From Foodborne Disease Outbreak (FBDO) to Investigation: The Plant Toxin Trap, Brittany, France, 2018
Abstract
1. Introduction
2. Results
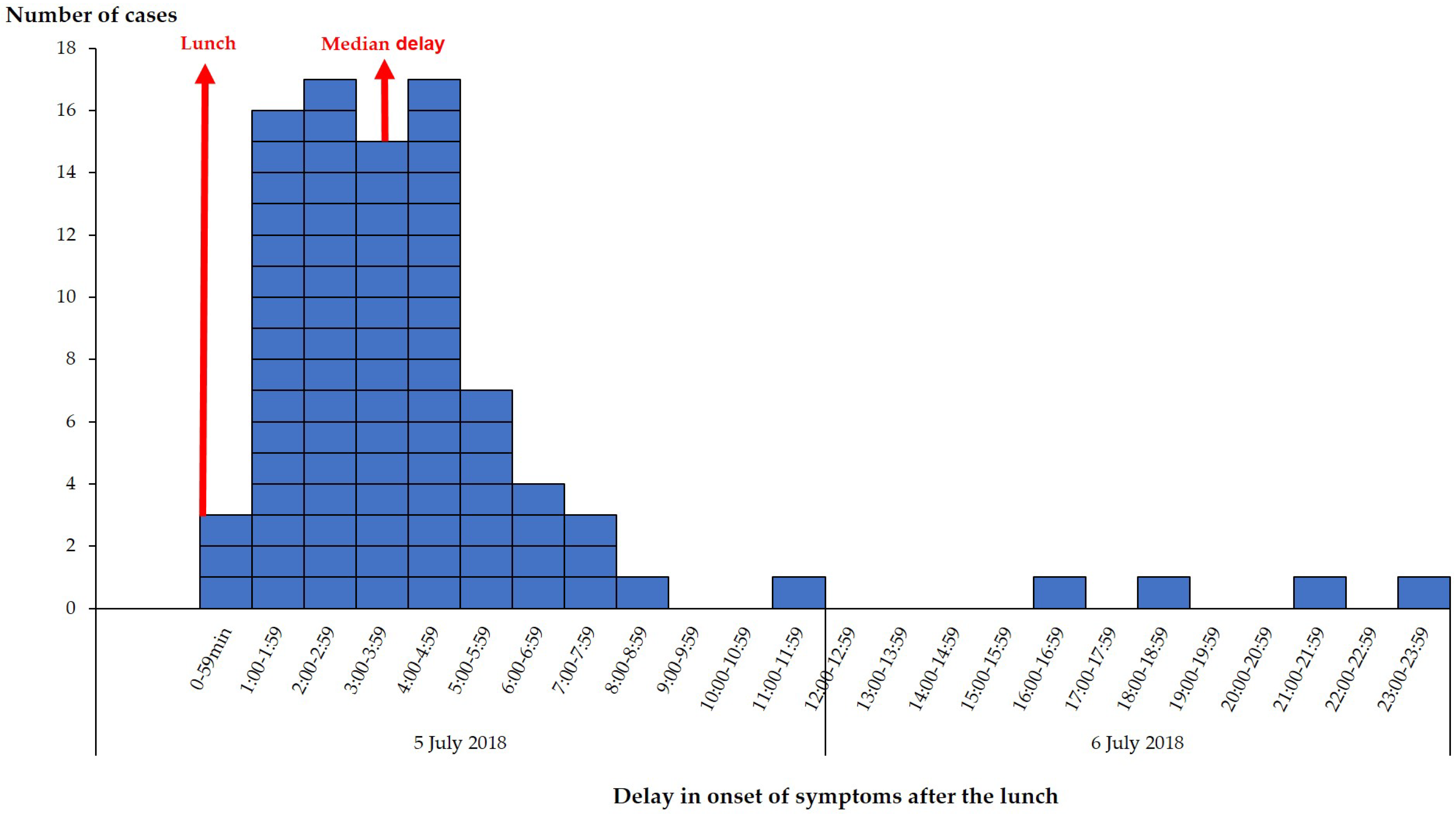
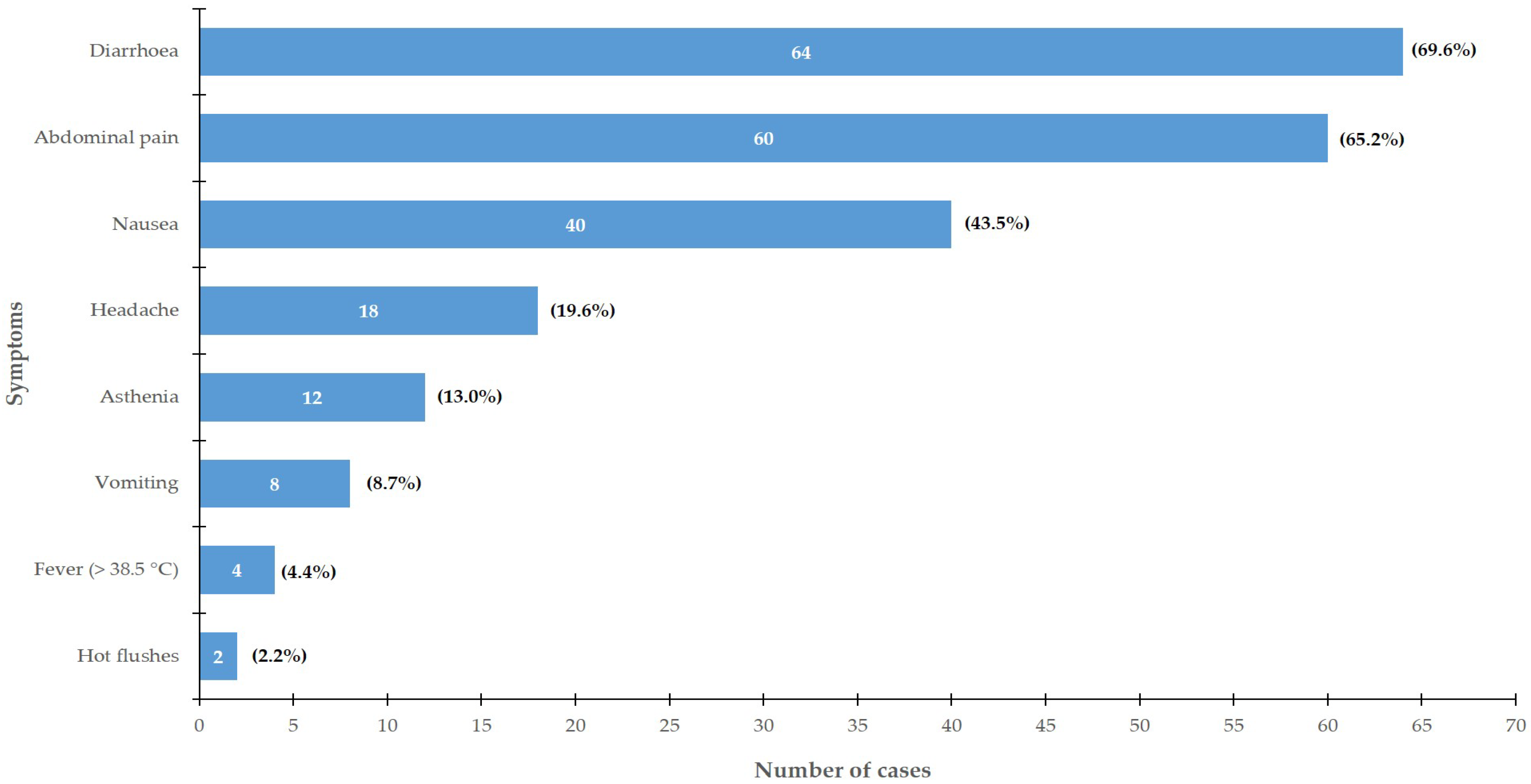
2.1. Epidemiological Investigation
2.2. Environmental Investigation
2.3. Laboratory Investigation
2.3.1. Round 1 Testing: FBDO Investigation
Stool Samples
Food Samples
2.3.2. Round 2 Testing: PHA and Exclusion of Differential Diagnoses
Stool Samples
Food Samples
2.4. Outbreak Control Measures
3. Discussion
4. Conclusions
5. Methods
5.1. Epidemiological Investigation
5.2. Environmental Investigation
5.3. Laboratory Investigation
5.3.1. Round 1 Testing: Investigation for FBDO
Stool Samples
Food Samples
5.3.2. Round 2 Testing: PHA and Exclusion of Differential Diagnoses
Stool Samples
Food Samples
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nizet, V.; Varki, A.; Aebi, M. Microbial Lectins: Hemagglutinins, Adhesins, and Toxins. In Essentials of Glycobiology, 3rd ed.; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G., Aebi, M., Darvill, A.G., Kinoshita, T., Packer, N.H., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017; pp. 481–491. [Google Scholar]
- Kumar, S.; Verma, A.K.; Das, M.; Jain, S.K.; Dwivedi, P.D. Clinical complications of kidney bean (Phaseolus vulgaris L.) consumption. Nutrition 2013, 29, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Miyake, K.; Tanaka, T.; McNeil, P.L. Lectin-based food poisoning: A new mechanism of protein toxicity. PLoS ONE 2007, 2, e687. [Google Scholar] [CrossRef] [PubMed]
- Al-Khaldi, S. Phytohaemagglutinin (Kidney Bean Lectin) In Bad Bug Book. Handbook of Food Borne Pathogenic Microorganisms and Natural Toxins, 2nd ed.; Lampel, K.A., Al-Khaldi, S., Cahill, S.M., Eds.; Food and Drug Administration: Silver Spring, MD, USA, 2012; pp. 254–256.
