Biothermodynamic Analysis of Norovirus: Mechanistic Model of Virus–Host Interactions and Virus–Virus Competition Based on Gibbs Energy
Abstract
1. Introduction
2. Methods
2.1. Data Sources
2.2. Atom Counting Method
2.3. Patel–Erickson Model
2.4. Battley Model
2.5. Biosynthesis Reactions
2.6. Thermodynamic Properties of Biosynthesis
2.7. Thermodynamic Properties of Binding
3. Results
4. Discussion
4.1. Chemical and Thermodynamic Properties of the Norovirus
4.2. Virus–Host Interactions of the Norovirus with Enterocytes
4.3. Virus–Virus Interaction of Norovirus with Rotavirus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saito, H.; Toho, M.; Tanaka, T.; Noda, M. Chapter 10—“PANtrap”: A Novel Detection Method for General Food Samples. In The Norovirus; Chan, P.K.S., Ed.; Academic Press: Amsterdam, The Netherlands, 2017; pp. 145–153. [Google Scholar] [CrossRef]
- Ahmed, S.M.; Hall, A.J.; Robinson, A.E.; Verhoef, L.; Premkumar, P.; Parashar, U.D.; Koopmans, M.; Lopman, B.A. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect. Dis. 2014, 14, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Lin, S.C.; Hsu, Y.H.; Chen, S.Y. Epidemiology, Clinical Features, and Unusual Complications of Norovirus Infection in Taiwan: What We Know after Rotavirus Vaccines. Pathogens 2022, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Popović, M.E.; Tadić, V.; Popović, M. (R)evolution of Viruses: Introduction to biothermodynamics of viruses. Virology 2025, 603, 110319. [Google Scholar] [CrossRef] [PubMed]
- Lucia, U.; Grisolia, G.; Deisboeck, T.S. Thermodynamics and SARS-CoV-2: Neurological effects in post-Covid 19 syndrome. Atti Della Accad. Peloritana Pericolanti 2021, 99, A3. [Google Scholar] [CrossRef]
- Kaniadakis, G.; Baldi, M.M.; Deisboeck, T.S.; Grisolia, G.; Hristopulos, D.T.; Scarfone, A.M.; Sparavigna, A.; Wada, T.; Lucia, U. The κ-statistics approach to epidemiology. Sci. Rep. 2020, 10, 19949. [Google Scholar] [CrossRef]
- Şimşek, B.; Özilgen, M.; Utku, F.Ş. How much energy is stored in SARS-CoV-2 and its structural elements? Energy Storage 2022, 4, e298. [Google Scholar] [CrossRef]
- Yilmaz, B.; Ercan, S.; Akduman, S.; Özilgen, M. Energetic and exergetic costs of COVID-19 infection on the body of a patient. Int. J. Exergy 2020, 32, 314–327. [Google Scholar] [CrossRef]
- Popovic, M.; Martin, J.H.; Head, R.J. COVID infection in 4 steps: Thermodynamic considerations reveal how viral mucosal diffusion, target receptor affinity and furin cleavage act in concert to drive the nature and degree of infection in human COVID-19 disease. Heliyon 2023, 9, e17174. [Google Scholar] [CrossRef]
- Gale, P. Thermodynamic equilibrium dose-response models for MERS-CoV infection reveal a potential protective role of human lung mucus but not for SARS-CoV-2. Microb. Risk Anal. 2020, 16, 100140. [Google Scholar] [CrossRef]
- Popovic, M. Why doesn’t Ebola virus cause pandemics like SARS-CoV-2? Microb. Risk Anal. 2022, 22, 100236. [Google Scholar] [CrossRef]
- Casasnovas, J.M.; Springer, T.A. Kinetics and thermodynamics of virus binding to receptor: Studies with rhinovirus, intercellular adhesion molecule-1 (ICAM-1), and surface plasmon resonance. J. Biol. Chem. 1995, 270, 13216–13224. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M. Formulas for death and life: Chemical composition and biothermodynamic properties of Monkeypox (MPV, MPXV, HMPXV) and Vaccinia (VACV) viruses. Therm. Sci. 2022, 26, 4855–4868. [Google Scholar] [CrossRef]
- Popović, M.E.; Šekularac, G.; Mihailović, M. Like a summer storm: Biothermodynamic analysis of Rotavirus A—Empirical formula, biosynthesis reaction and driving force of virus multiplication and antigen-receptor binding. Microb. Risk Anal. 2024, 26, 100291. [Google Scholar] [CrossRef]
- Gale, P. How virus size and attachment parameters affect the temperature sensitivity of virus binding to host cells: Predictions of a thermodynamic model for arboviruses and HIV. Microb. Risk Anal. 2020, 15, 100104. [Google Scholar] [CrossRef] [PubMed]
- Popović, M.E.; Stevanović, M.; Tadić, V. Biothermodynamic analysis of the Dengue virus: Empirical formulas, biosynthesis reactions and thermodynamic properties of antigen-receptor binding and biosynthesis. Microb. Risk Anal. 2024, 27–28, 100326. [Google Scholar] [CrossRef]
- Popovic, M.; Popovic, M.; Šekularac, G. Death from the Nile: Empirical formula, molar mass, biosynthesis reaction and Gibbs energy of biosynthesis of the West Nile virus. Microb. Risk Anal. 2023, 25, 100281. [Google Scholar] [CrossRef]
- Gale, P. Towards a thermodynamic mechanistic model for the effect of temperature on arthropod vector competence for transmission of arboviruses. Microb. Risk Anal. 2019, 12, 27–43. [Google Scholar] [CrossRef]
- Gale, P. Using thermodynamic parameters to calibrate a mechanistic dose-response for infection of a host by a virus. Microb. Risk Anal. 2018, 8, 1–13. [Google Scholar] [CrossRef]
- Popović, M.E.; Šekularac, G.M.; Tadić, V.M.; Pantović Pavlović, M.R. The silent assassin: Empirical formulas, molar masses, biosynthesis reactions, enthalpies, entropies and Gibbs energies of biosynthesis and Gibbs energies of binding of Coxsackieviruses A and B. Therm. Sci. 2024, 28, 4737–4757. [Google Scholar] [CrossRef]
- Popović, M.E.; Šekularac, G.; Popović, M. The wind of change: Gibbs energy of binding and infectivity evolution of Omicron BA.2.86 Pirola, EG.5.1, XBB.1.16 Arcturus, CH.1.1 and BN.1 variants of SARS-CoV-2. Microb. Risk Anal. 2024, 26, 100290. [Google Scholar] [CrossRef]
