Rapid Detection of Single- and Co-Contaminant Aflatoxins and Fumonisins in Ground Maize Using Hyperspectral Imaging Techniques
Abstract
1. Introduction
2. Results and Discussion
2.1. AF and FM Analysis
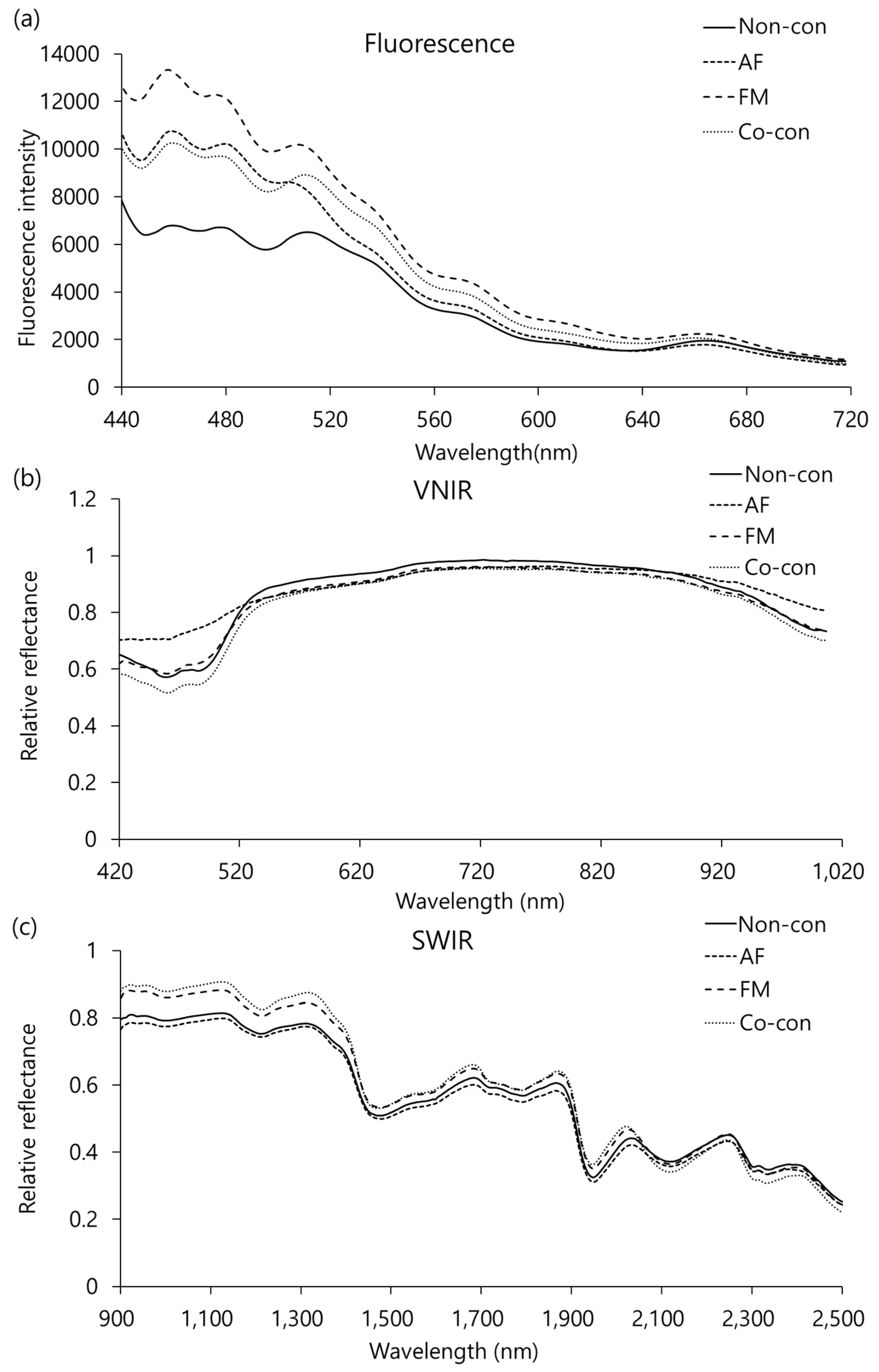
2.2. Spectral Analysis
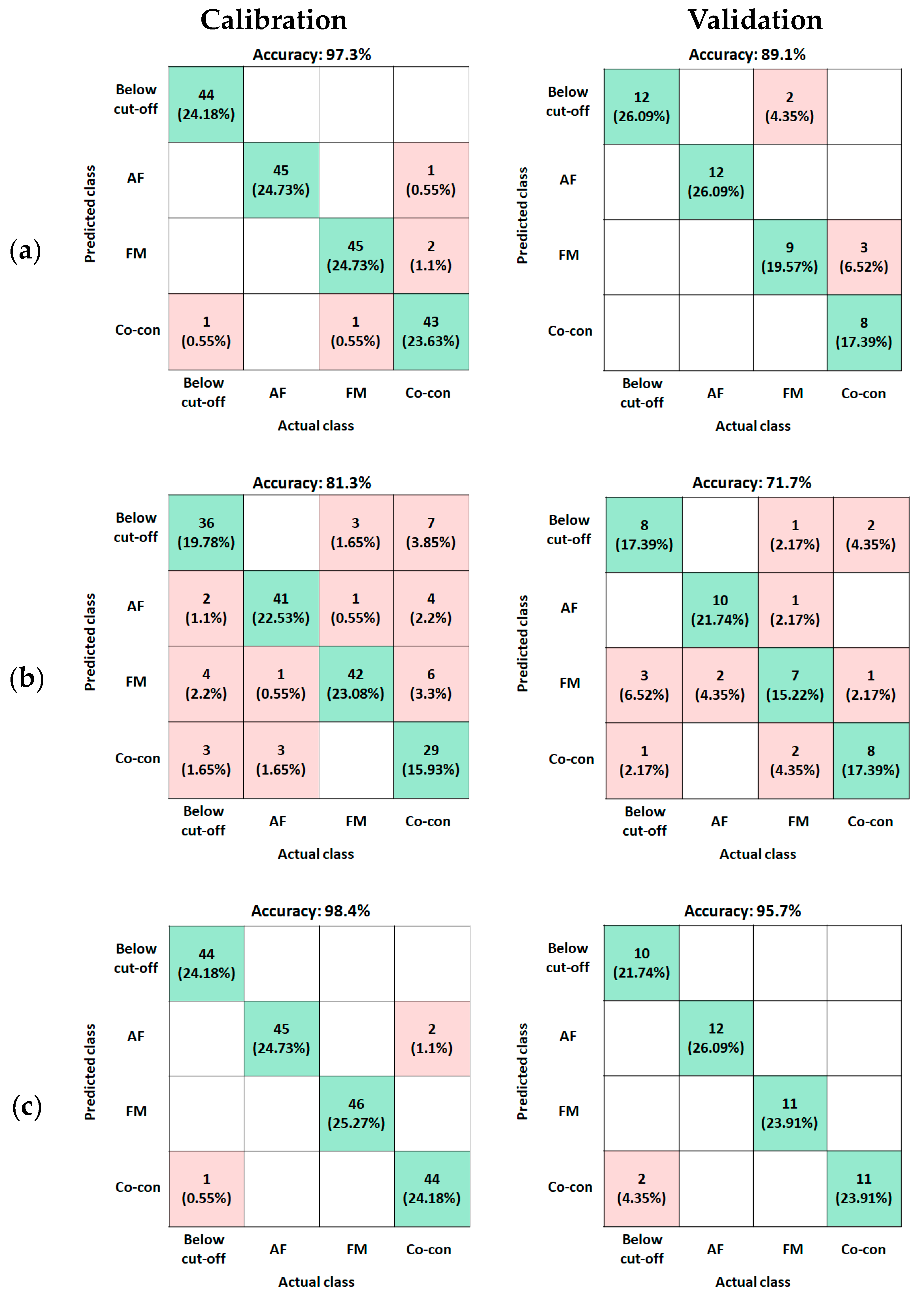
2.3. Classification Result of AF, FM, and AM+FM Co-Contamination
3. Conclusions
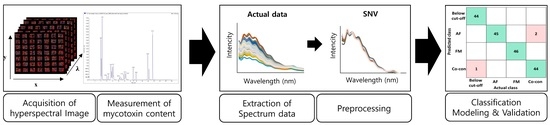
4. Materials and Methods
4.1. Sample Preparation
4.2. Mycotoxin Measurement
4.2.1. AF Analysis Using HPLC
4.2.2. FM Analysis Using LC-MS/MS
4.3. Spectroscopy and Spectral Acquisition
4.4. Spectral Data Preprocessing
4.5. Development and Evaluation of Classification Models
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Awuchi, C.G.; Ondari, E.N.; Nwozo, S.; Odongo, G.A.; Eseoghene, I.J.; Twinomuhwezi, H.; Ogbonna, C.U.; Upadhyay, A.K.; Adeleye, A.O.; Okpala, C.O.R. Mycotoxins’ Toxicological Mechanisms Involving Humans, Livestock and Their Associated Health Concerns: A Review. Toxins 2022, 14, 147. [Google Scholar]
- Eskola, M.; Kos, G.; Elliott, C.T.; Hajšlová, J.; Mayar, S.; Krska, R. Worldwide Contamination of Food-Crops with Mycotoxins: Validity of the Widely Cited ‘FAO Estimate’ of 25%. Crit. Rev. Food Sci. Nutr. 2020, 60, 2773–2789. [Google Scholar] [CrossRef] [PubMed]
- Kensler, T.W.; Roebuck, B.D.; Wogan, G.N.; Groopman, J.D. Aflatoxin: A 50-Year Odyssey of Mechanistic and Translational Toxicology. Toxicol. Sci. 2011, 120, S28–S48. [Google Scholar] [CrossRef] [PubMed]
- Cendoya, E.; Chiotta, M.L.; Zachetti, V.; Chulze, S.N.; Ramirez, M.L. Fumonisins and Fumonisin-Producing Fusarium Occurrence in Wheat and Wheat by Products: A Review. J. Cereal Sci. 2018, 80, 158–166. [Google Scholar]
- Varga, E.; Fodor, P.; Soros, C. Multi-Mycotoxin LC-MS/MS Method Validation and Its Application to Fifty-Four Wheat Flours in Hungary. Food Addit. Contam. Part A 2021, 38, 670–680. [Google Scholar] [CrossRef]
- Solfrizzo, M.; Gambacorta, L.; Bibi, R.; Ciriaci, M.; Paoloni, A.; Pecorelli, I. Multimycotoxin Analysis by LC-MS/MS in Cereal Food and Feed: Comparison of Different Approaches for Extraction, Purification, and Calibration. J. AOAC Int. 2018, 101, 647–657. [Google Scholar] [CrossRef]
