Effect of (−)-Epigallocatechin Gallate on Activation of JAK/STAT Signaling Pathway by Staphylococcal Enterotoxin A
Abstract
:1. Introduction
2. Results
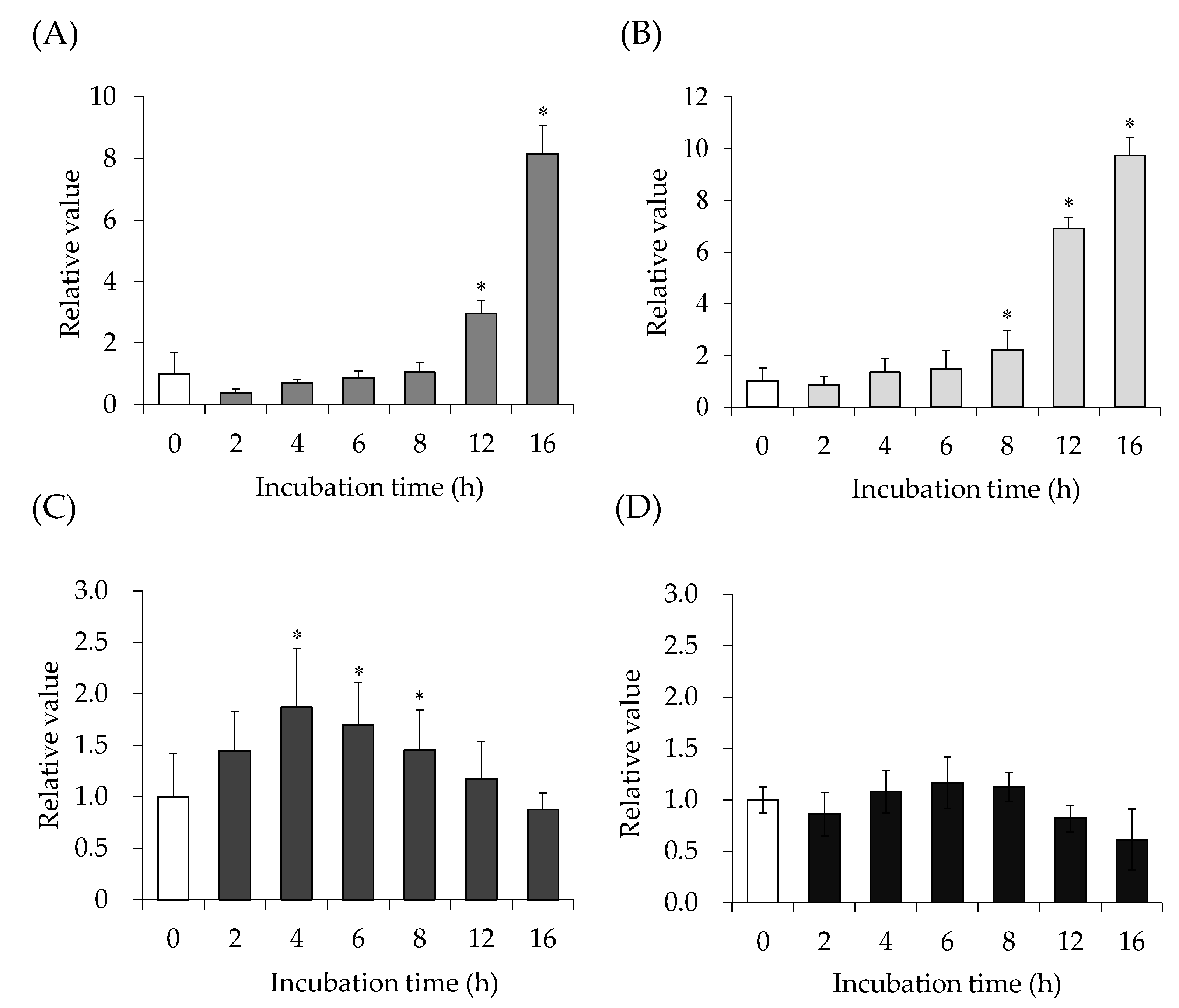
2.1. SEA-Induced JAK/STAT Signaling Pathway Gene Expression
2.2. Inhibitory Effect of Catechin on SEA-Induced JAK/STAT Signaling Pathway Gene Expression
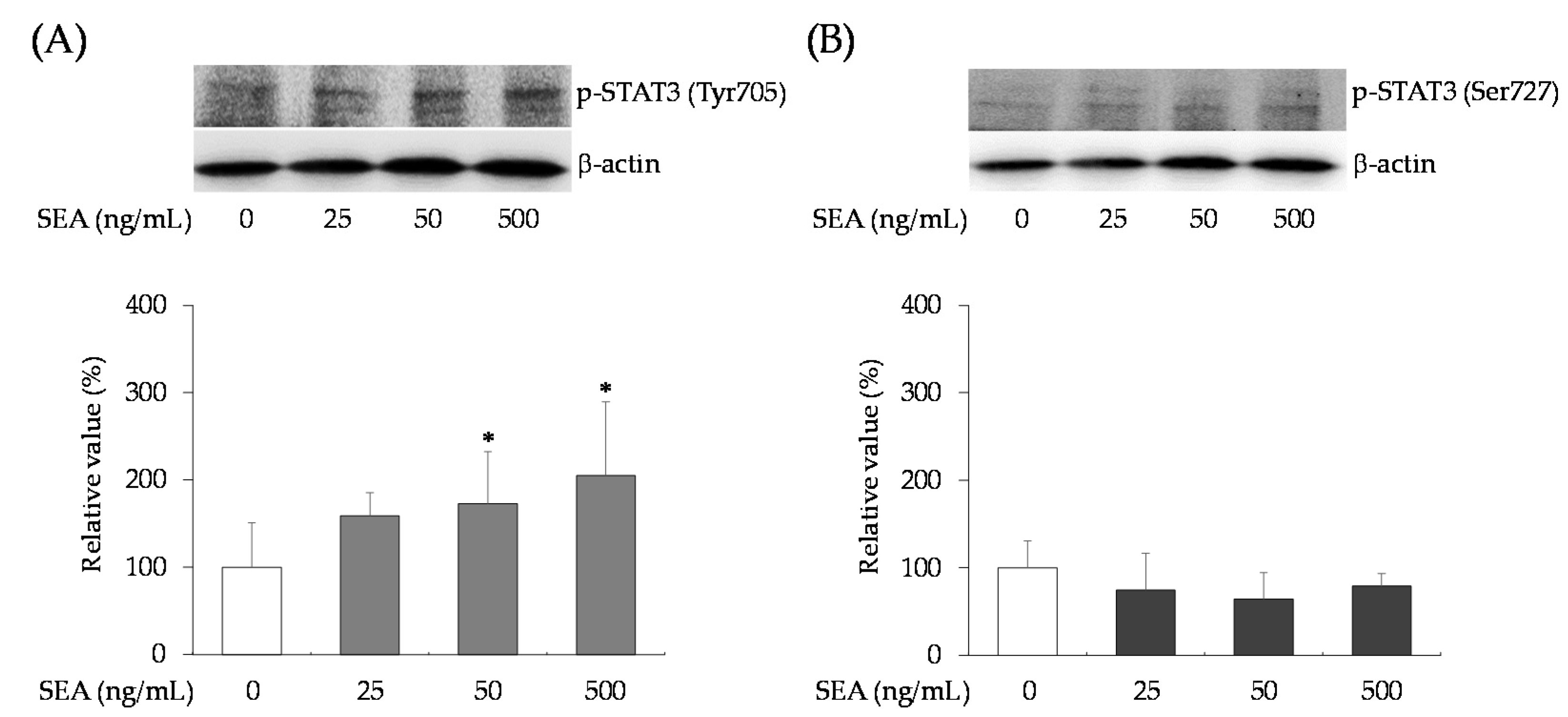
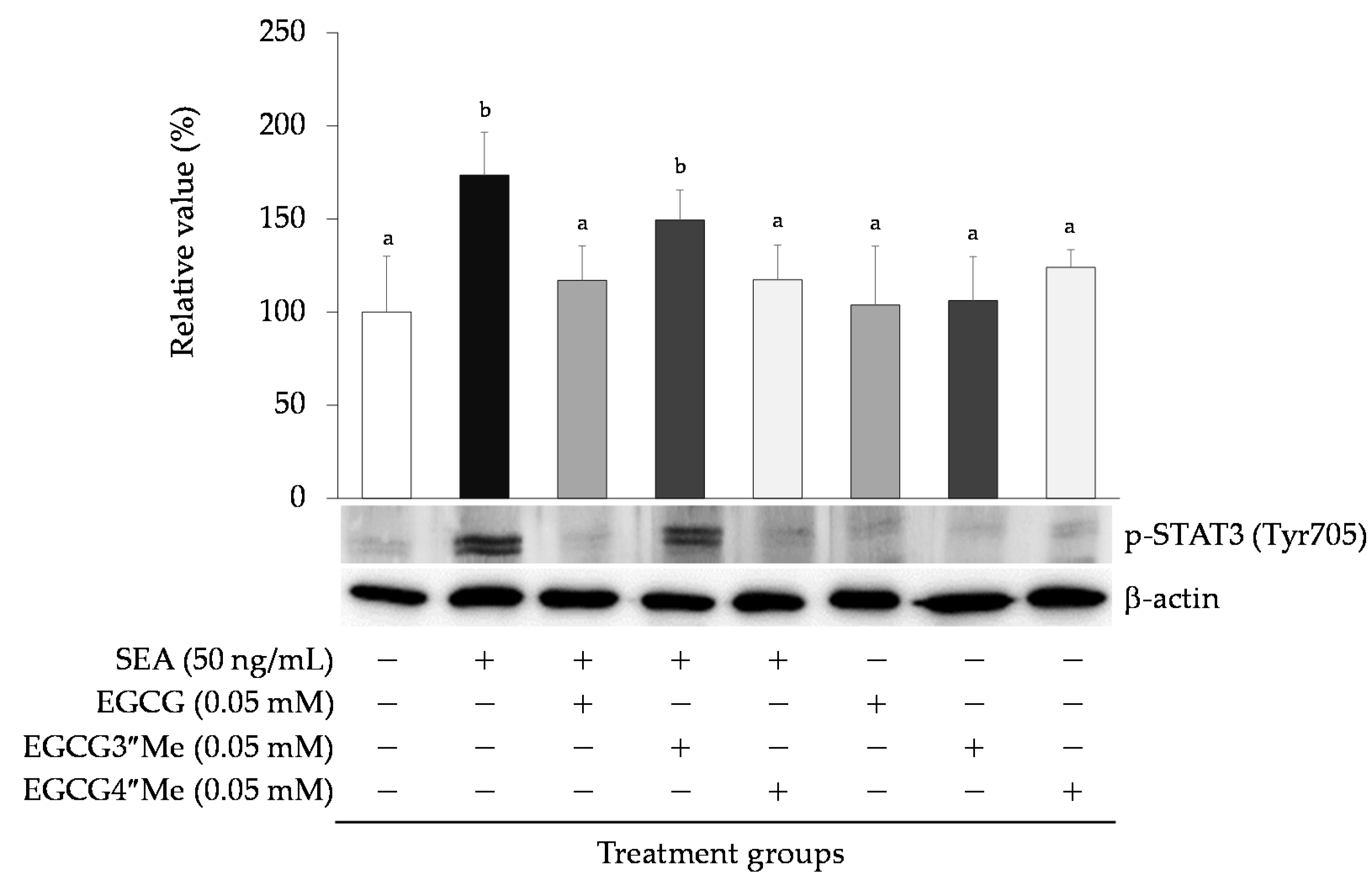
2.3. Effect of SEA on Phosphorylation of STAT3
2.4. Effects of Catechin on SEA-Induced Phosphorylated STAT3
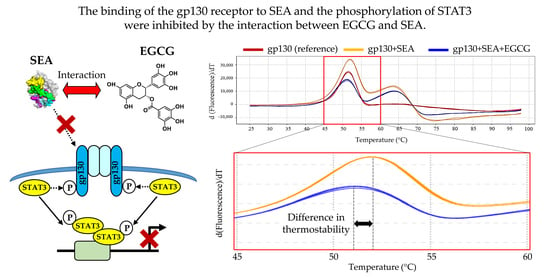
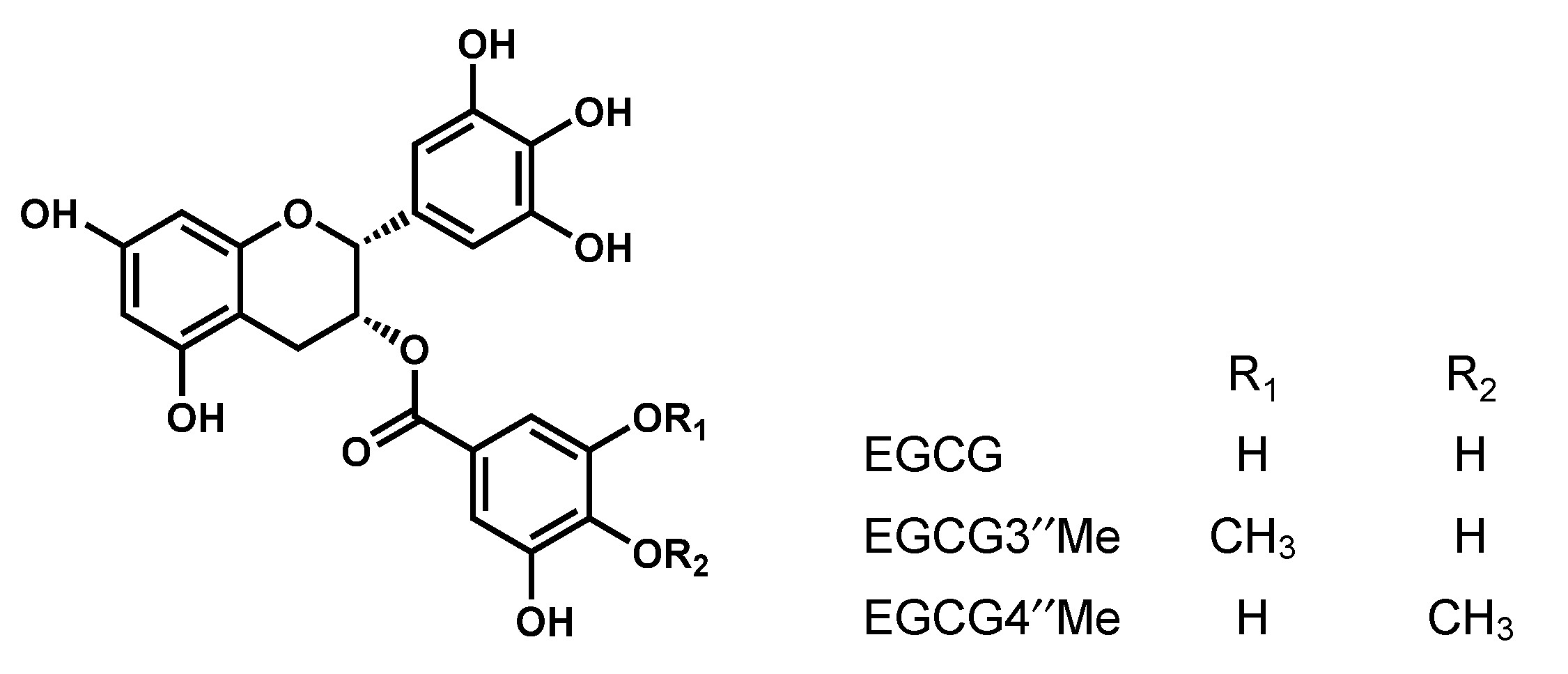
2.5. Interaction between SEA and Catechin
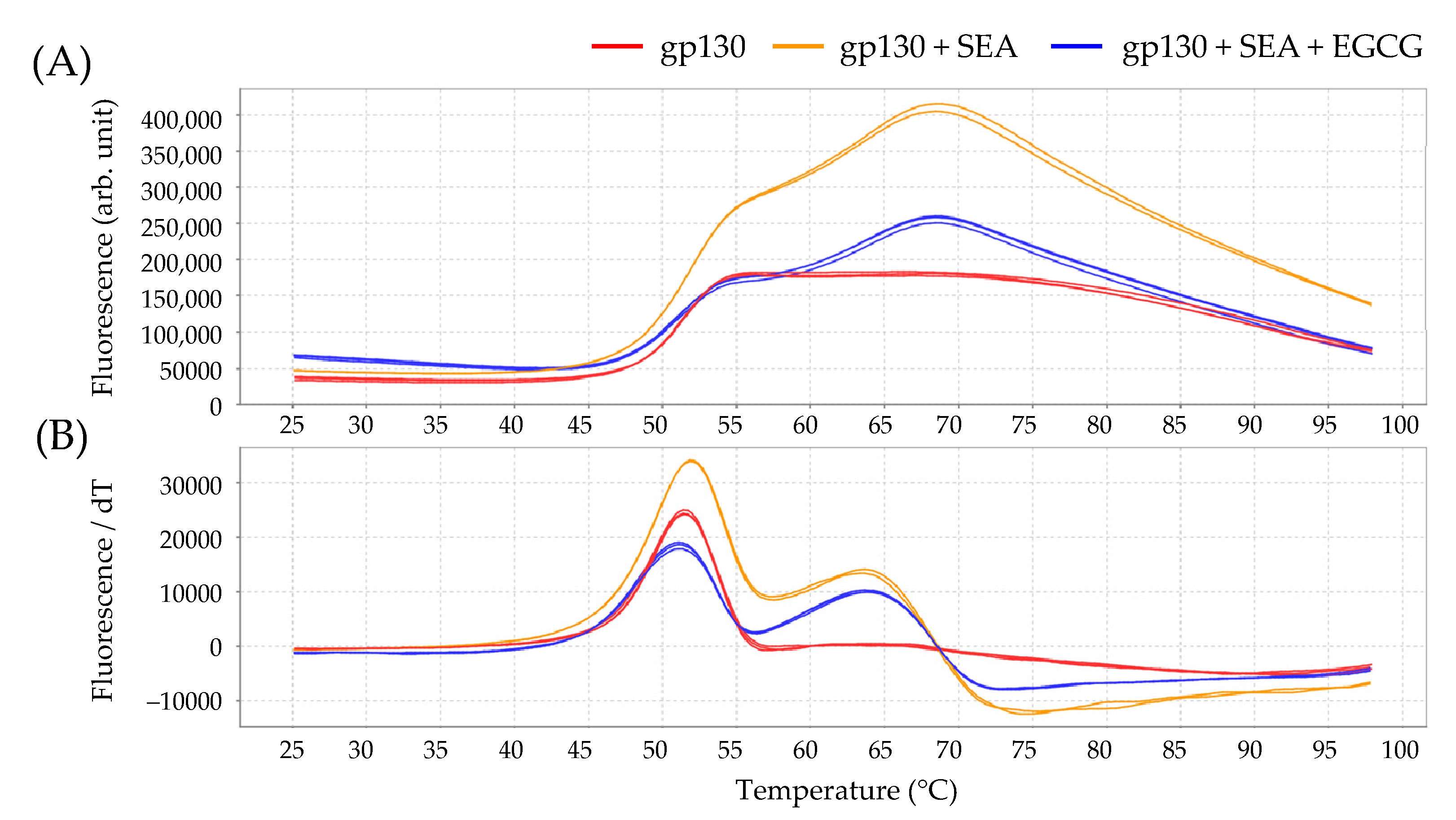
2.6. Interaction between SEA/EGCG and Gp130
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Isolation of Mouse Spleen Cells
4.3. JAK/STAT Signaling Pathway Gene Expression Using Real-Time RT-PCR
4.4. Detection of Phosphorylated STAT3 Using Western Blot Analysis
4.5. Analysis of Protein and Ligand Interactions by Thermal Shift Assay
4.6. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pinchuk, I.V.; Beswick, E.J.; Reyes, V.E. Staphylococcal enterotoxins. Toxins 2010, 2, 2177–2197. [Google Scholar] [CrossRef] [Green Version]
- Ono, H.K.; Nishizawa, M.; Yamamoto, Y.; Hu, D.L.; Nakane, A.; Shinagawa, K.; Omoe, K. Submucosal mast cells in the gastrointestinal tract are a target of staphylococcal enterotoxin type A. FEMS Immunol. Med. Microbiol. 2012, 64, 392–402. [Google Scholar] [CrossRef] [Green Version]