- Adamcová, A.; Laursen, K.H.; Ballin, N.Z. Lectin Activity in Commonly Consumed Plant-Based Foods: Calling for Method Harmonization and Risk Assessment. Foods 2021, 10, 2796. [Google Scholar] [CrossRef] [PubMed]
- Noah, N.D.; Bender, A.E.; Reaidi, G.B.; Gilbert, R.J. Food poisoning from raw red kidney beans. Br. Med. J. 1980, 281, 236–237. [Google Scholar]
- Rodhouse, J.C.; Haugh, C.A.; Roberts, D.; Gilbert, R.J. Red kidney bean poisoning in the UK: An analysis of 50 suspected incidents between 1976 and 1989. Epidemiol. Infect. 1990, 105, 485–491. [Google Scholar] [CrossRef]
- Vichova, P.; Jahodar, L. Plant poisonings in children in the Czech Republic, 1996−2001. Hum. Exp. Toxicol. 2003, 22, 467–472. [Google Scholar] [CrossRef]
- Ogawa, H.; Date, K. The “White Kidney Bean Incident” in Japan. Methods Mol. Biol. 2014, 1200, 39–45. [Google Scholar]
- Sun, Y.; Liu, J.; Huang, Y.; Li, M.; Lu, J.; Jin, N.; He, Y.; Fan, B. Phytohemagglutinin content in fresh kidney bean in China. Int. J. Food Prop. 2019, 22, 405–413. [Google Scholar] [CrossRef]
- Pilegaard, K.; Olesen, P.T. Forespørgsel om Mulig Forgiftning Forårsaget af “Gourmetbønner”/Snitbønner; J. nr 13/01534; The Danish Technical University: Lyngby, Denmark, 2013. [Google Scholar]
- Mayet, A.; Manet, G.; Decam, C.; Morisson, D.; Bédubourg, G.; de Santi, V.P.; Migliani, R. Epidemiology of food-borne disease outbreaks in the French armed forces: A review of investigations conducted from 1999 to 2009. J. Infect. 2011, 63, 370–374. [Google Scholar] [CrossRef]
- De Laval, F.; Nivoix, P.; Pommier de Santi, V.; Caballe, D.; Garnotel, E.; Maslin, J. Severe norovirus outbreak among soldiers in the field: Foodborne followed by person-to-person transmission. Clin. Infect. Dis. 2011, 53, 399–400. [Google Scholar] [CrossRef]
- Sanchez, M.A.; Corcostégui, S.P.; De Broucker, C.A.; Cabre, O.; Watier-Grillot, S.; Perelle, S.; de Santi, V.P. Norovirus GII.17 Outbreak Linked to an Infected Post-Symptomatic Food Worker in a French Military Unit Located in France. Food Environ. Virol. 2017, 9, 234–237. [Google Scholar] [CrossRef]
- Velut, G.; Delon, F.; Mérigaud, J.P.; Tong, C.; Duflos, G.; Boissan, F.; de Santi, V.P. Histamine food poisoning: A sudden, large outbreak linked to fresh yellowfin tuna from Reunion Island, France, April 2017. Euro Surveill. 2019, 24, 1800405. [Google Scholar] [CrossRef]
- Watier-Grillot, S.; Boni, M.; Tong, C.; Renoult, P.A.; Fournier, A.; Joie, L.; de Santi, V.P. Challenging Investigation of a Norovirus Foodborne Disease Outbreak During a Military Deployment in Central African Republic. Food Environ. Virol. 2017, 9, 498–501. [Google Scholar] [CrossRef]
- Cybulski, R.J.; Bateman, A.C.; Bourassa, L.; Bryan, A.; Beail, B.; Matsumoto, J.; Fang, F.C. Clinical Impact of a Multiplex Gastrointestinal Polymerase Chain Reaction Panel in Patients With Acute Gastroenteritis. Clin. Infect. Dis. 2018, 67, 1688–1696. [Google Scholar] [CrossRef]
- Watier-Grillot, S.; Costa, D.; Petit, C.; Razakandrainibe, R.; Larréché, S.; Tong, C.; de Santi, V.P. Cryptosporidiosis outbreaks linked to the public water supply in a military camp, France. PLoS Negl. Trop. Dis. 2022, 16, e0010776. [Google Scholar] [CrossRef]
- McClane, B.A.; Robertson, S.L.; Li, J. Clostridium perfringens. In Food Microbiology: Fundamentals and Frontiers, 4th ed.; Doyle, M.P., Buchanan, R.L., Eds.; ASM Press: Washington, DC, USA, 2013; pp. 465–489. [Google Scholar]
- Stiles, B.G.; Barth, G.; Barth, H.; Popoff, M.R. Clostridium perfringens epsilon toxin: A malevolent molecule for animals and man? Toxins 2013, 5, 2138–2160. [Google Scholar] [CrossRef]