- Nolan, P.; Auer, S.; Spehar, A.; Elliott, C.T.; Campbell, K. Current Trends in Rapid Tests for Mycotoxins. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2019, 36, 800–814. [Google Scholar] [PubMed]
- Lee, K.M.; Herrman, T.J. Determination and Prediction of Fumonisin Contamination in Maize by Surface–Enhanced Raman Spectroscopy (SERS). Food Bioprocess Technol. 2016, 9, 588–603. [Google Scholar] [CrossRef]
- Stasiewicz, M.J.; Falade, T.D.O.; Mutuma, M.; Mutiga, S.K.; Harvey, J.J.W.; Fox, G.; Pearson, T.C.; Muthomi, J.W.; Nelson, R.J. Multi-Spectral Kernel Sorting to Reduce Aflatoxins and Fumonisins in Kenyan Maize. Food Control 2017, 78, 203–214. [Google Scholar] [CrossRef]
- Jia, B.; Wang, W.; Ni, X.Z.; Chu, X.; Yoon, S.C.; Lawrence, K.C. Detection of Mycotoxins and Toxigenic Fungi in Cereal Grains Using Vibrational Spectroscopic Techniques: A Review. World Mycotoxin J. 2020, 13, 163–178. [Google Scholar] [CrossRef]
- Xing, F.; Yao, H.; Liu, Y.; Dai, X.; Brown, R.L.; Bhatnagar, D. Recent Developments and Applications of Hyperspectral Imaging for Rapid Detection of Mycotoxins and Mycotoxigenic Fungi in Food Products. Crit. Rev. Food Sci. Nutr. 2019, 59, 173–180. [Google Scholar] [PubMed]
- Su, W.H.; Sun, D.W. Fourier Transform Infrared and Raman and Hyperspectral Imaging Techniques for Quality Determinations of Powdery Foods: A Review. Compr. Rev. Food Sci. Food Saf. 2018, 17, 104–122. [Google Scholar] [CrossRef]
- Qin, J.; Vasefi, F.; Hellberg, R.S.; Akhbardeh, A.; Isaacs, R.B.; Yilmaz, A.G.; Hwang, C.; Baek, I.; Schmidt, W.F.; Kim, M.S. Detection of Fish Fillet Substitution and Mislabeling Using Multimode Hyperspectral Imaging Techniques. Food Control 2020, 114, 107234. [Google Scholar] [CrossRef]
- Qin, J.; Kim, M.S.; Chao, K.; Chan, D.E.; Delwiche, S.R.; Cho, B.K. Line-Scan Hyperspectral Imaging Techniques for Food Safety and Quality Applications. Appl. Sci. 2017, 7, 125. [Google Scholar]