- Banke, E.; Rödström, K.; Ekelund, M.; Dalla-Riva, J.; Lagerstedt, J.O.; Nilsson, S.; Degerman, E.; Lindkvist-Petersson, K.; Nilson, B. Superantigen activates the gp130 receptor on adipocytes resulting in altered adipocyte metabolism. Metabolism 2014, 63, 831–840. [Google Scholar] [CrossRef]
- Aaronson, D.S.; Horvath, C.M. A road map for those who don’t know JAK-STAT. Science 2002, 296, 1653–1655. [Google Scholar] [CrossRef] [PubMed]
- Atreya, R.; Neurath, M.F. Signaling molecules: The pathogenic role of the IL-6/STAT-3 trans signaling pathway in intestinal inflammation and in colonic cancer. Curr. Drug Targets 2008, 9, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Dumpati, R.; Ramatenki, V.; Vadija, R.; Vellanki, V.; Vuruputuri, U. Structural insights into suppressor of cytokine signaling 1 protein- identification of new leads for type 2 diabetes mellitus. J. Mol. Recognit. 2018, 31, e2706. [Google Scholar] [CrossRef] [PubMed]
- Liau, N.P.; Laktyushin, A.; Lucet, I.S.; Murphy, J.M.; Yao, S.; Whitlock, E.; Callaghan, K.; Nicola, N.A.; Kershaw, N.J.; Babon, J.J. The molecular basis of JAK/STAT inhibition by SOCS1. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Klopfenstein, N.; Brandt, S.L.; Castellanos, S.; Gunzer, M.; Blackman, A.; Serezani, C.H. SOCS-1 inhibition of type I interferon restrains Staphylococcus aureus skin host defense. PLoS Pathog. 2012, 17, e1009387. [Google Scholar]