- Gale, P. Using thermodynamic equilibrium models to predict the effect of antiviral agents on infectivity: Theoretical application to SARS-CoV-2 and other viruses. Microb. Risk Anal. 2022, 21, 100198. [Google Scholar] [CrossRef] [PubMed]
- Özilgen, M.; Yilmaz, B. COVID-19 disease causes an energy supply deficit in a patient. Int. J. Energy Res. 2021, 45, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Popović, M.; Stevanović, M.; Mihailović, M. Breaking news: Empirical formulas, molar masses, biosynthesis reactions, and thermodynamic properties of virus particles, biosynthesis and binding of Omicron JN.1 variant of SARS-CoV-2. J. Serbian Chem. Soc. 2024, 89, 305–320. [Google Scholar] [CrossRef]
- Popovic, M.; Pantović Pavlović, M.; Pavlović, M. Ghosts of the past: Elemental composition, biosynthesis reactions and thermodynamic properties of Zeta P.2, Eta B.1.525, Theta P.3, Kappa B.1.617.1, Iota B.1.526, Lambda C.37 and Mu B.1.621 variants of SARS-CoV-2. Microb. Risk Anal. 2023, 24, 100263. [Google Scholar] [CrossRef]
- Lucia, U.; Deisboeck, T.S.; Grisolia, G. Entropy-based pandemics forecasting. Front. Phys. 2020, 8, 274. [Google Scholar] [CrossRef]
- Villarreal, L.P. Are viruses alive? Sci. Am. 2004, 291, 100–105. Available online: https://www.jstor.org/stable/26060805 (accessed on 4 May 2025). [CrossRef]
- Moreno-Altamirano, M.M.B.; Kolstoe, S.E.; Sánchez-García, F.J. Virus control of cell metabolism for replication and evasion of host immune responses. Front. Cell. Infect. Microbiol. 2019, 9, 95. [Google Scholar] [CrossRef]
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How viral and intracellular bacterial pathogens reprogram the metabolism of host cells to allow their intracellular replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef]
- Sankaran, N.; Weiss, R.A. Viruses: Impact on Science and Society. Encycl. Virol. 2021, 1, 671–680. [Google Scholar] [CrossRef]
- Burrell, C.J.; Howard, C.R.; Murphy, F.A. History and Impact of Virology. Fenner White’s Med. Virol. 2017, 3–14. [Google Scholar] [CrossRef]
- Zuo, K.; Gao, W.; Wu, Z.; Zhang, L.; Wang, J.; Yuan, X.; Li, C.; Xiang, Q.; Lu, L.; Liu, H. Evolution of Virology: Science History through Milestones and Technological Advancements. Viruses 2024, 16, 374. [Google Scholar] [CrossRef] [PubMed]
- Enquist, L.W. Editors of the Journal of Virology Virology in the 21st century. J. Virol. 2009, 83, 5296–5308. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, S. A Review of Virology: Molecular Biology and Pathogenesis. J. Microbiol. Biol. Educ. JMBE 2011, 12, 213–214. [Google Scholar] [CrossRef]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 295, 12910–12934. [Google Scholar] [CrossRef]
- NCBI. NCBI Virus [Online] National Center for Biotechnology Information. 2024. Available online: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ (accessed on 3 August 2024).
- Ritsch, M.; Cassman, N.A.; Saghaei, S.; Marz, M. Navigating the Landscape: A Comprehensive Review of Current Virus Databases. Viruses 2023, 15, 1834. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef]
- Acheson, N.H. Fundamentals of Molecular Virology; John Wiley & Sons: Hoboken, NJ, USA, 2011; ISBN 978-0-470-90059-8. [Google Scholar]
- Cann, A. Principles of Molecular Virology; Academic Press: Amsterdam, The Netherlands, 2001; ISBN 978-0-121-58533-4. [Google Scholar]
- Lostroh, P. Molecular and Cellular Biology of Viruses; Garland Science: New York, NY, USA, 2019; ISBN 978-0-815-34523-7. [Google Scholar]
- Wimmer, E. The test-tube synthesis of a chemical called poliovirus. The simple synthesis of a virus has far-reaching societal implications. EMBO Rep. 2006, 7, S3–S9. [Google Scholar] [CrossRef]
- Popović, M.E.; Pantović Pavlović, M.; Popović, M. Eris—Another brick in the wall: Empirical formulas, molar masses, biosynthesis reactions, enthalpy, entropy and Gibbs energy of Omicron EG.5 Eris and EG.5.1 variants of SARS-CoV-2. Microb. Risk Anal. 2023, 25, 100280. [Google Scholar] [CrossRef]
- Sanchez, E.L.; Lagunoff, M. Viral activation of cellular metabolism. Virology 2015, 479–480, 609–618. [Google Scholar] [CrossRef]
- Polcicova, K.; Badurova, L.; Tomaskova, J. Metabolic reprogramming as a feast for virus replication. Acta Virol. 2020, 64, 201–215. [Google Scholar] [CrossRef]
- Goyal, P.; Rajala, M.S. Reprogramming of glucose metabolism in virus infected cells. Mol Cell Biochem 2023, 478, 2409–2418. [Google Scholar] [CrossRef] [PubMed]
- Louten, J. Virus Replication. Essent. Hum. Virol. 2016, 49–70. [Google Scholar] [CrossRef]
- Nagy, P.; Pogany, J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 2012, 10, 137–149. [Google Scholar] [CrossRef]
- Chinchar, V.G. Replication of viruses. Encycl. Virol. 1999, 1471–1478. [Google Scholar] [CrossRef]
- Buzón, P.; Maity, S.; Christodoulis, P.; Wiertsema, M.J.; Dunkelbarger, S.; Kim, C.; Wuite, G.J.L.; Zlotnick, A.; Roos, W.H. Virus self-assembly proceeds through contact-rich energy minima. Sci. Adv. 2021, 7, eabg0811. [Google Scholar] [CrossRef]
- Lawton, J.A.; Estes, M.K.; Prasad, B.V. Mechanism of genome transcription in segmented dsRNA viruses. Adv. Virus Res. 2000, 55, 185–229. [Google Scholar] [CrossRef]
- Walsh, D.; Mathews, M.B.; Mohr, I. Tinkering with translation: Protein synthesis in virus-infected cells. Cold Spring Harb. Perspect. Biol. 2013, 5, a012351. [Google Scholar] [CrossRef]
- Von Stockar, U. Live cells as open non-equilibrium systems. In Biothermodynamics: The Role of Thermodynamics in Biochemical Engineering; von Stockar, U., Ed.; EPFL Press: Lausanne, Switzerland, 2013; pp. 475–534. [Google Scholar] [CrossRef]