- Lee, H.; Kim, M.S.; Lohumi, S.; Cho, B.K. Detection of Melamine in Milk Powder Using MCT-Based Short-Wave Infrared Hyperspectral Imaging System. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2018, 35, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Jia, B.; Yoon, S.C.; Zhuang, H.; Ni, X.; Guo, B.; Gold, S.E.; Fountain, J.C.; Glenn, A.E.; Lawrence, K.C.; et al. Spatio-Temporal Patterns of Aspergillus Flavus Infection and Aflatoxin B1 Biosynthesis on Maize Kernels Probed by SWIR Hyperspectral Imaging and Synchrotron FTIR Microspectroscopy. Food Chem. 2022, 382, 132340. [Google Scholar] [CrossRef] [PubMed]
- Chavez, R.A.; Cheng, X.; Herrman, T.J.; Stasiewicz, M.J. Single Kernel Aflatoxin and Fumonisin Contamination Distribution and Spectral Classification in Commercial Corn. Food Control 2022, 131, 108393. [Google Scholar] [CrossRef]
- Chu, X.; Wang, W.; Ni, X.; Li, C.; Li, Y. Classifying Maize Kernels Naturally Infected by Fungi Using Near-Infrared Hyperspectral Imaging. Infrared Phys. Technol. 2020, 105, 103242. [Google Scholar] [CrossRef]
- Chavez, R.A.; Cheng, X.; Stasiewicz, M.J. A Review of the Methodology of Analyzing Aflatoxin and Fumonisin in Single Corn Kernels and the Potential Impacts of These Methods on Food Security. Foods 2020, 9, 297. [Google Scholar] [CrossRef]
- Aoun, M.; Stafstrom, W.; Priest, P.; Fuchs, J.; Windham, G.L.; Williams, W.P.; Nelson, R.J. Low-Cost Grain Sorting Technologies to Reduce Mycotoxin Contamination in Maize and Groundnut. Food Control 2020, 118, 107363. [Google Scholar] [CrossRef]
- Shen, G.; Cao, Y.; Yin, X.; Dong, F.; Xu, J.; Shi, J.; Lee, Y.W. Rapid and Nondestructive Quantification of Deoxynivalenol in Individual Wheat Kernels Using Near-Infrared Hyperspectral Imaging and Chemometrics. Food Control 2022, 131, 108420. [Google Scholar] [CrossRef]
- Shen, G.; Kang, X.; Su, J.; Qiu, J.; Liu, X.; Xu, J.; Shi, J.; Mohamed, S.R. Rapid Detection of Fumonisin B1 and B2 in Ground Corn Samples Using Smartphone-Controlled Portable near-Infrared Spectrometry and Chemometrics. Food Chem. 2022, 384, 132487. [Google Scholar] [CrossRef] [PubMed]
- Borutova, R.; Aragon, Y.A.; Nährer, K.; Berthiller, F. Co-Occurrence and Statistical Correlations between Mycotoxins in Feedstuffs Collected in the Asia-Oceania in 2010. Anim. Feed Sci. Technol. 2012, 178, 190–197. [Google Scholar] [CrossRef]