- Wang, Z.; Zhu, F. The expression profiles of immune genes in Mus musculus macrophages during Staphylococcus aureus infection. PLoS ONE 2018, 13, e0190490. [Google Scholar]
- Banerjee, S.; Biehl, A.; Gadina, M.; Hasni, S.; Schwartz, D.M. JAK–STAT signaling as a target for inflammatory and autoimmune diseases: Current and future prospects. Drugs 2017, 77, 521–546. [Google Scholar] [CrossRef]
- Feldman, K.S. Recent progress in ellagitannin chemistry. Phytochemistry 2004, 66, 1984–2000. [Google Scholar] [CrossRef]
- Friedman, M. Overview of antibacterial, antitoxin, antiviral, and antifungal activities of tea flavonoids and teas. Mol. Nutr. Food Res. 2007, 51, 116–134. [Google Scholar] [CrossRef] [PubMed]
- Sano, M.; Suzuki, M.; Miyase, T.; Yoshino, K.; Maeda-Yamamoto, M. Novel antiallergic catechin derivatives isolated from oolong tea. J. Agric. Food Chem. 1999, 47, 1906–1910. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Yoshino, K.; Maeda-Yamamoto, M.; Miyase, T.; Sano, M. Inhibitory effects of tea catechins and O-methylated derivatives of (−)-epigallocatechin-3-O-gallate on mouse type IV allergy. J. Agric. Food Chem. 2000, 48, 5633–5649. [Google Scholar] [CrossRef] [PubMed]