- Demirel, Y. Nonequilibrium Thermodynamics: Transport and Rate Processes in Physical, Chemical and Biological Systems, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2014; ISBN 978-0-444-59581-2. [Google Scholar]
- Sandler, S.I. Chemical, Biochemical, and Engineering Thermodynamics, 5th ed.; Wiley: Hoboken, NJ, USA, 2017; ISBN 978-0-470-50479-6. [Google Scholar]
- Bar-Even, A.; Flamholz, A.; Noor, E.; Milo, R. Thermodynamic constraints shape the structure of carbon fixation pathways. Biochim. Biophys. Acta 2012, 1817, 1646–1659. [Google Scholar] [CrossRef]
- Yang, X.; Heinemann, M.; Howard, J.; Huber, G.; Iyer-Biswas, S.; Le Treut, G.; Lynch, M.; Montooth, K.L.; Needleman, D.J.; Pigolotti, S.; et al. Physical bioenergetics: Energy fluxes, budgets, and constraints in cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2026786118. [Google Scholar] [CrossRef]
- Taha, A.; Patón, M.; Penas, D.R.; Banga, J.R.; Rodríguez, J. Optimal evaluation of energy yield and driving force in microbial metabolic pathway variants. PLoS Comput. Biol. 2023, 19, e1011264. [Google Scholar] [CrossRef]
- Popovic, M. Atom counting method for determining elemental composition of viruses and its applications in biothermodynamics and environmental science. Comput. Biol. Chem. 2022, 96, 107621. [Google Scholar] [CrossRef] [PubMed]
- Popović, M.; Popović, M.; Šekularac, G.; Pantović Pavlović, M. Omicron BA.2.86 Pirola nightmare: Empirical formulas and thermodynamic properties (enthalpy, entropy and Gibbs energy) of nucleocapsid, virus particle and biosynthesis of BA.2.86 Pirola variant of SARS-CoV-2. J. Serbian Chem. Soc. 2024, 89, 807–822. [Google Scholar] [CrossRef]
- Head, R.J.; Popovic, M.; Martin, J.H. Has the human biological interaction with SARS-CoV2 variants entered a pliant “Faustian bargain”? Pharmacol. Res. Perspect. 2024, 12, e1244. [Google Scholar] [CrossRef] [PubMed]
- Head, R.J.; Lumbers, E.R.; Jarrott, B.; Tretter, F.; Smith, G.; Pringle, K.G.; Islam, S.; Martin, J.H. Systems analysis shows that thermodynamic physiological and pharmacological fundamentals drive COVID-19 and response to treatment. Pharmacol. Res. Perspect. 2022, 10, e00922. [Google Scholar] [CrossRef] [PubMed]
- Katen, S.; Zlotnick, A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009, 455, 395–417. [Google Scholar] [CrossRef]
- Bruinsma, R.F.; Gelbart, W.M.; Reguera, D.; Rudnick, J.; Zandi, R. Viral self-assembly as a thermodynamic process. Phys. Rev. Lett. 2003, 90, 248101. [Google Scholar] [CrossRef]
- Alexander, C.G.; Jürgens, M.C.; Shepherd, D.A.; Freund, S.M.; Ashcroft, A.E.; Ferguson, N. Thermodynamic origins of protein folding, allostery, and capsid formation in the human hepatitis B virus core protein. Proc. Natl. Acad. Sci. USA 2013, 110, E2782–E2791. [Google Scholar] [CrossRef]
- Popović, M.E.; Pantović Pavlović, M.; Šekularac, G. Chemical and thermodynamic properties of Bombyx mori (domestic silk moth): Empirical formula, driving force, and biosynthesis, catabolism and metabolism reactions. Therm. Sci. 2023, 27, 4893–4910. [Google Scholar] [CrossRef]
- Popovic, M. Strain Wars 5: Gibbs energies of binding of BA.1 through BA.4 variants of SARS-CoV-2. Microb. Risk Anal. 2022, 22, 100231. [Google Scholar] [CrossRef]
- Skene, K.R. Systems theory, thermodynamics and life: Integrated thinking across ecology, organization and biological evolution. Bio Syst. 2024, 236, 105123. [Google Scholar] [CrossRef]
- Skene, K.R. Life’s a Gas: A Thermodynamic Theory of Biological Evolution. Entropy 2015, 17, 5522–5548. [Google Scholar] [CrossRef]
- Popovic, M.E.; Mihailović, M.; Pavlović, S. Upcoming epidemic storm: Empirical formulas, biosynthesis reactions, thermodynamic properties and driving forces of multiplication of the omicron XBB.1.9.1, XBF and XBB.1.16 (Arcturus) variants of SARS-CoV-2. Microb. Risk Anal. 2023, 25, 100273. [Google Scholar] [CrossRef]
- Popovic, M. Omicron BA.2.75 subvariant of SARS-CoV-2 is expected to have the greatest infectivity compared with the competing BA.2 and BA.5, due to most negative Gibbs energy of binding. BioTech 2022, 11, 45. [Google Scholar] [CrossRef]
- Robilotti, E.; Deresinski, S.; Pinsky, B.A. Norovirus. Clin. Microbiol. Rev. 2015, 28, 134–164. [Google Scholar] [CrossRef]
- Desselberger, U. Caliciviridae Other Than Noroviruses. Viruses 2019, 11, 286. [Google Scholar] [CrossRef]
- Greer, A.L.; Drews, S.J.; Fisman, D.N. Why “Winter” Vomiting Disease? Seasonality, Hydrology, and Norovirus Epidemiology in Toronto, Canada. EcoHealth 2009, 6, 192–199. [Google Scholar] [CrossRef]
- Kirby, A.E.; Streby, A.; Moe, C.L. Vomiting as a Symptom and Transmission Risk in Norovirus Illness: Evidence from Human Challenge Studies. PLoS ONE 2016, 11, e0143759. [Google Scholar] [CrossRef]
- Tan, M.; Tian, Y.; Zhang, D.; Wang, Q.; Gao, Z. Aerosol Transmission of Norovirus. Viruses 2024, 16, 151. [Google Scholar] [CrossRef]
- Randazzo, W.; D’Souza, D.H.; Sanchez, G. Norovirus: The Burden of the Unknown. Adv. Food Nutr. Res. 2018, 86, 13–53. [Google Scholar] [CrossRef]
- Carlson, K.B.; Dilley, A.; O’Grady, T.; Johnson, J.A.; Lopman, B.; Viscidi, E. A narrative review of norovirus epidemiology, biology, and challenges to vaccine development. NPJ Vaccines 2024, 9, 94. [Google Scholar] [CrossRef]
- Capece, G.; Gignac, E. Norovirus. [Updated 14 August 2023]. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK513265/ (accessed on 30 January 2024).