- Grenier, B.; Oswald, I.P. Mycotoxin Co-Contamination of Food and Feed: Meta-Analysis of Publications Describing Toxicological Interactions. World Mycotoxin J. 2011, 4, 285–313. [Google Scholar] [CrossRef]
- Alassane-Kpembi, I.; Schatzmayr, G.; Taranu, I.; Marin, D.; Puel, O.; Oswald, I.P. Mycotoxins Co-Contamination: Methodological Aspects and Biological Relevance of Combined Toxicity Studies. Crit. Rev. Food Sci. Nutr. 2017, 57, 3489–3507. [Google Scholar] [CrossRef] [PubMed]
- Tao, F.; Yao, H.; Zhu, F.; Hruska, Z.; Liu, Y.; Rajasekaran, K.; Bhatnagar, D. A Rapid and Nondestructive Method for Simultaneous Determination of Aflatoxigenic Fungus and Aflatoxin Contamination on Corn Kernels. J. Agric. Food Chem. 2019, 67, 5230–5239. [Google Scholar] [CrossRef]
- Ottoboni, M.; Pinotti, L.; Tretola, M.; Giromini, C.; Fusi, E.; Rebucci, R.; Grillo, M.; Tassoni, L.; Foresta, S.; Gastaldello, S.; et al. Combining E-Nose and Lateral Flow Immunoassays (LFIAs) for Rapid Occurrence/Co-Occurrence Aflatoxin and Fumonisin Detection in Maize. Toxins 2018, 10, 416. [Google Scholar] [CrossRef]
- Wu, Q.; Xu, J.; Xu, H. Interactions of Aflatoxin B1 and Related Secondary Metabolites with Native Cyclodextrins and Their Potential Utilization. Food Control 2018, 94, 102–107. [Google Scholar] [CrossRef]
- Hernández-Sánchez, N.; Lleó, L.; Ammari, F.; Cuadrado, T.R.; Roger, J.M. Fast Fluorescence Spectroscopy Methodology to Monitor the Evolution of Extra Virgin Olive Oils Under Illumination. Food Bioprocess Technol. 2017, 10, 949–961. [Google Scholar] [CrossRef]
- Ojeda-Amador, R.M.; Fregapane, G.; Salvador, M.D. Composition and Properties of Virgin Pistachio Oils and Their By-Products from Different Cultivars. Food Chem. 2018, 240, 123–130. [Google Scholar] [CrossRef]
- Kurek, M.A.; Sokolova, N. Optimization of Bread Quality with Quinoa Flour of Different Particle Size and Degree of Wheat Flour Replacement. Food Sci. Technol. 2020, 40, 307–314. [Google Scholar] [CrossRef]
- Dachoupakan Sirisomboon, C.; Wongthip, P.; Sirisomboon, P. Potential of near Infrared Spectroscopy as a Rapid Method to Detect Aflatoxins in Brown Rice. J. Near Infrared Spectrosc. 2019, 27, 232–240. [Google Scholar] [CrossRef]