- Neumann, T.; Krüger, M.; Weisemann, J.; Mahrhold, S.; Stern, D.; Dorner, M.B.; Dorner, B.G. Innovative and Highly Sensitive Detection of Clostridium perfringens Enterotoxin Based on Receptor Interaction and Monoclonal Antibodies. Toxins 2021, 13, 266. [Google Scholar] [CrossRef]
- Brynestad, S.; Synstad, B.; Granum, P.E. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 1997, 143, 2109–2115. [Google Scholar] [CrossRef]
- Miyamoto, K.; Chakrabarti, G.; Morino, Y.; McClane, B.A. Organization of the plasmid cpe locus in Clostridium perfringens type A isolates. Infect. Immun. 2002, 70, 4261–4272. [Google Scholar] [CrossRef]
- Sparks, S.G.; Carman, R.J.; Sarker, M.R.; McClane, B.A. Genotyping of enterotoxigenic Clostridium perfringens fecal isolates associated with antibiotic-associated diarrhea and food poisoning in North America. J. Clin. Microbiol. 2001, 39, 883–888. [Google Scholar] [CrossRef]
- Kitadokoro, K.; Nishimura, K.; Kamitani, S.; Fukui-Miyazaki, A.; Toshima, H.; Abe, H.; Horiguchi, Y. Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J. Biol. Chem. 2011, 286, 19549–19555. [Google Scholar] [CrossRef] [PubMed]
- Mancheño, J.M.; Tateno, H.; Goldstein, I.J.; Martínez-Ripoll, M.; Hermoso, J.A. Structural analysis of the Laetiporus sulphureus hemolytic pore-forming lectin in complex with sugars. J. Biol. Chem. 2005, 280, 17251–17259. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Natural Toxins in Food. Available online: https://www.who.int/news-room/fact-sheets/detail/natural-toxins-in-food (accessed on 29 May 2023).
- Titball, R.W.; Hunter, S.E.; Martin, K.L.; Morris, B.C.; Shuttleworth, A.D.; Rubidge, T.; Kelly, D.C. Molecular cloning and nucleotide sequence of the alpha-toxin (phospholipase C) of Clostridium perfringens. Infect. Immun. 1989, 57, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Van Damme-Jongsten, M.; Wernars, K.; Notermans, S. Cloning and sequencing of the Clostridium perfringens enterotoxin gene. Antonie Van Leeuwenhoek 1989, 56, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Tweten, R.K. Cloning and expression in Escherichia coli of the perfringolysin O (theta-toxin) gene from Clostridium perfringens and characterization of the gene product. Infect. Immun. 1988, 56, 3228–3234. [Google Scholar] [CrossRef] [PubMed]
- Goswami, P.P.; Girish, K.S.; Chaudhuri, P.; Tiwari, V.; Akare, S.J.; Harbola, P.C. Cloning and sequencing of beta toxin gene of Clostridium perfringens type C. Indian J. Exp. Biol. 2002, 40, 109–110. [Google Scholar]
- Alouf, J.E.; Jolivet-Reynaud, C. Purification and characterization of Clostridium perfringens delta-toxin. Infect. Immun. 1981, 31, 536–546. [Google Scholar] [CrossRef]
- Hunter, S.E.; Clarke, I.N.; Kelly, D.C.; Titball, R.W. Cloning and nucleotide sequencing of the Clostridium perfringens epsilon-toxin gene and its expression in Escherichia coli. Infect. Immun. 1992, 60, 102–110. [Google Scholar] [CrossRef]
- Perelle, S.; Gibert, M.; Boquet, P.; Popoff, M.R. Characterization of Clostridium perfringens iota-toxin genes and expression in Escherichia coli. Infect. Immun. 1993, 61, 5147–5156, Erratum in Infect. Immun. 1995, 63, 4967. [Google Scholar] [CrossRef]
- Keyburn, A.L.; Yan, X.X.; Bannam, T.L.; Van Immerseel, F.; Rood, J.I.; Moore, R.J. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet. Res. 2010, 41, 21. [Google Scholar] [CrossRef]
- Amimoto, K.; Noro, T.; Oishi, E.; Shimizu, M. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 2007, 153, 1198–1206. [Google Scholar] [CrossRef]
- International Organization for Standardization (ISO). ISO/IEC 17025:2017 Standard. General Requirements for the Competence of Testing and Calibration Laboratories. Available online: https://www.iso.org/standard/66912.html (accessed on 29 May 2023).