- Siebert, K.J.; Troukhanova, N.V.; Lynn, P.Y. Nature of polyphenol−protein interactions. J. Agric. Food Chem. 1996, 44, 80–85. [Google Scholar] [CrossRef]
- Shimamura, Y.; Utsumi, M.; Hirai, C.; Nakano, S.; Ito, S.; Tsuji, A.; Ishii, T.; Hosoya, T.; Kan, T.; Ohashi, N.; et al. Binding of catechins to staphylococcal enterotoxin A. Molecules 2018, 23, 1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimamura, Y.; Utsumi, M.; Hirai, C.; Kurokawa, A.; Kan, T.; Ohashi, N.; Masuda, S. Effect of (−)-epigallocatechin gallate to staphylococcal enterotoxin A on toxin activity. Molecules 2020, 25, 1867. [Google Scholar] [CrossRef]
- Kelso, A. Cytokines: Principles and prospects. Immunol. Cell Biol. 1998, 76, 300–317. [Google Scholar] [CrossRef]
- Rasooly, R.; Do, P.M.; Friedman, M. Inhibition of biological activity of staphylococcal enterotoxin A (SEA) by apple juice and apple polyphenols. J. Agric. Food Chem. 2010, 58, 5421–5426. [Google Scholar] [CrossRef] [PubMed]
- Ihle, J.N. STATs: Signal transducers and activators of transcription. Cell 1996, 84, 331–334. [Google Scholar] [CrossRef] [Green Version]
- Vu, B.G.; Stach, C.S.; Kulhankova, K.; Salgado-Pabón, W.; Klingelhutz, A.J.; Schlievert, P.M. Chronic superantigen exposure induces systemic inflammation, elevated bloodstream endotoxin, and abnormal glucose tolerance in rabbits: Possible role in diabetes. MBio 2015, 6, e02554-14. [Google Scholar] [CrossRef] [Green Version]