- da Conceição, R.R.P.; Simeone, M.L.F.; Queiroz, V.A.V.; de Medeiros, E.P.; de Araújo, J.B.; Coutinho, W.M.; da Silva, D.D.; de Araújo Miguel, R.; de Paula Lana, U.G.; de Resende Stoianoff, M.A. Application of Near-Infrared Hyperspectral (NIR) Images Combined with Multivariate Image Analysis in the Differentiation of Two Mycotoxicogenic Fusarium Species Associated with Maize. Food Chem. 2021, 344, 128615. [Google Scholar] [CrossRef] [PubMed]
- Dachoupakan Sirisomboon, C.; Putthang, R.; Sirisomboon, P. Application of near Infrared Spectroscopy to Detect Aflatoxigenic Fungal Contamination in Rice. Food Control 2013, 33, 207–214. [Google Scholar] [CrossRef]
- Mellon, J.E.; Cotty, P.J.; Dowd, M.K. Influence of Lipids with and without Other Cottonseed Reserve Materials on Aflatoxin B1 Production by Aspergillus Flavus. J. Agric. Food Chem. 2000, 48, 3611–3615. [Google Scholar] [CrossRef]
- Cheng, X.; Vella, A.; Stasiewicz, M.J. Classification of Aflatoxin Contaminated Single Corn Kernels by Ultraviolet to near Infrared Spectroscopy. Food Control 2019, 98, 253–261. [Google Scholar] [CrossRef]
- He, X.; Yan, C.; Jiang, X.; Shen, F.; You, J.; Fang, Y. Classification of Aflatoxin B1 Naturally Contaminated Peanut Using Visible and Near-Infrared Hyperspectral Imaging by Integrating Spectral and Texture Features. Infrared Phys. Technol. 2021, 114, 103652. [Google Scholar] [CrossRef]
- Tyska, D.; Mallmann, A.O.; Vidal, J.K.; de Almeida, C.A.A.; Gressler, L.T.; Mallmann, C.A. Multivariate Method for Prediction of Fumonisins B1 and B2 and Zearalenone in Brazilian Maize Using near Infrared Spectroscopy (NIR). PLoS ONE 2021, 16, e0244957. [Google Scholar] [CrossRef]
- De Girolamo, A.; von Holst, C.; Cortese, M.; Cervellieri, S.; Pascale, M.; Longobardi, F.; Catucci, L.; Porricelli, A.C.R.; Lippolis, V. Rapid Screening of Ochratoxin A in Wheat by Infrared Spectroscopy. Food Chem. 2019, 282, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Wang, M.; Wu, J.; Tao, F.; Chen, Q.; Wang, Q.; Ouyang, Q.; Shi, J.; Zou, X. Quantitative Assessment of Zearalenone in Maize Using Multivariate Algorithms Coupled to Raman Spectroscopy. Food Chem. 2019, 286, 282–288. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Baek, I.; Lee, K.-M.; Qin, J.; Kim, G.; Shin, B.K.; Chan, D.; Herrman, T.J.; Cho, S.; Kim, M.S. Investigation of Reflectance, Fluorescence, and Raman Hyperspectral Imaging Techniques for Rapid Detection of Aflatoxins in Ground Maize. Food Control 2021, 132, 108479. [Google Scholar] [CrossRef]