- Wehrle, E.; Didier, A.; Moravek, M.; Dietrich, R.; Märtlbauer, E. Detection of Bacillus cereus with enteropathogenic potential by multiplex real-time PCR based on SYBR Green I. Mol. Cell Probes 2010, 24, 124–130. [Google Scholar] [CrossRef]
- Kim, J.B.; Kim, J.M.; Park, Y.B.; Han, J.A.; Lee, S.H.; Kwak, H.S.; Oh, D.H. Evaluation of various PCR assays for the detection of emetic toxin producing Bacillus cereus. J. Microbiol. Biotechnol. 2010, 20, 1107–1113. [Google Scholar]
- Shi, L.; Arntfield, S.D.; Nickerson, M. Changes in levels of phytic acid, lectins and oxalates during soaking and cooking of Canadian pulses. Food Res. Int. 2018, 107, 660–668. [Google Scholar] [CrossRef]
- Liener, I.E.; Hill, E.G. The effect of heat treatment on the nutritive value and hemagglutinating activity of soybean oil meal. J. Nutr. 1953, 49, 609–620. [Google Scholar] [CrossRef]
- Féraudet-Tarisse, C.; Goulard-Huet, C.; Nia, Y.; Devilliers, K.; Marcé, D.; Dambrune, C.; Simon, S. Highly Sensitive and Specific Detection of Staphylococcal Enterotoxins SEA, SEG, SEH, and SEI by Immunoassay. Toxins 2021, 13, 130. [Google Scholar] [CrossRef]
- Cornillot, E.; Saint-Joanis, B.; Daube, G.; Katayama, S.; Granum, P.E.; Canard, B.; Cole, S.T. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 1995, 15, 639–647. [Google Scholar] [CrossRef]
| Food Items | Outbreak Cases (n = 92) | Control Cases (n = 113) | OR [95% CI] | p Value | aOR [95% CI] | p Value |
|---|---|---|---|---|---|---|
| n (%) | n (%) | |||||
| Starters | ||||||
| Mexican salad with rice | 1 (1.1) | 0 (0) | NA | NA | - | - |
| Celery remoulade | 3 (3.3) | 4 (3.5) | 0.9 [0.2–4.2] | 1.0 | - | - |
| Grated carrots | 4 (4.3) | 12 (10.6) | 0.4 [0.1–1.2] | 0.09 | - | - |
| Green bean salad | 2 (2.2) | 5 (4.4) | 0.5 [0.1–2.5] | 0.5 | - | - |
| Mushroom salad | 3 (3.3) | 3 (2.7) | 1.2 [0.2–6.3] | 1.0 | - | - |
| Pasta salad with parsley | 1 (1.1) | 2 (1.8) | 0.6 [0.05–6.8] | 1.0 | - | - |
| Rabbit pâté | 4 (4.3) | 0 (0) | NA | NA | - | - |
| Cured ham | 1 (1.1) | 6 (5.3) | 0.2 [0.02–1.7] | 0.13 | - | - |
| Boiled eggs with mayonnaise | 4 (4.3) | 8 (7.1) | 0.6 [0.2–2.0] | 0.4 | - | - |
| Guacamole/surimi verrines | 6 (6.5) | 2 (1.8) | 3.9 [0.8–19.7] | 0.14 | - | - |
| Avocado salad | 1 (1.1) | 0 (0) | NA | NA | - | - |
| Main courses | ||||||
| Chili con carne | 83 (90.2) | 26 (23) | 30.9 [11.7–69.8] | <0.0001 | 32.8 [13.8–77.8] | <0.0001 |
| Chili sin carne (vegetarian chili) | 2 (2.2) | 0 (0) | NA | NA | - | - |
| Pan-fried corn and red kidney beans | 32 (34.8) | 13 (11.5) | 4.1 [2.0–8.4] | <0.0001 | 4.8 [1.8–13.1] | 0.002 |
| Mexican pan-fried vegetables | 8 (8.7) | 5 (4.4) | 2.1 [0.6–6.5] | 0.26 | - | - |
| Fish of the day | 0 (0) | 8 (7.1) | NA | NA | - | - |
| Hake fish | 0 (0) | 1 (0.9) | NA | NA | - | - |
| Boiled ham | 0 (0) | 1 (0.9) | NA | NA | - | - |
| Baked ham | 2 (2.2) | 2 (1.8) | 1.2 [2.0–8.9] | 1.0 | - | - |
| Hamburger | 0 (0) | 8 (7.1) | NA | NA | - | - |
| Butcher’s minced steak | 0 (0) | 9 (8.0) | NA | NA | - | - |
| T-bone steak | 0 (0) | 2 (1.8) | NA | NA | - | - |
