Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA
Abstract
1. Introduction
2. Results
2.1. Identification of A Putative RTX Toxin, FtxA, Encoded by F. alocis Reference Strain ATCC 35896
2.2. Isolation of F. alocis Clinical Strains and PCR Screening for ftxA
2.3. Isolation of F. alocis Clinical Strains and PCR Screening for ftxA
2.4. Expression and Purification of Purified Recombinant FtxA
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Isolation of F. alocis Strains, and Culturing Conditions
5.2. DNA Isolation and Polymerase Chain Reaction-Based Characterization
5.3. Whole-Genome Sequencing
5.4. In Silico Analysis and Protein Domain Prediction
5.5. Multilocus Sequence Typing (MLST) and Phylogenetic Analysis
5.6. Cloning and Expression of Recombinant FtxA
5.7. Ethical Considerations
5.8. Image Procession
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Aruni, W.; Chioma, O.; Fletcher, H.M. Filifactoralocis: The Newly Discovered Kid on the Block with Special Talents. J. Dent. Res. 2014, 93, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, D.; Afacan, B.; Emingil, G.; Bostanci, N.; Belibasakis, G.N. Salivary Microbiome Shifts in Response to Periodontal Treatment Outcome. Proteom. Clin. Appl. 2020, 14, e2000011. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Martin, I.; Doolittle-Hall, J.; Teles, R.P.; Patel, M.; Belibasakis, G.N.; Hammerle, C.H.F.; Jung, R.E.; Teles, F.R.F. Exploring the microbiome of healthy and diseased peri-implant sites using Illumina sequencing. J. Clin. Periodontol. 2017, 44, 1274–1284. [Google Scholar] [CrossRef]
- Zehnder, M.; Rechenberg, D.K.; Thurnheer, T.; Luthi-Schaller, H.; Belibasakis, G.N. FISHing for gutta-percha-adhered biofilms in purulent post-treatment apical periodontitis. Mol. Oral Microbiol. 2017, 32, 226–235. [Google Scholar] [CrossRef]
- Aruni, A.W.; Roy, F.; Fletcher, H.M. Filifactoralocis has virulence attributes that can enhance its persistence under oxidative stress conditions and mediate invasion of epithelial cells by Porphyromonas gingivalis. Infect. Immun. 2011, 79, 3872–3886. [Google Scholar] [CrossRef] [PubMed]
