDCRQTCQYPVG | Conkunitzin | XIV | [32] | ||
| Chelyconus | purpurascens | Contryphan-P | GCVLLPWC | O2 | [35] | |
| Gastridium | geographus | Contryphan-G | GCPWEPWC | O2 | [32] | |
| Conopressin-G | CFIRNCPKGX | Conopressin | [28,33] | |||
| Contulakin-G | QSEEGGSNATKKPYIL | C | [33] | |||
| G5.1 | QGWCCKENIACCV | T | V | [28,33] | ||
| Scratcher peptide | KFLSGGFKIVCHRYCAKGIAKEFCNCPD | XIV | [28,33] | |||
| Conantokin-G | GE(Gla)(Gla)LQ(Gla)NQ(Gla)LIR(Gla)KSN | B1 | [28,33] | |||
| Conophysin-G | THPCMSCSFGQCVGPQICCGLGGCEMGTAEANKCIEEDDDQTPCQVLGDHCDLNNLDIEGHCVADGICCVDDTCAIHSSC | Conophysin | [28] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Gastridium | geographus | σ-GVIIIA | GCTRTCGGOKCTGTCTCTNSSKCGCRYNVHPSG(Btr)GCGCACSX | S | VIII | [28,33] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Cysteine Framework | References |
|---|---|---|---|---|---|
| Conus | marmoreus | α-Mr1.1 | GCCSHPACSVNNPDICX | I | [81] |
| Mr1.8a | ECCTHPACHVSNPELCX | I | [81] | ||
| Mr1.8 | ROECCTHOACHVSNPELCS | I | [81] | ||
| Cylinder | victoriae | α-VcIA | GCCSDPRCNYDHPEICX | I | [28] |
| ammiralis | Ai1.2 | PECCSDPRCNSTHPELCG | I | [80] | |
| Ai1.1 | QECCSYPACNLDHPELC | I | [80] | ||
| P_113 | AGINDVCKSWRDCPQGADCYVDVGLRCRWPSDHSCTANNQCSVDSCINGICKANIGGRCLSDRDCPKGATCKSQE | VIII | [80] | ||
| P_165 | DCPVTGGPNPYHHCMIACMADGTKEYCRCHYCKDCVDSNGDKPAC | XXII | [80] | ||
| P_114 | AGINDVCKSWRDCPQGADCYVDVGLRCRWPSDHSCTANNQCSVDSCINGICKANIGGRCLSNKDCPEGATCKSQGWLNFEKKCET | XXXIII | [80] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Cysteine Framework | References |
|---|---|---|---|---|---|
| Conus | marmoreus | Mr1.9 | VCCPFGGCHELCTADDX | I | [81] |
| Mr3.11 | CCRIACNLKCNOCCX | III | [81] | ||
| Mr3.15 | VCCPHGGCHQICQCCGC | III | [81] | ||
| Mr3.18 | CCHRNWCDHLCSCCGS | III | [81] | ||
| MrIIIF | VCCPFGGCHELCLCCDX | III | [81] | ||
| Mr1e | CCHSSWCKHLC | I | [28,33,81] | ||
| Mr3.8 | CCHWNWCDHLCSCCGS | III | [81] | ||
| MrIIIB | SKQCCHLAACRFGCTOCCW | III | [81] | ||
| MrIIID | CCRLSCGLGCHOCCX | III | [81] | ||
| MrIIIE | VCCPFGGCHELCYCCDX | III | [81] | ||
| MrIIIG | DCCOLPACPFGCNOCCX | III | [81] | ||
| Cylinder | textile | TxIIIC | CCRTCFGCTOCCX | III | [16] |
| Tx3f | RCCKFPCPDSCRYLCCX | III | [16] | ||
| Tx3a | CCSWDVCDHPSCTCCGX | III | [16] | ||
| Tx3h | KFCCDSNWCHISDCECCYX | III | [16] | ||
| ammiralis | P_148 | RCCRFPCPDTCRHLCC | III | [80] | |
| P_099 | FCCDSDWCHLPECLCCN | III | [80] | ||
| P_147 | CCMTCFGCTPCC | III | [80] | ||
| P_063 | CCSWDVCDHPSCTCCS | III | [80] | ||