| Ribs | 2 (2.2) | 12 (10.6) | 0.2 [0.04–0.9] | 0.02 | - | - |
| Poultry cutlet | 0 (0) | 6 (5.3) | NA | NA | - | - |
| Sliced poultry | 1 (1.1) | 1 (0.9) | 1.2 [0.1–19.9] | 1.0 | - | - |
| Bacon pizza | 0 (0) | 7 (6.2) | NA | NA | - | - |
| Pasta recipe 1 | 0 (0) | 12 (10.6) | NA | NA | - | - |
| Pasta recipe 2 | 3 (3.3) | 6 (5.3) | 0.6 [0.15–2.5] | 0.7 | - | - |
| Parsleyed courgettes | 6 (6.5) | 8 (7.1) | 0.9 [0.3–2.7] | 0.9 | - | - |
| French fries | 8 (8.7) | 29 (25.7) | 0.3 [0.1–0.6] | 0.002 | - | - |
| Boiled rice | 30 (32.6) | 25 (22.1) | 1.7 [0.9–3.2] | 0.09 | - | - |
| Testing Parameter | Testing Method | Result |
|---|---|---|
| Round 1 testing | ||
| 22 gastrointestinal pathogens a | BioFire® FilmArray® Gastrointestinal Panel (BioMérieux Laboratories, Craponne, France) b,c | Negative |
| Salmonella spp. | Culture (BioMérieux Laboratories specific agars, Craponne, France) b,c | Negative |
| Shigella spp. | Negative | |
| Yesinia enterolytica | Negative | |
| Staphylococcus aureus | Negative | |
| Campylobacter spp. | Negative | |
| CPE | PET-RPLA® Toxin Detection Kit (Oxoid Ltd., Hampshire, United Kingdom) b | Positive for four stools Stool 1: positive at 1/64 dilution Stool 2: positive at 1/32 dilution Stools 3,4: positive without dilution d |
| Clostridium perfringens | Culture (BioMérieux Laboratories specific agars, Craponne, France) b,c | Positive for one stool (Stool 1) |
| Molecular biology and strain typing e,f | Positive for one stool (Stool 1) Strain producing CPE, Alpha and Theta toxins | |
| Round 2 testing | ||
| Bacillus cereus | Molecular biology e | Negative |
| Food Item | Testing Parameter | Testing Method | Results |
|---|---|---|---|
| Round 1 testing | |||
| Chili con carne a Chili sin carne a Red kidney beans b Frozen ground beef b Frozen sliced onions b Frozen pepper strips b Chili powder b Mexican spice mix b | C. perfringens | Culture (NF EN ISO 7937 standard) c | Negative |
| B. cereus | Culture (NF EN ISO 7932 standard) c | ||
| S. aureus | Culture (NF EN ISO 6888-2 standard) c | ||
| E. coli | Culture (BRD 07/11—12/05 certificate) c,d | ||
| Salmonella spp. | Culture (3M 01/8—06/01 certificate) c,d | ||
| Chili con carne a | C. perfringens | Molecular biology e | Negative |
| Round 2 testing | |||
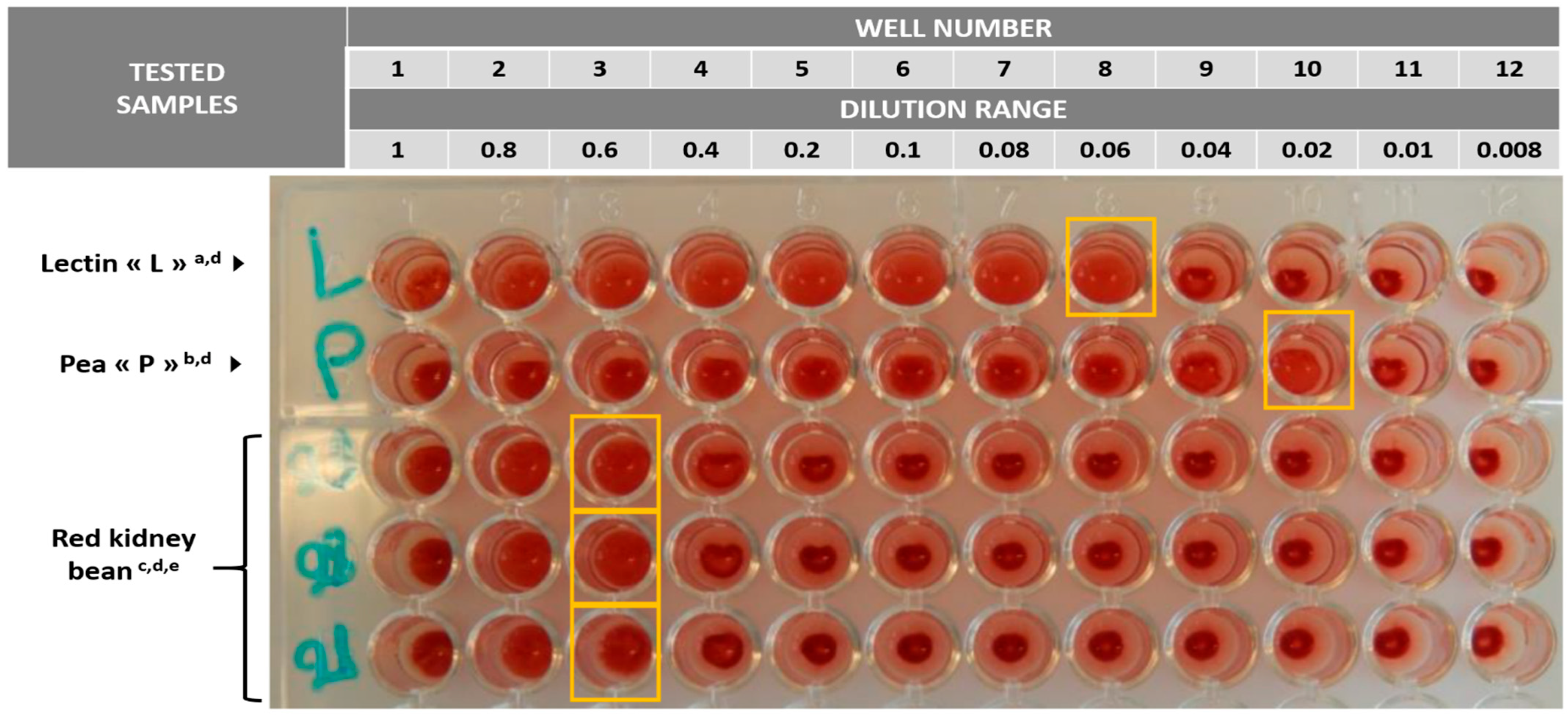
| Chili con carne a | PHA | Haemagglutination using rabbit blood cells g | 400 HAU/g k |
| Chili sin carne a | <400 HAU/g k | ||
| Red kidney beans b | 66,667 HAU/g l | ||
| Red kidney beans b,f (ground) | Cross-reactivity between CPE and PHA | PET-RPLA® Toxin Detection Kit (Oxoid Ltd., Hampshire, United Kingdom) h | Positive at 1/64 dilution |
| Red kidney beans b,f (soaking water) | Negative | ||
| Red kidney beans b,f (ground) | RIDASCREEN® Clostridium perfringens Enterotoxin (R-Biopharm AG, Darmstadt, Germany) h | Negative | |
| Red kidney beans b,f (soaking water) | Negative | ||
| Chili con carne a | B. cereus | Molecular biology e | Negative |