- Aruni, A.W.; Roy, F.; Sandberg, L.; Fletcher, H.M. Proteome variation among Filifactor alocis strains. Proteomics 2012, 12, 3343–3364. [Google Scholar] [CrossRef] [PubMed]
- Uriarte, S.M.; Edmisson, J.S.; Jimenez-Flores, E. Human neutrophils and oral microbiota: A constant tug-of-war between a harmonious and a discordant coexistence. Immunol. Rev. 2016, 273, 282–298. [Google Scholar] [CrossRef]
- Armstrong, C.L.; Klaes, C.K.; Vashishta, A.; Lamont, R.J.; Uriarte, S.M. Filifactoralocis manipulates human neutrophils affecting their ability to release neutrophil extracellular traps induced by PMA. Innate Immun. 2018, 24, 210–220. [Google Scholar] [CrossRef]
- Edmisson, J.S.; Tian, S.; Armstrong, C.L.; Vashishta, A.; Klaes, C.K.; Miralda, I.; Jimenez-Flores, E.; Le, J.; Wang, Q.; Lamont, R.J.; et al. Filifactoralocis modulates human neutrophil antimicrobial functional responses. Cell Microbiol. 2018, 20, e12829. [Google Scholar] [CrossRef]
- Miralda, I.; Vashishta, A.; Uriarte, S.M. Neutrophil Interaction with Emerging Oral Pathogens: A Novel View of the Disease Paradigm. Adv. Exp. Med. Biol. 2019, 1197, 165–178. [Google Scholar] [CrossRef]
- Linhartova, I.; Bumba, L.; Masin, J.; Basler, M.; Osicka, R.; Kamanova, J.; Prochazkova, K.; Adkins, I.; Hejnova-Holubova, J.; Sadilkova, L.; et al. RTX proteins: A highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 2010, 34, 1076–1112. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Alves, J.M.; Kitten, T.; Brown, A.; Chen, Z.; Ozaki, L.S.; Manque, P.; Ge, X.; Serrano, M.G.; Puiu, D.; et al. Genome of the opportunistic pathogen Streptococcus sanguinis. J. Bacteriol. 2007, 189, 3166–3175. [Google Scholar] [CrossRef]
- Uhlin, B.E.; Oscarsson, J.; Wai, S.N. Haemolysins. In Pathogenic Escherichia Coli: Molecular and Cellular Microbiology; Morabito, S., Ed.; Horizon Press: Londn, UK, 2014; pp. 161–180. [Google Scholar]
- Johansson, A. Aggregatibacteractinomycetemcomitans leukotoxin: A powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef] [PubMed]
- Ristow, L.C.; Welch, R.A. RTX Toxins Ambush Immunity’s First Cellular Responders. Toxins 2019, 11, 720. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Sharp, A.M. News from the interface: The molecular structures of triacylglyceride lipases. Trends Biochem. Sci. 1993, 18, 20–25. [Google Scholar] [CrossRef]