- Lamy, S.; Akla, N.; Ouanouki, A.; Lord-Dufour, S.; Béliveau, R. Diet-derived polyphenols inhibit angiogenesis by modulating the interleukin-6/STAT3 pathway. Exp. Cell Res. 2012, 318, 1586–1596. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Kunnumakkara, A.B.; Harikumar, K.B.; Gupta, S.R.; Tharakan, S.T.; Koca, C.; Dey, S.; Sung, B. Signal transducer and activator of transcription-3, inflammation, and cancer: How intimate is the relationship? Ann. N. Y. Acad. Sci. 2009, 1171, 59–76. [Google Scholar] [CrossRef] [Green Version]
- Li, W.Q.; Dehnade, F.; Zafarullah, M. Oncostatin M-induced matrix metalloproteinase and tissue inhibitor of metalloproteinase-3 genes expression in chondrocytes requires Janus kinase/STAT signaling pathway. J. Immunol. 2001, 166, 3491–3498. [Google Scholar] [CrossRef] [Green Version]
- Chakravarti, N.; Myers, J.N.; Aggarwal, B.B. Targeting constitutive and interleukin-6-inducible signal transducers and activators of transcription 3 pathway in head and neck squamous cell carcinoma cells by curcumin (diferuloylmethane). Int. J. Cancer 2006, 119, 1268–1275. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Sethi, G.; Vadhan-Raj, S.; Bueso-Ramos, C.; Takada, Y.; Gaur, U.; Nair, A.S.; Shishodia, S. Resveratrol inhibits proliferation, induces apoptosis, and overcomes chemoresistance through down-regulation of STAT3 and nuclear factor-kappaB-regulated antiapoptotic and cell survival gene products in human multiple myeloma cells. Blood 2007, 109, 2293–2302. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wung, B.S.; Hsu, M.C.; Wu, C.C.; Hsieh, C.W. Resveratrol suppresses IL-6-induced ICAM-1 gene expression in endothelial cells: Effects on the inhibition of STAT3 phosphorylation. Life Sci. 2005, 78, 389–397. [Google Scholar] [CrossRef]
- You, L.; Wang, Z.; Li, H.; Shou, J.; Jing, Z.; Xie, J.; Sui, X.; Pan, H.; Han, W. The role of STAT3 in autophagy. Autophagy 2015, 11, 729–739. [Google Scholar] [CrossRef] [Green Version]
- Jonchère, B.; Bélanger, A.; Guette, C.; Barré, B.; Coqueret, O. STAT3 as a new autophagy regulator. Jak-Stat 2013, 2, 667–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decker, T.; Kovarik, P. Serine phosphorylation of STATs. Oncogene 2000, 19, 2628–2637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Z.; Darnell, J.E., Jr. Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997, 25, 2062–2067. [Google Scholar] [CrossRef] [Green Version]
- Chung, J.; Uchida, E.; Grammer, T.C.; Blenis, J. STAT3 serine phosphorylation by ERK-dependent and-independent pathways negatively modulates its tyrosine phosphorylation. Mol. Cell. Biol. 1997, 17, 6508–6516. [Google Scholar] [CrossRef] [Green Version]