- Hall, A.J. Noroviruses: The perfect human pathogens? J. Infect. Dis. 2012, 205, 1622–1624. [Google Scholar] [CrossRef] [PubMed]
- Esposito, S.; Ascolese, B.; Senatore, L.; Codecà, C. Pediatric norovirus infection. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Barclay, L.; Park, G.W.; Vega, E.; Hall, A.; Parashar, U.; Vinjé, J.; Lopman, B. Infection control for norovirus. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2014, 20, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Overbey, K.N.; Hamra, G.B.; Nachman, K.E.; Rock, C.; Schwab, K.J. Quantitative microbial risk assessment of human norovirus infection in environmental service workers due to healthcare-associated fomites. J. Hosp. Infect. 2021, 117, 52–64. [Google Scholar] [CrossRef]
- CDC. About Norovirus [Online], U.S. Centers for Disease Control and Prevention. 2024. Available online: https://www.cdc.gov/norovirus/about/index.html (accessed on 8 June 2024).
- HSA Norovirus: What to Do If You Catch It and Helping to Stop the Spread [Online] United Kingdom Health Security Agency. 2024. Available online: https://ukhsa.blog.gov.uk/2022/11/17/norovirus-what-to-do-if-you-catch-it-and-helping-to-stop-the-spread/ (accessed on 8 June 2024).
- Winder, N.; Gohar, S.; Muthana, M. Norovirus: An Overview of Virology and Preventative Measures. Viruses 2022, 14, 2811. [Google Scholar] [CrossRef]
- Yunus, M.A. Molecular Mechanisms for Norovirus Genome Replication. IntechOpen 2021. [Google Scholar] [CrossRef]
- Modrow, S.; Falke, D.; Truyen, U.; Schätzl, H. Viruses with Single-Stranded, Positive-Sense RNA Genomes. Mol. Virol. 2013, 185–349. [Google Scholar] [CrossRef]
- Ryu, W.S. Other Positive-Strand RNA Viruses. Mol. Virol. Hum. Pathog. Viruses 2017, 177–184. [Google Scholar] [CrossRef]
- Rani, M.; Rajyalakshmi, S.; Pakalapaty, S.; Kammilli, N. Norovirus Structure and Classification. IntechOpen 2021. [Google Scholar] [CrossRef]
- Graziano, V.R.; Wei, J.; Wilen, C.B. Norovirus Attachment and Entry. Viruses 2019, 11, 495. [Google Scholar] [CrossRef]
- Song, C.; Takai-Todaka, R.; Miki, M.; Haga, K.; Fujimoto, A.; Ishiyama, R.; Oikawa, K.; Yokoyama, M.; Miyazaki, N.; Iwasaki, K.; et al. Dynamic rotation of the protruding domain enhances the infectivity of norovirus. PLoS Pathog. 2020, 16, e1008619. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Chen, Y.; Jiang, X.; Xia, M.; Yang, Y.; Tan, M.; Li, X.; Rao, Z. A Unique Human Norovirus Lineage with a Distinct HBGA Binding Interface. PLoS Pathog. 2015, 11, e1005025. [Google Scholar] [CrossRef] [PubMed]
- Pogan, R.; Weiss, V.U.; Bond, K.; Dülfer, J.; Krisp, C.; Lyktey, N.; Müller-Guhl, J.; Zoratto, S.; Allmaier, G.; Jarrold, M.F.; et al. N-terminal VP1 Truncations Favor T = 1 Norovirus-Like Particles. Vaccines 2020, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Salmen, W.; Chen, R.; Zhou, Y.; Neill, F.; Crowe, J.E.; Jr Atmar, R.L.; Estes, M.K.; Prasad, B.V.V. Atomic structure of the predominant GII.4 human norovirus capsid reveals novel stability and plasticity. Nat. Commun. 2022, 13, 1241. [Google Scholar] [CrossRef]
- Prasad, B.V.; Schmid, M.F. Principles of virus structural organization. Adv. Exp. Med. Biol. 2012, 726, 17–47. [Google Scholar] [CrossRef]
- Venkataram Prasad, B.V.; Shanker, S.; Muhaxhiri, Z.; Choi, J.M.; Atmar, R.L.; Estes, M.K. Structural Biology of Noroviruses. Viral Gastroenteritis 2016, 329–354. [Google Scholar] [CrossRef]
- Smith, H.Q.; Smith, T.J. The Dynamic Capsid Structures of the Noroviruses. Viruses 2019, 11, 235. [Google Scholar] [CrossRef]
- Devant, J.M.; Hansman, G.S. Structural heterogeneity of a human norovirus vaccine candidate. Virology 2021, 553, 23–34. [Google Scholar] [CrossRef]
- Devant, J.M.; Hofhaus, G.; Bhella, D.; Hansman, G.S. Heterologous expression of human norovirus GII.4 VP1 leads to assembly of T=4 virus-like particles. Antivir. Res. 2019, 168, 175–182. [Google Scholar] [CrossRef]
- Hardy, M.E. Norovirus protein structure and function. FEMS Microbiol. Lett. 2005, 253, 1–8. [Google Scholar] [CrossRef]
- Jung, J.; Grant, T.; Thomas, D.R.; Diehnelt, C.W.; Grigorieff, N.; Joshua-Tor, L. High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations. Proc. Natl. Acad. Sci. USA 2019, 116, 12828–12832. [Google Scholar] [CrossRef] [PubMed]