- Kim, M.S.; Chao, K.; Chan, D.E.; Jun, W.; Lefcourt, A.M.; Delwiche, S.R.; Kang, S.; Lee, K. Line-Scan Hyperspectral Imaging Platform for Agro-Food Safety and Quality Evaluation: System Enhancement and Characterization. Trans. ASABE 2011, 54, 703–711. [Google Scholar] [CrossRef]
- Kim, M.S.; Chen, Y.R.; Mehl, P.M. Hyperspectral Reflectance and Fluorescence Imaging System for Food Quality and Safety. Trans. Am. Soc. Agric. Eng. 2001, 44, 721–729. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial Least Squares for Discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Wold, S.; Sjostrom, M.; Eriksson, L. PLS-Regression: A Basic Tool of Chemometrics. Chemometrics and Intelligent Laboratory Systems. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Z.; Lu, W.; Zhang, X. Determination of the Spread Parameter in the Gaussian Kernel for Classification and Regression. Neurocomputing 2003, 55, 643–663. [Google Scholar] [CrossRef]
- Vapnik, V.; Lerner, A.Y. Pattern Recognition Using Generalized Portrait Method. Autom. Remont Contr. 1963, 24, 888–900. [Google Scholar] [CrossRef]
- Lin, H.; Chen, Q.; Zhao, J.; Zhou, P. Determination of Free Amino Acid Content in Radix Pseudostellariae Using near Infrared (NIR) Spectroscopy and Different Multivariate Calibrations. J. Pharm. Biomed. Anal. 2009, 50, 803–808. [Google Scholar] [CrossRef]
| Parameter | No. of Samples | Min (mg/kg) | Max (mg/kg) | Median (mg/kg) | Mean (mg/kg) | SD (mg/kg) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Below cut-off | 57 | AF < 0.01, FM < 1 | |||||||||
| AF contaminated | 57 | 0.021 | 0.585 | 0.056 | 0.158 | 0.183 | |||||
| FM contaminated | 57 | 1.1 | 16.6 | 4.2 | 4.8 | 2.8 | |||||
| Co-contaminated | 57 | AF | FM | AF | FM | AF | FM | AF | FM | AF | FM |
| 0.012 | 1.1 | 0.136 | 15 | 0.042 | 4.4 | 0.049 | 5.1 | 0.028 | 2.8 | ||
| Preprocessing Method | Raw | Max Normalization | Range Normalization | SNV | SG2 | ||