- Jobin, M.C.; Martinez, G.; Motard, J.; Gottschalk, M.; Grenier, D. Cloning, purification, and enzymatic properties of dipeptidyl peptidase IV from the swine pathogen Streptococcus suis. J. Bacteriol. 2005, 187, 795–799. [Google Scholar] [CrossRef]
- Osicka, R.; Prochazkova, K.; Sulc, M.; Linhartova, I.; Havlicek, V.; Sebo, P. A novel “clip-and-link” activity of repeat in toxin (RTX) proteins from gram-negative pathogens. Covalent protein cross-linking by an Asp-Lys isopeptide bond upon calcium-dependent processing at an Asp-Pro bond. J. Biol. Chem. 2004, 279, 24944–24956. [Google Scholar] [CrossRef]
- Belibasakis, G.N.; Maula, T.; Bao, K.; Lindholm, M.; Bostanci, N.; Oscarsson, J.; Ihalin, R.; Johansson, A. Virulence and Pathogenicity Properties of Aggregatibacter actinomycetemcomitans. Pathogens 2019, 8, 222. [Google Scholar] [CrossRef]
- Masi, M.; Wandersman, C. Multiple signals direct the assembly and function of a type 1 secretion system. J. Bacteriol. 2010, 192, 3861–3869. [Google Scholar] [CrossRef]
- O’Brien, D.P.; Perez, A.C.S.; Karst, J.; Cannella, S.E.; Enguene, V.Y.N.; Hessel, A.; Raoux-Barbot, D.; Voegele, A.; Subrini, O.; Davi, M.; et al. Calcium-dependent disorder-to-order transitions are central to the secretion and folding of the CyaA toxin of Bordetella pertussis, the causative agent of whooping cough. Toxicon 2018, 149, 37–44. [Google Scholar] [CrossRef]
- Sizova, M.V.; Chilaka, A.; Earl, A.M.; Doerfert, S.N.; Muller, P.A.; Torralba, M.; McCorrison, J.M.; Durkin, A.S.; Nelson, K.E.; Epstein, S.S. High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae. Stand. Genom. Sci. 2015, 10, 37. [Google Scholar] [CrossRef]
- Elabdeen, H.R.; Mustafa, M.; Hasturk, H.; Klepac-Ceraj, V.; Ali, R.W.; Paster, B.J.; Van Dyke, T.; Bolstad, A.I. Subgingival microbial profiles of Sudanese patients with aggressive periodontitis. J. Periodontal. Res. 2015, 50, 674–682. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hsiao, W.W.; Li, K.L.; Liu, Z.; Jones, C.; Fraser-Liggett, C.M.; Fouad, A.F. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genom. 2012, 13, 345. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, T.; Sakamoto, M.; Takeuchi, Y.; Ohkuma, M.; Izumi, Y. Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J. Oral. Microbiol. 2010, 2. [Google Scholar] [CrossRef] [PubMed]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J.; et al. The α/β hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef]