- Wakahara, R.; Kunimoto, H.; Tanino, K.; Kojima, H.; Inoue, A.; Shintaku, H.; Nakajima, K. Phospho-Ser727 of STAT3 regulates STAT3 activity by enhancing dephosphorylation of phospho-Tyr705 largely through TC45. Genes Cells 2012, 17, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Ganapathy, S.; Hingorani, S.R.; Srivastava, R.K. EGCG inhibits growth, invasion, angiogenesis and metastasis of pancreatic cancer. Front. Biosci. 2008, 13, 440–452. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, S.; Marotte, H.; Kwan, K.; Ruth, J.H.; Campbell, P.L.; Rabquer, B.J.; Pakozdi, A.; Koch, A.E. Epigallocatechin-3-gallate inhibits IL-6 synthesis and suppresses transsignaling by enhancing soluble gp130 production. Proc. Natl. Acad. Sci. USA 2008, 105, 14692–14697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.N.; Fu, J.; Shankar, S.; Srivastava, R.K. EGCG enhances the therapeutic potential of gemcitabine and CP690550 by inhibiting STAT3 signaling pathway in human pancreatic cancer. PLoS ONE 2012, 7, e31067. [Google Scholar] [CrossRef] [PubMed]
- Aihara, Y.; Yoshida, A.; Furuta, T.; Wakimoto, T.; Akizawa, T.; Konishi, M.; Kan, T. Regioselective synthesis of methylated epigallocatechin gallate via nitrobenzenesulfonyl (Ns) protecting group. Bioorg. Med. Chem. Lett. 2009, 19, 4171–4174. [Google Scholar] [CrossRef] [PubMed]

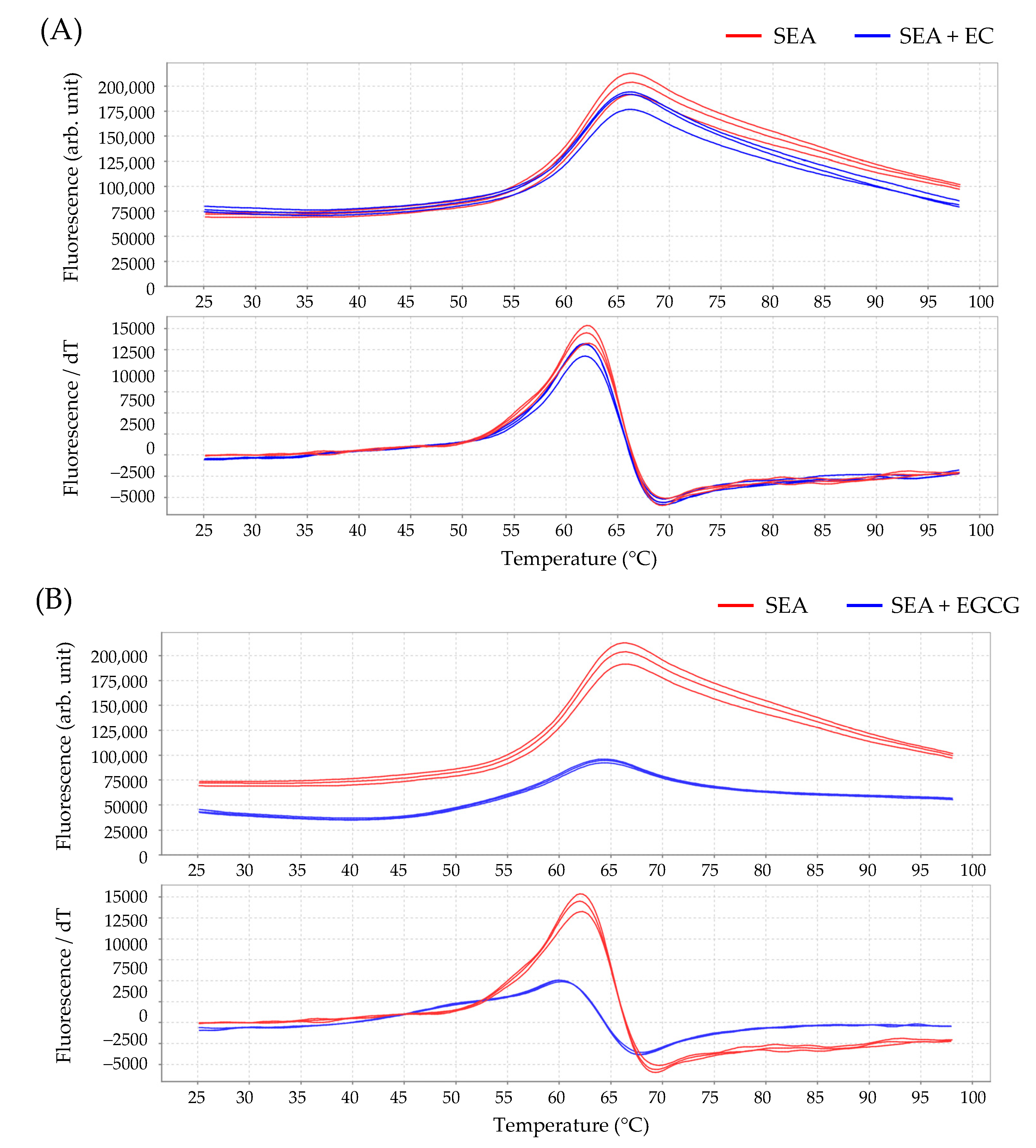
| Sample | Tm Value (°C) |
|---|---|