| P_122 | CCNDSECDYSCWPCCIFS | III | [80] | ||
| P_143 | CCSWDVCDHPSCTCC | III | [80] | ||
| P_149 | CCNAGFCRFGCTPCCWMTSFVIAASSSV | III | [80] | ||
| P_151 | VCCPFGGCHELCQCCE | III | [80] | ||
| P_170 | GILLPALRKFCCDSNWCHISDCECCY | III | [80] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Cysteine Framework | References |
|---|---|---|---|---|---|
| Conus | marmoreus | Mr6.22 | CIDGGEMCDPFSSDCCSGWCIFFFCT | VI/VII | [81] |
| Mr6.8 | CIDGGEICDIFFPNCCSGWCIILVCA | VI/VII | [81] | ||
| MaIr332 | CLDGGEICGILFPSCCSGWCIVLVCA | VI/VII | [81] | ||
| MaIr34 | ECLEADYYCVLPFVGNGMCCSGICVFVCIAQKY | VI/VII | [81] | ||
| MaIr137 | DDECEPPGDFCGFFKIGPPCCSGWCFLWCA | VI/VII | [81] | ||
| Mr6.17 | ACRQKWEYCIVPILGFVYCCPGLICGPFVCV | VI/VII | [81] | ||
| μ-MrVIA | ACRKKWEYCIVPIIGFIYCCPGLICGPFVCV | VI/VII | [81] | ||
| μ-MrVIB | ACSKKWEYCIVPILGFVYCCPGLICGPFVCV | VI/VII | [81] | ||
| Cylinder | textile | TxO1 | CLDAGEVCDIFFPTCCGYCILLFCA | VI/VII | [16] |
| δ-TxVIA | WCKQSGEMCNLLDQNCCDGYCIVLVCT | VI/VII | [16] | ||
| TxO4 | YDCEPPGNFCGMIKIGPOCCSG(Btr)CFFACA | VI/VII | [16] | ||
| victoriae | VcVIB | GKPCHEEGQLCDPFLQNCCLGWNCVFVCI | VI/VII | [28] | |
| ammiralis | P_020 | CVDQFDPCDMIRHTCCVGVCFLMACI | VI/VII | [80] | |
| D_045 | CKQADEPCSILSLDQCCSGVCFGICI | VI/VII | [80] | ||
| D_050 | ECQEKWDYCPIPFFGSRYCCYGLFCTLFFCA | VI/VII | [80] | ||
| D_047 | WCKQSGEMCNFTDQNCCDGYCILLFCT | VI/VII | [80] | ||
| P_019 | CTQSGELCDVIDPDCCNKFCIIFFCI | VI/VII | [80] | ||
| P_162 | CYDGGTSCNTGNQCCSGWCIFVCL | VI/VII | [80] | ||
| P_076 | VKPCRKEGQLCDPIFQNCCRGWNCVFVCI | VI/VII | [80] | ||
| P_121 | DDCEPPGNFCGMIKIGPPCCSGWCFFACA | VI/VII | [80] | ||
| Ai6.1 | WCKQSGEMCNLLDQNCCEGYCIVLVCT | VI/VII | [80] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Cysteine Framework | References |
|---|---|---|---|---|---|
| Conus | marmoreus | Mr6.13 | DCLPIGSLCHSSEQCCSGWCSPKRVC | VI/VII | [28,81] |
| Mr6.14 | SCDQTGEPCVLNEQCCYGWCTNHGTCY | VI/VII | [28,81] | ||
| Mr6.15 | SCVPIGRPCASNEQCCTRWCTPRRIC | VI/VII | [81] | ||
| Mr6.12 | GCKATWMSCSSGWECCSMSCDMYCX | VI/VII | [81] | ||
| MaI51 | QCEDVWMPCTSNWECCSLDCEMYCTQIX | VI/VII | [81] | ||
| Cylinder | ammiralis | P_053 | LCPDYTEPCSHAHECCSWNCHNGHCT | VI/VII | [80] |
| P_167 | NCSDDWQYCESPSDCCSWDCDVVCS | VI/VII | [80] | ||
| D_054 | CRMTPLC | [80] | |||
| P_163 | ECPWKPWC | [80] | |||
| P_067 | WWDGDCRTWNAPCNPGVECCFGRCSHRRCVFW | VI/VII | [80] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Cysteine Framework | References |
|---|---|---|---|---|---|
| Conus | marmoreus | MrVA | NACCIVRQCC | V | [81] |