- Udatha, D.B.; Madsen, K.M.; Panagiotou, G.; Olsson, L. Multiple nucleophilic elbows leading to multiple active sites in a single module esterase from Sorangium cellulosum. J. Struct. Biol. 2015, 190, 314–327. [Google Scholar] [CrossRef]
- Kim, H.Y.; Lim, Y.; An, S.J.; Choi, B.K. Characterization and immunostimulatory activity of extracellular vesicles from Filifactor alocis. Mol. Oral. Microbiol. 2020, 35, 1–9. [Google Scholar] [CrossRef]
- Wang, Q.; Jotwani, R.; Le, J.; Krauss, J.L.; Potempa, J.; Coventry, S.C.; Uriarte, S.M.; Lamont, R.J. Filifactoralocis infection and inflammatory responses in the mouse subcutaneous chamber model. Infect. Immun. 2014, 82, 1205–1212. [Google Scholar] [CrossRef]
- Vashishta, A.; Jimenez-Flores, E.; Klaes, C.K.; Tian, S.; Miralda, I.; Lamont, R.J.; Uriarte, S.M. Putative Periodontal Pathogens, Filifactor alocis and Peptoanaerobacter stomatis, Induce Differential Cytokine and Chemokine Production by Human Neutrophils. Pathogens 2019, 8, 59. [Google Scholar] [CrossRef]
- Cato, E.P.; Moore, L.V.; Moore, W.E.C. Fusobacteriumalocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int. J. Syst. Bacteriol. 1985, 35, 475–477. [Google Scholar] [CrossRef]
- Jalava, J.; Eerola, E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: Proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int. J. Syst. Bacteriol. 1999, 49, 1375–1379. [Google Scholar] [CrossRef][Green Version]
- Möller, A.J. Microbiological examination of root canals and periapical tissues of human teeth. Methodological studies. Odontol. Tidskr. 1966, 74, 1–380. [Google Scholar]
- Claesson, R.; Sjögren, U.; Esberg, A.; Brundin, M.; Granlund, M. Actinomycesradicidentis and Actinomyces haliotis, coccoid Actinomyces species isolated from the human oral cavity. Anaerobe 2017, 48, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Bahrani-Mougeot, F.K.; Paster, B.J.; Coleman, S.; Ashar, J.; Barbuto, S.; Lockhart, P.B. Diverse and novel oral bacterial species in blood following dental procedures. J. Clin. Microbiol. 2008, 46, 2129–2132. [Google Scholar] [CrossRef]