| SEA (control) | 62.08 ± 0.07 |
| SEA + EC | 61.86 ± 0.05 |
| SEA + EGCG | 60.13 ± 0.09 * |
| Sample | Tm Value (°C) |
|---|---|
| gp130 (control) | 50.80 ± 0.05 |
| gp130 + SEA | 51.96 ± 0.01 * |
| gp130 + SEA+ EGCG | 51.09 ± 0.02 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimamura, Y.; Noaki, R.; Kurokawa, A.; Utsumi, M.; Hirai, C.; Kan, T.; Masuda, S. Effect of (−)-Epigallocatechin Gallate on Activation of JAK/STAT Signaling Pathway by Staphylococcal Enterotoxin A. Toxins 2021, 13, 609. https://doi.org/10.3390/toxins13090609
Shimamura Y, Noaki R, Kurokawa A, Utsumi M, Hirai C, Kan T, Masuda S. Effect of (−)-Epigallocatechin Gallate on Activation of JAK/STAT Signaling Pathway by Staphylococcal Enterotoxin A. Toxins. 2021; 13(9):609. https://doi.org/10.3390/toxins13090609
Chicago/Turabian StyleShimamura, Yuko, Rina Noaki, Ami Kurokawa, Mio Utsumi, Chikako Hirai, Toshiyuki Kan, and Shuichi Masuda. 2021. "Effect of (−)-Epigallocatechin Gallate on Activation of JAK/STAT Signaling Pathway by Staphylococcal Enterotoxin A" Toxins 13, no. 9: 609. https://doi.org/10.3390/toxins13090609
APA StyleShimamura, Y., Noaki, R., Kurokawa, A., Utsumi, M., Hirai, C., Kan, T., & Masuda, S. (2021). Effect of (−)-Epigallocatechin Gallate on Activation of JAK/STAT Signaling Pathway by Staphylococcal Enterotoxin A. Toxins, 13(9), 609. https://doi.org/10.3390/toxins13090609