| Chili con carne a | B. cereus enterotoxins | Immunoassay (Duopath® Cereus Enterotoxins, Merck KGaA, Darmstadt, Germany) c,i | Negative |
| Chili con carne a | S. aureus enterotoxins (SEA, SEB, SEG, SEH and SEI) | Immunoassay j | Negative |
| Chili con carne a | CPE | Immunoassay j | Negative |
| Causal Agent | For | Against |
|---|---|---|
| C. perfringens | Clinical pattern (lower digestive tract symptoms predominate) [6] | Incubation period (mean: 6 to 12 h after ingestion) [6] |
| Hazardous ingredients incorporated in chili con carne (minced beef, vegetables, spices) | Negative results for culture and molecular biology on food samples (including chili con carne) | |
| Positive result on all stool samples for CPE testing | The strain found in one stool sample is not typically involved in C. perfringens FBDO | |
| Positive result for one stool sample for culture and molecular biology | Negative results for CPE testing in chili con carne * | |
| PHA | Incubation period (mean: 1 to 3 h after ingestion) [6] | None |
| Clinical pattern (lower digestive tract symptoms predominate) [6] | ||
| High haemagglutination activity measured in raw red kidney beans incorporated into chili con carne (66,667 HAU/g dry weight) | ||
| Red kidney beans incorporated into chili con carne underwent an inadequate cooking process to effectively destroy PHA (cooking at +80 °C overnight, uneven heat distribution during subsequent cooking steps) | ||
| Cases reported the presence of hard beans (i.e., undercooked) in the chili con carne | ||
| Residual haemagglutination activity found in chili con carne can possibly cause symptoms (400 HAU/g wet weight; >400 HAU/g dry weight) | ||
| Positive results for cross-reactivity between PHA and CPE with PET-RPLA® Toxin Detection Kit, retrospectively explaining the positive results obtained on stool sample for CPE testing | ||
| Other enterotoxin-producing bacteria ** | ||
| S. aureus | Incubation period (mean: 2 to 4 h after ingestion) [4] | Clinical pattern (symptoms most commonly described during S. aureus food poisoning are upper digestive tract disorders, whereas lower digestive tract disorders were mainly reported in this epidemic) [4] |
| Negative results for culture on food samples | ||
| Negative results for S. aureus enterotoxin testing on food samples (chili con carne) | ||
| B. cereus | Clinical pattern (lower digestive tract symptoms predominate) [4] | Incubation period (mean: 6 to 12 h after ingestion for diarrheic toxin) [4] |
| Negative results for culture on food samples | ||
| Negative results for molecular biology on stool and food samples | ||
| Negative results for B. cereus enterotoxin testing on food samples (chili con carne) | ||
| Toxin | Coding Gene | Primers | Sequence | Size (pb) | T° of Annealing | References |
|---|---|---|---|---|---|---|
| Alpha toxin | cpa | PL3 | AAG TTA CCT TTG CTG CAT AAT CCC | 236 | 50 | [28] |
| PL7 | ATA GAT ACT CCA TAT CAT CCT GCT | |||||
| Enterotoxin | cpe | P145 | GAA AGA TCT GTA TCT ACA ACT GCT GGT CC | 425 | 50 | [29] |
| P146 | GCT GGC TAA GAT TCT ATA TTT TTG TCC AGT | |||||