| Mr5.6 | NGCCRAGDCCS | V | [81] | ||
| χ/λ-CMrX | GICCGVSFCYPC | [81] | |||
| Mr10.2 | ACCVYKICYPC | X | [81] | ||
| Gla-MrIII | FCCRTQ(Gla)VCC(Gla)AIKNX | V | [81] | ||
| Mr5.1b | CCPGWELCC(Gla)WDDGW | V | [28,81] | ||
| Mr5.4a | CCQVMPQCCEWN | V | [81] | ||
| Mr5.4b | CCQIVPQCCEWN | V | [81] | ||
| Mr5.8 | CCQIVPQCCEWVSD | V | [81] | ||
| χ-MrIA | NGVCCGYKLCHPC | X | [81] | ||
| Cylinder | textile | Tx-D0111 | QCCWYFDISCCITV | V | [16] |
| TxXIIIA | TSDCCFYHNCCC | V | [16] | ||
| victoriae | VcVA | CCPGKPCCRIX | V | [28] | |
| Vc5.3 | VNCCGIDESCCS | V | [28] | ||
| ammiralis | P_175 | RCCSIHDNSCCGL | V | [80] | |
| P_071 | NMCCGFKPYCCNW | V | [80] | ||
| P_038 | HDMPLASFHGNAMRILQMLSNNRYCCIFDHSCCLWP | V | [80] | ||
| P_159 | SGCCVIDRNCC | V | [80] | ||
| P_070 | TSDCCFYHNCCC | V | [80] | ||
| P_166 | GCCSYFDVSCCLWP | V | [80] | ||
| P_037 | PCCSIHDSSCCGL | V | [80] | ||
| P_039 | FCCRPVTPCCA | V | [80] | ||
| P_072 | QACCGFKMCVPC | I | [80] | ||
| P_097 | NLQILCCKHTLSCCT | V | [80] |
| Clades | Conus Species | Conotoxin/ Conopeptide | Mature Sequence | Conopeptide Class | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Conus | marmoreus | χ/λ-CMrVIA | VCCGYKLCHPC | X | [81] | |
| Contryphan-M2 | ESECPWHPWCX | Contryphan | [81] | |||
| ω-Contryphan-M | NESECPWHPWCX | Contryphan | [81] | |||
| Conomarphin-Mr1 | DWEYHAHPKPNSFWT | Conomarphin | [81] | |||
| Conomarphin-Mr2 | DWVNHAHPQPNSIWS | Conomarphin | [81] | |||
| Cylinder | textile | Textile Convulsant Peptide (TCP) | NCPYCVVYCCPPAYCEASGCRPPX | O1 | [16] | |
| victoriae | Elevenin-Vc1 | RRIDCKVFVFAPICRGVAA | Elevenin | [21] | ||
| Prohormone-4-Vc1 | IGFPGFSTPPR | Prohormone | [21] | |||
| P_088 | TTVEKNKPGVLDIPVKSNSDDDSIFRYGRRDMQSPLLSERLRF | Conorfamide | [80] | |||
| P_087 | SFGGEHVCWLGDPNHPQGVCGPQVANIVEIRCEEKEAEQGGANNARANTGRTSSLMKRRGFLSLLKKRGKRDEGSPLQRSGRGIVCECCKHHCTKEEFTEYCH | Insulin | XII | [80] | ||
| ammiralis | D_086 | HSGILLAWSGPRNRFVRFG | Conorfamide | [80] | ||
| D_002 | AQDYSTAELCRINSNDCSVPFSWIPCQQHFLAACDRHDTCYLCGAHFNFTQDDCDNAFLRDMTALCANGTDDEGFCLQ | Conodipine | XXXIII | [80] | ||
| D_030 | ASCPENSCDFASPFQCGEMQTCIRLFQVCDGLWHCENGFDEDLAVCAAVLRPLECAIWEFLEEQSDWILPELFNNADSDLVAPVLHGAYSMGDLQSILNLTAQNIENIRNSTRGAIEGDERPLLALGMPEGAWNDVRYLLEELYKLGLDVWTE | Prohormone-4 | XXII | [80] | ||
| ammiralis | P_016 | INCKVFVYAPICRGVAA | Conoporin | [80] | ||
| P_158 | HPTKACMNCTFGQCVGPQVCCGAGGCEMGTAEANRCSEEDEDPIPCLVIGAHCSLNNPGNIHGNCVAHGICCVDDTCAIHFGCL | Conophysin | [80] | |||