|---|---|---|---|---|---|---|---|
| Fluorescence | Calibration (%) | Accuracy | 50.5 | 92.9 | 92.9 | 97.3 | 100 |
| Validation (%) | Accuracy | 39.1 | 80.5 | 87.0 | 89.7 | 67.4 | |
| Precision | 29.1 | 81.1 | 87.1 | 90.2 | 71.7 | ||
| Recall | 40.9 | 80.1 | 86.6 | 88.6 | 67.6 | ||
| F1 score | 29.6 | 80.0 | 86.2 | 88.7 | 67.9 | ||
| VNIR | Calibration (%) | Accuracy | 87.9 | 85.9 | 81.3 | 99.5 | 79.1 |
| Validation (%) | Accuracy | 63.0 | 71.7 | 71.7 | 67.4 | 47.8 | |
| Precision | 62.5 | 71.2 | 72.6 | 67.7 | 48.8 | ||
| Recall | 63.1 | 71.4 | 71.6 | 67.0 | 47.5 | ||
| F1 score | 62.7 | 71.1 | 71.9 | 67.1 | 47.6 | ||
| SWIR | Calibration (%) | Accuracy | 97.8 | 92.0 | 87.4 | 100 | 98.4 |
| Validation (%) | Accuracy | 89.1 | 95.7 | 87.0 | 91.3 | 95.7 | |
| Precision | 90.8 | 96.2 | 91.2 | 92.1 | 96.2 | ||
| Recall | 88.8 | 95.8 | 86.7 | 91.3 | 95.8 | ||
| F1 score | 89.0 | 95.6 | 86.8 | 91.5 | 95.6 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.-K.; Baek, I.; Lee, K.-M.; Kim, G.; Kim, S.; Kim, S.-Y.; Chan, D.; Herrman, T.J.; Kim, N.; Kim, M.S. Rapid Detection of Single- and Co-Contaminant Aflatoxins and Fumonisins in Ground Maize Using Hyperspectral Imaging Techniques. Toxins 2023, 15, 472. https://doi.org/10.3390/toxins15070472
Kim Y-K, Baek I, Lee K-M, Kim G, Kim S, Kim S-Y, Chan D, Herrman TJ, Kim N, Kim MS. Rapid Detection of Single- and Co-Contaminant Aflatoxins and Fumonisins in Ground Maize Using Hyperspectral Imaging Techniques. Toxins. 2023; 15(7):472. https://doi.org/10.3390/toxins15070472
Chicago/Turabian StyleKim, Yong-Kyoung, Insuck Baek, Kyung-Min Lee, Geonwoo Kim, Seyeon Kim, Sung-Youn Kim, Diane Chan, Timothy J. Herrman, Namkuk Kim, and Moon S. Kim. 2023. "Rapid Detection of Single- and Co-Contaminant Aflatoxins and Fumonisins in Ground Maize Using Hyperspectral Imaging Techniques" Toxins 15, no. 7: 472. https://doi.org/10.3390/toxins15070472
APA StyleKim, Y.-K., Baek, I., Lee, K.-M., Kim, G., Kim, S., Kim, S.-Y., Chan, D., Herrman, T. J., Kim, N., & Kim, M. S. (2023). Rapid Detection of Single- and Co-Contaminant Aflatoxins and Fumonisins in Ground Maize Using Hyperspectral Imaging Techniques. Toxins, 15(7), 472. https://doi.org/10.3390/toxins15070472