| Theta toxin | pfOA | P1685 | TCC ATC AGA TCT TTT TGA TGA CA | 495 | 55 | [30] |
| P1686 | TGT GCA ACA TAG GCT CCA CTA T | |||||
| Beta1 toxin | cpb1 | P1677 | TCA ATT GAA AGC GAA TAT GCT G | 621 | 55 | [31] |
| P1678 | CTA TGG ACG CTC CCC CTA TT | |||||
| Beta2 toxin | cpb2 | 465 | TTT TCT ATA TAT AAT CTT ATT TGT CTA GCA | 277 | 50 | |
| 466 | AGT TTG TAC ATG GGA TGA TGA ACT AGC ACA | |||||
| Delta toxin | cpd | 934 | CTA AAT GCA AAT TAT GCT GTT | 400 | 50 | [32] |
| 935 | TGT TTC TTC AAT TTT ACT ATC TGG | |||||
| Epsilon toxin | etx | 497 | GTC CCT TCA CAA GAT ATA CTA GTA CC | 172 | 50 | [33] |
| 498 | CCT AGG AAA AGC TAA ATA ACT AGG | |||||
| Iota toxin A component | iap | 499 | TAA TTT TAA CTA GTT CAT TTC CTA GTT A | 317 | 50 | [34] |
| 500 | TTT TTG TAT TCT TTT TCT CTA GGA TT | |||||
| Iota toxin B component | ibp | 501 | CTT ATG AAA AAA ATG GCT ATA CTA | 324 | 50 | |
| 502 | GTT TTA CTA TTT GTA GTA GCC CTA GAA A | |||||
| TpeL toxin | tpeL | 1557 | ATA TAG AGT CAA GCA GTG GAG | 464 | 50 | [35] |
| 1558 | GGA ATA CCA CTT GAT ATA CCT | |||||
| NetB toxin | netB | P1644 | TTT GTT GAG ACT AAG GAC GGT T | 266 | 55 | [36] |
| P1645 | TCG CCA TTG AGT AGT TTC CC |
| Toxin | Coding Gene | Primers | Sequence | Size (pb) | T° of Annealing | References |
|---|---|---|---|---|---|---|
| Enterotoxin | nhe | nheA-F | TAC GCT AAG GAG GGG CA | 500 | 55 | [38] |
| nheA-R | GTT TTT ATT GCT TCA TCG GCT | |||||
| Emetic toxin (cereulide) | ces | ces-F | ATC ATA AAG GTG CGA ACA AGA | 188 | 55 | [39] |
| ces-R | AAG ATC AAC CGA ATG CAA CTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Watier-Grillot, S.; Larréché, S.; Mazuet, C.; Baudouin, F.; Feraudet-Tarisse, C.; Holterbach, L.; Dia, A.; Tong, C.; Bourget, L.; Hery, S.; et al. From Foodborne Disease Outbreak (FBDO) to Investigation: The Plant Toxin Trap, Brittany, France, 2018. Toxins 2023, 15, 457. https://doi.org/10.3390/toxins15070457
Watier-Grillot S, Larréché S, Mazuet C, Baudouin F, Feraudet-Tarisse C, Holterbach L, Dia A, Tong C, Bourget L, Hery S, et al. From Foodborne Disease Outbreak (FBDO) to Investigation: The Plant Toxin Trap, Brittany, France, 2018. Toxins. 2023; 15(7):457. https://doi.org/10.3390/toxins15070457
Chicago/Turabian StyleWatier-Grillot, Stéphanie, Sébastien Larréché, Christelle Mazuet, Frédéric Baudouin, Cécile Feraudet-Tarisse, Lise Holterbach, Aïssata Dia, Christelle Tong, Laure Bourget, Sophie Hery, and et al. 2023. "From Foodborne Disease Outbreak (FBDO) to Investigation: The Plant Toxin Trap, Brittany, France, 2018" Toxins 15, no. 7: 457. https://doi.org/10.3390/toxins15070457
APA StyleWatier-Grillot, S., Larréché, S., Mazuet, C., Baudouin, F., Feraudet-Tarisse, C., Holterbach, L., Dia, A., Tong, C., Bourget, L., Hery, S., Pottier, E., Bouilland, O., Tanti, M., Merens, A., Simon, S., Diancourt, L., Chesnay, A., & Pommier de Santi, V. (2023). From Foodborne Disease Outbreak (FBDO) to Investigation: The Plant Toxin Trap, Brittany, France, 2018. Toxins, 15(7), 457. https://doi.org/10.3390/toxins15070457