- Creutznacher, R.; Maass, T.; Dülfer, J.; Feldmann, C.; Hartmann, V.; Lane, M.S.; Knickmann, J.; Westermann, L.T.; Thiede, L.; Smith, T.J.; et al. Distinct dissociation rates of murine and human norovirus P-domain dimers suggest a role of dimer stability in virus-host interactions. Commun. Biol. 2022, 5, 563. [Google Scholar] [CrossRef] [PubMed]
- Tubiana, T.; Boulard, Y.; Bressanelli, S. Dynamics and asymmetry in the dimer of the norovirus major capsid protein. PLoS ONE 2017, 12, e0182056. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Jiang, X. The p domain of norovirus capsid protein forms a subviral particle that binds to histo-blood group antigen receptors. J. Virol. 2005, 79, 14017–14030. [Google Scholar] [CrossRef]
- Tan, M.; Hegde, R.S.; Jiang, X. The P domain of norovirus capsid protein forms dimer and binds to histo-blood group antigen receptors. J. Virol. 2004, 78, 6233–6242. [Google Scholar] [CrossRef]
- Vongpunsawad, S.; Venkataram Prasad, B.V.; Estes, M.K. Norwalk Virus Minor Capsid Protein VP2 Associates within the VP1 Shell Domain. J. Virol. 2013, 87, 4818–4825. [Google Scholar] [CrossRef]
- Snowden, J.S.; Hurdiss, D.L.; Adeyemi, O.O.; Ranson, N.A.; Herod, M.R.; Stonehouse, N.J. Dynamics in the murine norovirus capsid revealed by high-resolution cryo-EM. PLoS Biol. 2020, 18, e3000649. [Google Scholar] [CrossRef]
- Tan, M.; Jiang, X. Norovirus Capsid Protein-Derived Nanoparticles and Polymers as Versatile Platforms for Antigen Presentation and Vaccine Development. Pharmaceutics 2019, 11, 472. [Google Scholar] [CrossRef]
- Strong, D.W.; Thackray, L.B.; Smith, T.J.; Virgin, H.W. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol. 2012, 86, 2950–2958. [Google Scholar] [CrossRef]
- Chen, Y.L.; Chang, P.J.; Huang, C.T. Small P particles formed by the Taiwan-native norovirus P domain overexpressed in Komagataella pastoris. Appl. Microbiol. Biotechnol. 2018, 102, 9707–9718. [Google Scholar] [CrossRef]
- Campillay-Véliz, C.P.; Carvajal, J.J.; Avellaneda, A.M.; Escobar, D.; Covián, C.; Kalergis, A.M.; Lay, M.K. Human Norovirus Proteins: Implications in the Replicative Cycle, Pathogenesis, and the Host Immune Response. Front. Immunol. 2020, 11, 961. [Google Scholar] [CrossRef] [PubMed]
- Lochridge, V.P.; Jutila, K.L.; Graff, J.W.; Hardy, M.E. Epitopes in the P2 domain of norovirus VP1 recognized by monoclonal antibodies that block cell interactions. J. Gen. Virol. 2005, 86 Pt 1, 2799–2806. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Huang, P.; Meller, J.; Zhong, W.; Farkas, T.; Jiang, X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: Evidence for a binding pocket. J. Virol. 2003, 77, 12562–12571. [Google Scholar] [CrossRef]
- Cao, S.; Lou, Z.; Tan, M.; Chen, Y.; Liu, Y.; Zhang, Z.; Zhang, X.C.; Jiang, X.; Li, X.; Rao, Z. Structural basis for the recognition of blood group trisaccharides by norovirus. J. Virol. 2007, 81, 5949–5957. [Google Scholar] [CrossRef]
- Ludwig-Begall, L.F.; Mauroy, A.; Thiry, E. Noroviruses-The State of the Art, Nearly Fifty Years after Their Initial Discovery. Viruses 2021, 13, 1541. [Google Scholar] [CrossRef]
- Perry, J.W.; Wobus, C.E. Endocytosis of murine norovirus 1 into murine macrophages is dependent on dynamin II and cholesterol. J. Virol. 2010, 84, 6163–6176. [Google Scholar] [CrossRef]
- Perry, J.W.; Taube, S.; Wobus, C.E. Murine norovirus-1 entry into permissive macrophages and dendritic cells is pH-independent. Virus Res. 2009, 143, 125–129. [Google Scholar] [CrossRef]
- Le Pendu, J.; Rydell, G.E.; Nasir, W.; Larson, G. Chapter 3.3—Human Norovirus Receptors; Gastroenteritis, V., Svensson, L., Desselberger, U., Greenberg, H.B., Estes, M.K., Eds.; Academic Press: Amsterdam, The Netherlands, 2016. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- NCBI NCBI Database [Online]. National Center for Biotechnology Information. 2024. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 24 May 2024).
- NCBI NC_044932.1—Norovirus GII GII.NA2[PNA2], Complete Sequence [Online] National Center for Biotechnology Information. 2024. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NC_044932.1 (accessed on 24 May 2024).
- Tohma, K.; Saito, M.; Mayta, H.; Zimic, M.; Lepore, C.J.; Ford-Siltz, L.A.; Gilman, R.H.; Parra, G.I. Complete Genome Sequence of a Nontypeable GII Norovirus Detected in Peru. Genome Announc. 2018, 6, e00095-18. [Google Scholar] [CrossRef]
- NCBI AIV43156—VP1, Norovirus GII.4 [Online] National Center for Biotechnology Information. 2024. Available online: https://www.ncbi.nlm.nih.gov/protein/AIV43156.2 (accessed on 24 May 2024).