- Siqueira, J.F., Jr.; Rocas, I.N. Detection of Filifactor alocis in endodontic infections associated with different forms of periradicular diseases. Oral. Microbiol. Immunol. 2003, 18, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef]
- Bogomolovas, J.; Simon, B.; Sattler, M.; Stier, G. Screening of fusion partners for high yield expression and purification of bioactive viscotoxins. Protein Expr. Purif. 2009, 64, 16–23. [Google Scholar] [CrossRef] [PubMed]
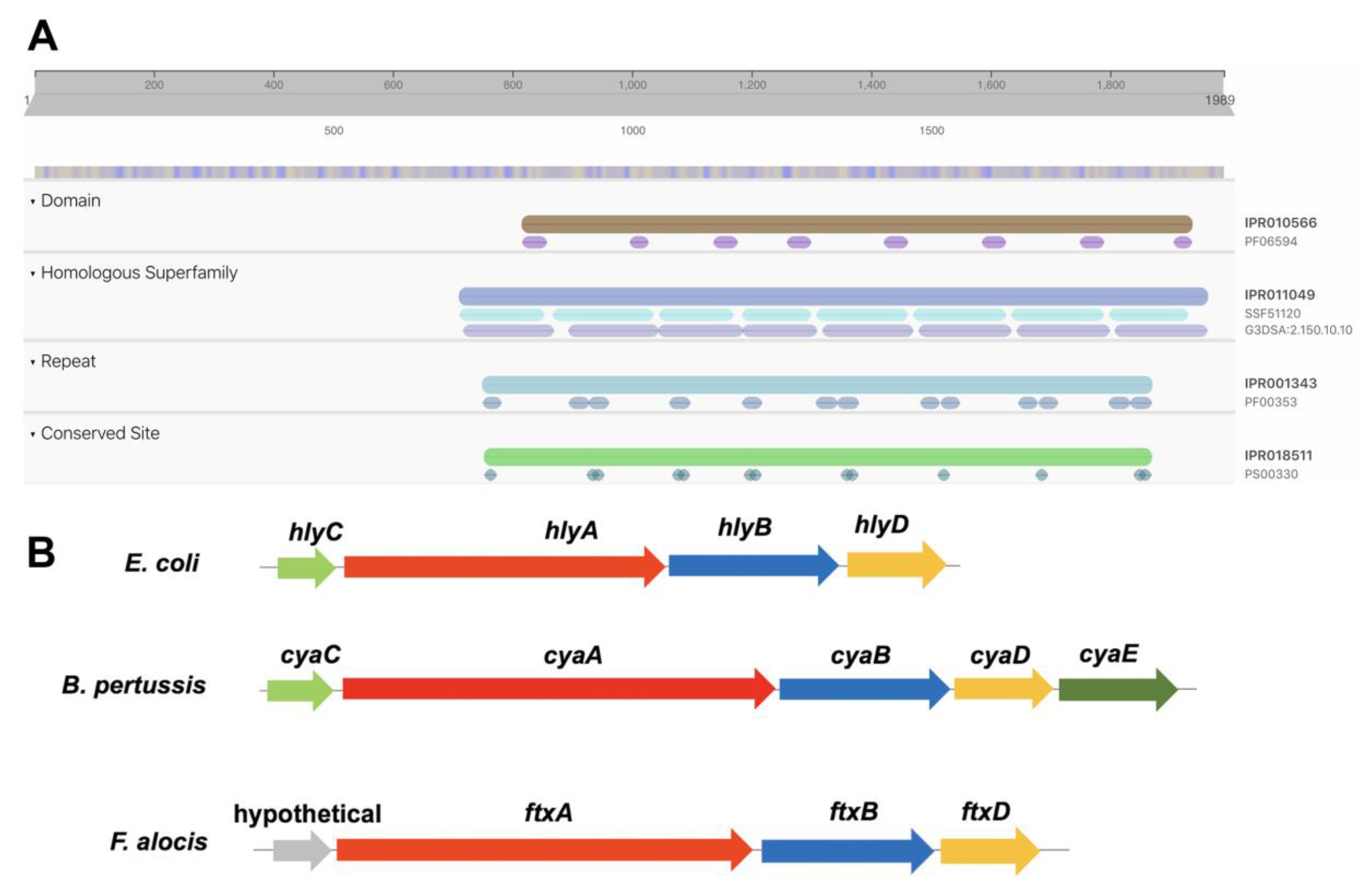
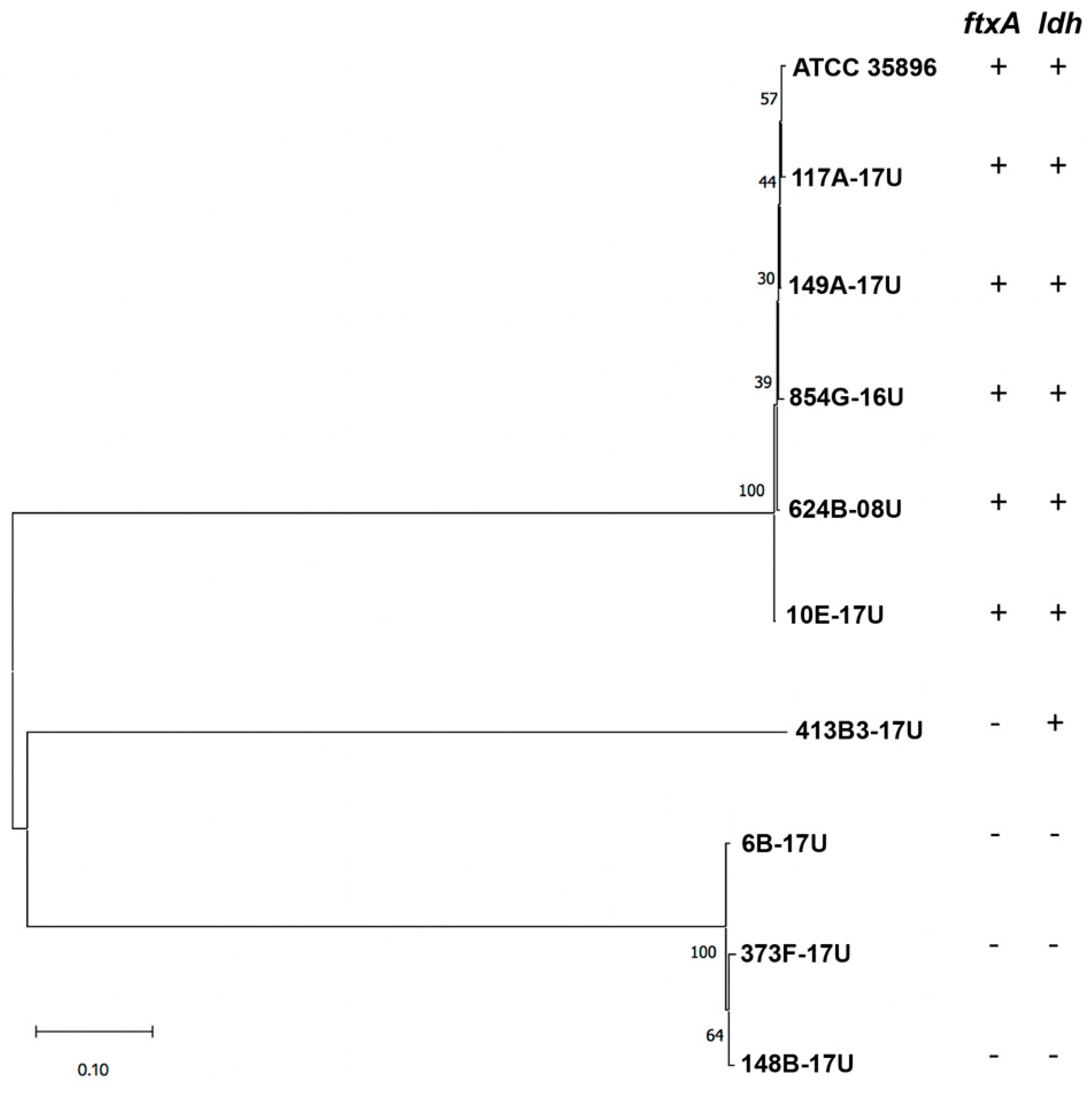
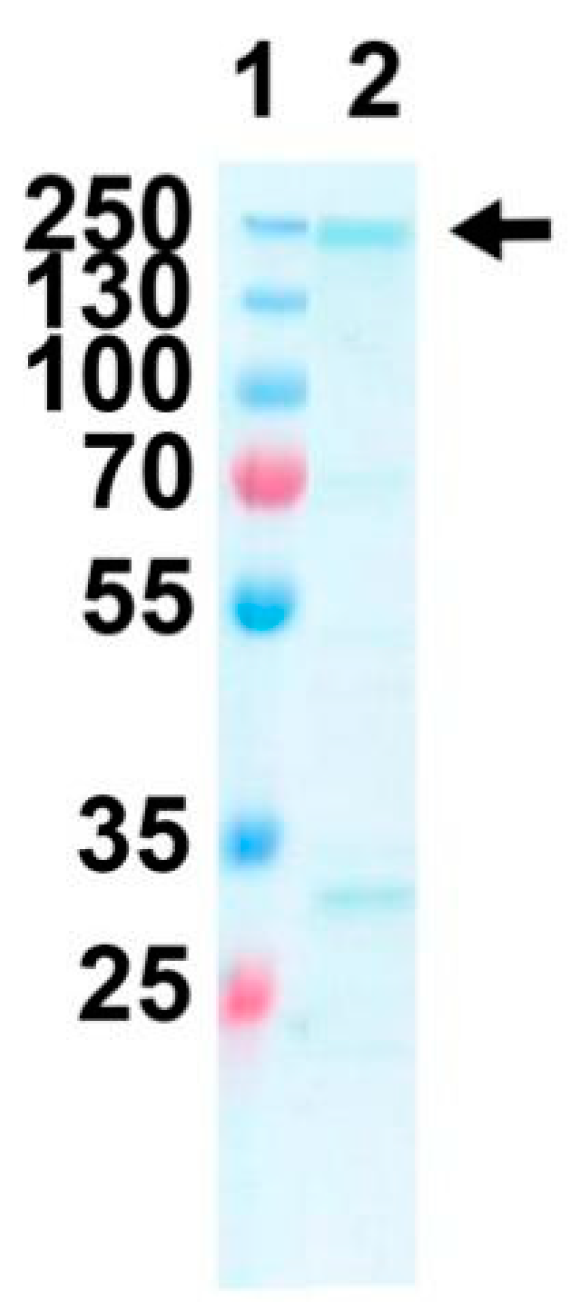
| Strain | Source | ftxA-Positive (+) or Negative (−) | ldh-Positive (+) or Negative (−) |
|---|---|---|---|
| 854G-16U | apical periodontitis, fistula | + | + |
| 117A-17U | periodontitis | + | + |
| 149A-17U | periodontitis | + | + |
| 624B-08U | acute necrotizing ulcerative gingivitis (ANUG) | + | + |
| 373F-17U | peri-implantitis | − | − |
| 6B-17U | root canal | − | − |
| 10E-17U | root canal | + | + |
| 413B3-17U | periodontitis | − | + |
| 148B-17U | periodontitis | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oscarsson, J.; Claesson, R.; Bao, K.; Brundin, M.; Belibasakis, G.N. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins 2020, 12, 687. https://doi.org/10.3390/toxins12110687
Oscarsson J, Claesson R, Bao K, Brundin M, Belibasakis GN. Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins. 2020; 12(11):687. https://doi.org/10.3390/toxins12110687
Chicago/Turabian StyleOscarsson, Jan, Rolf Claesson, Kai Bao, Malin Brundin, and Georgios N. Belibasakis. 2020. "Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA" Toxins 12, no. 11: 687. https://doi.org/10.3390/toxins12110687
APA StyleOscarsson, J., Claesson, R., Bao, K., Brundin, M., & Belibasakis, G. N. (2020). Phylogenetic Analysis of Filifactor alocis Strains Isolated from Several Oral Infections Identified a Novel RTX Toxin, FtxA. Toxins, 12(11), 687. https://doi.org/10.3390/toxins12110687