| CFIRNCPSGG | Conopressin | [80] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Stephanoconus | imperialis | Im1.1 | DYCCHRGPCMVWC | A | I | [9] |
| Im22.1 | NCKKNILRTYCSNKICGEATKNTNGELQCTMYCRCANGCFRGQYIDWPNQQTNLLFC | E | XXIII | [9] | ||
| Im11.8 | CSDNIGATCSDRFDCCGSMCCIGGQCVVTFAECS | I1 | XI | [9] | ||
| Im11.9 | CHMDCSKMTCCSGICCFYCGRPMCPGT | I2 | XI | [9] | ||
| Im23b | IPYCGQTGAECYSWCIKQDLSKDWCCDFVKTIARLPPAHICSQ | K | XXIII | [9] | ||
| Im23.4 | VPYCGQTGAECYSWCKEQHLIRCCDFVKYVGMNPPADKC | K | XXIII | [9] | ||
| Im23a | IPYCGQTGAECYSWCIKQDLSKDWCCDFVKDIRMNPPADKCP | K | XXIII | [9] | ||
| Vituliconus | planorbis | Pl1.1 | GIRGNCCMFHTCPIDYSRFYCP | A | I | [87] |
| Pl169 | TVIMHNCCTRSFCKRIYPDLCS | A | I | [87] | ||
| Pl170 | GIGGSCCVIRSCAIKFSTLCG | A | I | [87] | ||
| Pl172 | TCYGVCLEDKKPEEHCWEEVTKTVRGEPGDVQFC | A | [87] | |||
| α/κ-PlXIVA | FPRPRICNLACRAGIGHKYPFCHCRX | J | XIV | [87] | ||
| Pl058 | FPRPRICNLACRAGIGYKYPFCHCR | J | [87] | |||
| Pl022 | SFDCCPQYDYCCW | T | [87] | |||
| κ-Y-Pl1 | ARFLHPFQYYTLYRYLTRFLHRYPIYYIRY | Conopeptide Y | [87] | |||
| Pl069 | FQSWPLTNPDLKAAFVKGSAQRVAHGYG | NSf-1 | [87] | |||
| Pl074 | AFKQYNWQRMPYGT | NSf-1 | [87] |
| Clades | Conus Species | Conotoxins | Mature Sequence | Gene Superfamily | Cysteine Framework | References |
|---|---|---|---|---|---|---|
| Stephanoconus | imperialis | Im28.1 | LHCHEISDLTPWILCSPEPLCGGKGCCAQEVCDCSGPVCTCPPCL | D | XXVIII | [9] |
| Rhizoconus | vexillum | α-VxXXA | DVQDCQVSTPGSKWGRCCLNRVCGPMCCPASHCYCVYHRGRGHGCSC | D | XX | [86] |
| α-VxXXB | DDESECIINTRDSPWGRCCRTRMCGSMCCPRNGCTCVYHWRRGHGCSCPG | D | XX | [86] | ||
| α-VxXXC | DLRQCTRNAPGSTWGRCCLNPMCGNFCCPRSGCTCAYNWRRGIYCSC | D | XX | [86] | ||
| capitaneus | Cp20.3 | EVQECQVDTPGSSWGKCCMTRMCGTMCCSRSVCTCVYHWRRGHGCSCPG | D | XX | [86] | |
| Cp20.5 | DNEAECQIDTPGSSWGKCCMTRMCGTMCCSRSVCTCVYHWRRGHGCSCPG | D | XX | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ratibou, Z.; Inguimbert, N.; Dutertre, S. Predatory and Defensive Strategies in Cone Snails. Toxins 2024, 16, 94. https://doi.org/10.3390/toxins16020094
Ratibou Z, Inguimbert N, Dutertre S. Predatory and Defensive Strategies in Cone Snails. Toxins. 2024; 16(2):94. https://doi.org/10.3390/toxins16020094
Chicago/Turabian StyleRatibou, Zahrmina, Nicolas Inguimbert, and Sébastien Dutertre. 2024. "Predatory and Defensive Strategies in Cone Snails" Toxins 16, no. 2: 94. https://doi.org/10.3390/toxins16020094
APA StyleRatibou, Z., Inguimbert, N., & Dutertre, S. (2024). Predatory and Defensive Strategies in Cone Snails. Toxins, 16(2), 94. https://doi.org/10.3390/toxins16020094