- Chan, M.C.; Lee, N.; Hung, T.N.; Kwok, K.; Cheung, K.; Tin, E.K.; Lai, R.W.; Nelson, E.A.; Leung, T.F.; Chan, P.K. Rapid emergence and predominance of a broadly recognizing and fast-evolving norovirus GII.17 variant in late 2014. Nat. Commun. 2015, 6, 10061. [Google Scholar] [CrossRef]
- Prasad, B.V.; Hardy, M.E.; Dokland, T.; Bella, J.; Rossmann, M.G.; Estes, M.K. X-ray crystallographic structure of the Norwalk virus capsid. Science 1999, 286, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Meller, J.; Jiang, X. C-terminal arginine cluster is essential for receptor binding of norovirus capsid protein. J. Virol. 2006, 80, 7322–7331. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.A.; Wilen, C.B.; Dai, Y.N.; Orchard, R.C.; Kim, A.S.; Stegeman, R.A.; Hsieh, L.L.; Smith, T.J.; Virgin, H.W.; Fremont, D.H. Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. USA 2018, 115, E9201–E9210. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.T.; Minogue, S.C.; Snowden, J.S.; Arden, W.K.C.; Rowlands, D.J.; Stonehouse, N.J.; Wobus, C.E.; Herod, M.R. Amino acid substitutions in norovirus VP1 dictate cell tropism via an attachment process dependent on membrane mobility. Biorxiv Prepr. Serv. Biol. 2023. [Google Scholar] [CrossRef]
- Popovic, M.; Tadić, V.; Mihailović, M. From genotype to phenotype with biothermodynamics: Empirical formulas, biosynthesis reactions and thermodynamic properties of preproinsulin, proinsulin and insulin molecules. J. Biomol. Struct. Dyn. 2023, 42, 10388–10400. [Google Scholar] [CrossRef]
- Popović, M.E.; Stevanović, M.; Pantović Pavlović, M. Biothermodynamics of hemoglobin and red blood cells: Analysis of structure and evolution of hemoglobin and red blood cells, based on molecular and empirical formulas, biosynthesis reactions, and thermodynamic properties of formation and biosynthesis. J. Mol. Evol. 2024, 92, 776–798. [Google Scholar] [CrossRef]
- Popović, M.E.; Popović, M.; Pei, D. Biothermodynamic Analysis of Caenorhabditis elegans: Model of Growth and Metabolism Based on Empirical Formulas, Metabolism Reactions, and Thermodynamic Properties of Living Matter and Metabolism. Biophysica 2025, 5, 19. [Google Scholar] [CrossRef]
- Patel, S.A.; Erickson, L.E. Estimation of heats of combustion of biomass from elemental analysis using available electron concepts. Biotechnol. Bioeng. 1981, 23, 2051–2067. [Google Scholar] [CrossRef]
- Battley, E.H. The development of direct and indirect methods for the study of the thermodynamics of microbial growth. Thermochim. Acta 1998, 309, 17–37. [Google Scholar] [CrossRef]
- Popovic, M. Animal bioenergetics: Thermodynamic and kinetic analysis of growth and metabolism of Anguilla anguilla. Zoology 2024, 163, 126158. [Google Scholar] [CrossRef]
- Grisolia, G.; Fino, D.; Lucia, U. Thermodynamic optimisation of the biofuel production based on mutualism. Energy Rep. 2020, 6, 1561–1571. [Google Scholar] [CrossRef]
- Grisolia, G.; Fino, D.; Lucia, U. Biomethanation of rice straw: A sustainable perspective for the valorisation of a field residue in the energy sector. Sustainability 2022, 14, 5679. [Google Scholar] [CrossRef]
- Barros, N.; Popovic, M.; Molina-Valero, J.; Lestido-Cardama, Y.; Pérez-Cruzado, C. Unravelling the thermodynamic properties of soil ecosystems in mature beech forests. Sci. Rep. 2024, 14, 16644. [Google Scholar] [CrossRef] [PubMed]
- Battley, E.H. The thermodynamics of microbial growth. In Handbook of Thermal Analysis and Calorimetry, Vol. 4: From Macromolecules to Man; Kemp, E.B., Ed.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 219–235. [Google Scholar] [CrossRef]
- Battley, E.H. On the enthalpy of formation of Escherichia coli K-12 cells. Biotechnol. Bioeng. 1992, 39, 5–12. [Google Scholar] [CrossRef]
- Thornton, W.M.X.V. The relation of oxygen to the heat of combustion of organic compounds. Lond. Edinb. Dublin Philos. Mag. J. Sci. 1917, 33, 196–203. [Google Scholar] [CrossRef]
- Battley, E.H. An empirical method for estimating the entropy of formation and the absolute entropy of dried microbial biomass for use in studies on the thermodynamics of microbial growth. Thermochim. Acta 1999, 326, 7–15. [Google Scholar] [CrossRef]
- Battley, E.H.; Stone, J.R. A comparison of values for the entropy and the entropy of formation of selected organic substances of biological importance in the solid state, as determined experimentally or calculated empirically. Thermochim. Acta 2000, 349, 153–161. [Google Scholar] [CrossRef]
- Atkins, P.W.; de Paula, J. Physical Chemistry for the Life Sciences, 2nd ed.; W.H. Freeman and Company: New York, NY, USA, 2011; ISBN 978-1-429-23114-5. [Google Scholar]
- Atkins, P.W.; de Paula, J. Physical Chemistry: Thermodynamics, Structure, and Change, 10th ed.; W.H. Freeman and Company: New York, NY, USA, 2014; ISBN 978-1-429-29019-7. [Google Scholar]
- Özilgen, M. Review on biothermoydnamics applications: Timeline, challenges, and opportunities. Int. J. Energy Res. 2017, 41, 1513–1533. [Google Scholar] [CrossRef]
- Brown, T.; LeMay, H.; Bursten, B.; Murphy, C.; Woodward, P.; Stoltzfus, M. Chemistry: The Central Science, 14th ed.; Pearson: London, UK, 2017; ISBN 978-0-134-41423-2. [Google Scholar]
- McMurry, J.; Ballantine, D.; Hoeger, C.; Peterson, V. Fundamentals of General, Organic, and Biological Chemistry (MasteringChemistry), 8th ed.; Pearson: London, UK, 2016. [Google Scholar]
- Assael, M.J.; Maitland, G.C.; Maskow, T.; von Stockar, U.; Wakeham, W.A.; Will, S. Commonly Asked Questions in Thermodynamics, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2022; ISBN 978-0-367-33891-6. [Google Scholar] [CrossRef]
- Von Stockar, U. Biothermodynamics of live cells: Energy dissipation and heat generation in cellular structures. In Biothermodynamics: The role of thermodynamics in Biochemical Engineering; von Stockar, U., Ed.; EPFL Press: Lausanne, Switzerland, 2013; pp. 475–534. [Google Scholar] [CrossRef]
- von Stockar, U. The role of thermodynamics in biochemical engineering. J. Non Equilib. Thermodyn. 2013, 38, 225–240. [Google Scholar] [CrossRef]
- Battley, E.H. Calculation of thermodynamic properties of protein in Escherichia coli K-12 grown on succinic acid, energy changes accompanying protein anabolism, and energetic role of ATP in protein synthesis. Biotechnol. Bioeng. 1992, 40, 280–288. [Google Scholar] [CrossRef]
- Popovic, M. XBB.1.5 Kraken cracked: Gibbs energies of binding and biosynthesis of the XBB.1.5 variant of SARS-CoV-2. Microbiol. Res. 2023, 270, 127337. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M. The SARS-CoV-2 Hydra, a monster from the 21st century: Thermodynamics of the BA.5.2 and BF.7 variants. Microb. Risk Anal. 2023, 23, 100249. [Google Scholar] [CrossRef] [PubMed]
- Popovic, M. SARS-CoV-2 strain wars continues: Chemical and thermodynamic characterization of live matter and biosynthesis of Omicron BN.1, CH.1.1 and XBC variants. Microb. Risk Anal. 2023, 24, 100260. [Google Scholar] [CrossRef] [PubMed]
- Battley, E.H. A comparison of energy changes accompanying growth processes by Saccharomyces cerevisiae. J. Therm. Anal. Calorim. 2011, 104, 193–200. [Google Scholar] [CrossRef]
- Battley, E.H. The sources of thermal energy exchange accompanying microbial anabolism. J. Therm. Anal. Calorim. 2007, 87, 105–111. [Google Scholar] [CrossRef]
- Semerciöz, A.S.; Soyalp, K.; Ulu, G.; Özilgen, M. Effects of energy storage by the seaweeds on their ecosystem. Energy Storage 2021, 3, e266. [Google Scholar] [CrossRef]
- Sorgüven, E.; Özilgen, M. Energy utilization, carbon dioxide emission, and exergy loss in flavored yogurt production process. Energy 2012, 40, 214–225. [Google Scholar] [CrossRef]
- Lucia, U.; Fino, D.; Wensel, P.; Grisolia, G. Thermodynamic approach to biofuels from microalgae and cyanobacteria: The role of electrochemical potential. Atti Della Accad. Peloritana Pericolanti Cl. Sci. Fis. Mat. E Nat. 2022, 100, 1. [Google Scholar] [CrossRef]
- Riedel, S.; Hobden, J.A.; Miller, S.; Morse, S.A.; Mietzner, T.A.; Detrick, B.; Mitchell, T.G.; Sakanari, J.A.; Hotez, P.; Mejia, R. Jawetz, Melnick and Adelberg’s Medical Microbiology, 28th ed.; McGraw-Hill: New York, NY, USA, 2019; ISBN 978-1-260-01202-6. [Google Scholar]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into Protein-Ligand Interactions: Mechanisms, Models, and Methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Rusnati, M.; Chiodelli, P.; Bugatti, A.; Urbinati, C. Bridging the past and the future of virology: Surface plasmon resonance as a powerful tool to investigate virus/host interactions. Crit. Rev. Microbiol. 2015, 41, 238–260. [Google Scholar] [CrossRef]
- Beatty, J.D.; Beatty, B.G.; Vlahos, W.G. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J. Immunol. Methods 1987, 100, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Karst, S.M.; Wobus, C.E. A working model of how noroviruses infect the intestine. PLoS Pathog. 2015, 11, e1004626. [Google Scholar] [CrossRef] [PubMed]
- Gelderblom, H.R. Structure and Classification of Viruses. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Summers, W.C. Virus Infection. Encycl. Microbiol. 2009, 546–552. [Google Scholar] [CrossRef]
- Davey, N.E.; Travé, G.; Gibson, T.J. How viruses hijack cell regulation. Trends Biochem. Sci. 2011, 36, 159–169. [Google Scholar] [CrossRef]
- Thaker, S.K.; Ch’ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef]
- Girdhar, K.; Powis, A.; Raisingani, A.; Chrudinová, M.; Huang, R.; Tran, T.; Sevgi, K.; Dogus Dogru, Y.; Altindis, E. Viruses and Metabolism: The Effects of Viral Infections and Viral Insulins on Host Metabolism. Annu. Rev. Virol. 2021, 8, 373–391. [Google Scholar] [CrossRef]
- Mayer, K.A.; Stöckl, J.; Zlabinger, G.J.; Gualdoni, G.A. Hijacking the Supplies: Metabolism as a Novel Facet of Virus-Host Interaction. Front. Immunol. 2019, 10, 1533. [Google Scholar] [CrossRef]
- Goodwin, C.M.; Xu, S.; Munger, J. Stealing the Keys to the Kitchen: Viral Manipulation of the Host Cell Metabolic Network. Trends Microbiol. 2015, 23, 789–798. [Google Scholar] [CrossRef]
- Popovic, M. Biothermodynamics of Viruses from Absolute Zero (1950) To—Virothermodynamics (2022). Vaccines 2022, 10, 2112. [Google Scholar] [CrossRef]
- Popovic, M. Omicron BA.2.75 Sublineage (Centaurus) Follows the Expectations of the Evolution Theory: Less Negative Gibbs Energy of Biosynthesis Indicates Decreased Pathogenicity. Microbiol. Res. 2022, 13, 937–952. [Google Scholar] [CrossRef]
- Popovic, M. Comparative study of entropy and information change in closed and open thermodynamic systems. Thermochim. Acta 2014, 598, 77–81. [Google Scholar] [CrossRef]
- Popovic, M.; Stenning, G.B.; Göttlein, A.; Minceva, M. Elemental composition, heat capacity from 2 to 300 K and derived thermodynamic functions of 5 microorganism species. J. Biotechnol. 2021, 331, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Balmer, R.T. Modern Engineering Thermodynamics; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar] [CrossRef]
- Ozilgen, M.; Sorguven Oner, E. Biothermodynamics: Principles and Applications, 1st ed.; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar] [CrossRef]
- Popovic, M. Never ending story? Evolution of SARS-CoV-2 monitored through Gibbs energies of biosynthesis and antigen-receptor binding of Omicron BQ.1, BQ.1.1, XBB and XBB.1 variants. Microb. Risk Anal. 2023, 23, 100250. [Google Scholar] [CrossRef] [PubMed]
- von Stockar, U. Biothermodynamics of live cells: A tool for biotechnology and biochemical engineering. J. Non Equilib. Thermodyn. 2010, 35, 415–475. [Google Scholar] [CrossRef]
- Von Stockar, U.; Maskow, T.; Liu, J.; Marison, I.W.; Patino, R. Thermodynamics of microbial growth and metabolism: An analysis of the current situation. J. Biotechnol. 2006, 121, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Von Stockar, U.; van der Wielen, L.A. Thermodynamics in biochemical engineering. J. Biotechnol. 1997, 59, 25–37. [Google Scholar] [CrossRef]
- Chu, P.Y.; Huang, H.W.; Boonchan, M.; Tyan, Y.C.; Louis, K.L.; Lee, K.M.; Motomura, K.; Ke, L.Y. Mass Spectrometry-Based System for Identifying and Typing Norovirus Major Capsid Protein VP1. Viruses 2021, 13, 2332. [Google Scholar] [CrossRef]
- Feng, K.; Divers, E.; Ma, Y.; Li, J. Inactivation of a human norovirus surrogate, human norovirus virus-like particles, and vesicular stomatitis virus by gamma irradiation. Appl. Environ. Microbiol. 2011, 77, 3507–3517. [Google Scholar] [CrossRef]
- Molla, A.; Paul, A.V.; Wimmer, E. Cell-free, de novo synthesis of poliovirus. Science 1991, 254, 1647–1651. [Google Scholar] [CrossRef]
- Mehndiratta, M.M.; Mehndiratta, P.; Pande, R. Poliomyelitis: Historical facts, epidemiology, and current challenges in eradication. Neurohospitalist 2014, 4, 223–229. [Google Scholar] [CrossRef]
- Kapikian, A.Z.; Shope, R.E. Rotaviruses, Reoviruses, Coltiviruses, and Orbiviruses. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 63. Available online: https://www.ncbi.nlm.nih.gov/books/NBK8558/ (accessed on 4 May 2025).
- Martín-Acebes, M.A.; Saiz, J.C. West Nile virus: A re-emerging pathogen revisited. World J. Virol. 2012, 1, 51–70. [Google Scholar] [CrossRef] [PubMed]
- Azuma, K.; Yanagi, U.; Kagi, N.; Kim, H.; Ogata, M.; Hayashi, M. Environmental factors involved in SARS-CoV-2 transmission: Effect and role of indoor environmental quality in the strategy for COVID-19 infection control. Environ. Health Prev. Med. 2020, 25, 66. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.C.W.; Shan Kwan, H.; Chan, P.K.S. Structure and Genotypes of Noroviruses. Norovirus 2017, 51–63. [Google Scholar] [CrossRef]
- Degueldre, C. Single virus inductively coupled plasma mass spectroscopy analysis: A comprehensive study. Talanta 2021, 228, 122211. [Google Scholar] [CrossRef]
- Popovic, M.; Šekularac, G.; Stevanović, M. Thermodynamics of microbial consortia: Enthalpies and Gibbs energies of microorganism live matter and macromolecules of E. coli, G. oxydans, P. fluorescens, S. thermophilus and P. chrysogenum. J. Biotechnol. 2024, 379, 6–17. [Google Scholar] [CrossRef]
- Battley, E.H. A theoretical study of the thermodynamics of microbial growth using Saccharomyces cerevisiae and a different free energy equation. Q. Rev. Biol. 2013, 88, 69–96. [Google Scholar] [CrossRef]
- Karst, S.M. Pathogenesis of noroviruses, emerging RNA viruses. Viruses 2010, 2, 748–781. [Google Scholar] [CrossRef]
- Mateu, M.G. Introduction: The structural basis of virus function. Sub Cell. Biochem. 2013, 68, 3–51. [Google Scholar] [CrossRef]
- Melano, I.; Kuo, L.L.; Lo, Y.C.; Sung, P.W.; Tien, N.; Su, W.C. Effects of Basic Amino Acids and Their Derivatives on SARS-CoV-2 and Influenza-A Virus Infection. Viruses 2021, 13, 1301. [Google Scholar] [CrossRef]
- Payne, S. Virus Interactions With the Cell. Viruses 2017, 23–35. [Google Scholar] [CrossRef]
- Tyl, M.D.; Betsinger, C.N.; Cristea, I.M. Virus-host protein interactions as footprints of human cytomegalovirus replication. Curr. Opin. Virol. 2022, 52, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ouyang, Q. Nonequilibrium Thermodynamics in Biochemical Systems and Its Application. Entropy 2021, 23, 271. [Google Scholar] [CrossRef] [PubMed]
- von Stockar, U.; Liu, J. Does microbial life always feed on negative entropy? Thermodynamic analysis of microbial growth. Biochim. Biophys. Acta 1999, 1412, 191–211. [Google Scholar] [CrossRef]
- Popović, M.E.; Stevanović, M.; Pantović Pavlović, M. Return of the forgotten nightmare: Bordetella pertussis uses a more negative Gibbs energy of metabolism to outcompete its host organism. Microb. Risk Anal. 2024, 26, 100292. [Google Scholar] [CrossRef]
- Nagy, P.D.; Lin, W. Taking over Cellular Energy-Metabolism for TBSV Replication: The High ATP Requirement of an RNA Virus within the Viral Replication Organelle. Viruses 2020, 12, 56. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Qin, C.; Xie, T.; Yeh WWSavas, A.C.; Feng, P. Metabolic Enzymes in Viral Infection and Host Innate Immunity. Viruses 2023, 16, 35. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bonora, M.; Patergnani, S.; Rimessi, A.; De Marchi, E.; Suski, J.M.; Bononi, A.; Giorgi, C.; Marchi, S.; Missiroli, S.; Poletti, F.; et al. ATP synthesis and storage. Purinergic Signal. 2012, 8, 343–357. [Google Scholar] [CrossRef]
- Duponchel, S.; Fischer, M.G. Viva lavidaviruses! Five features of virophages that parasitize giant DNA viruses. PLoS Pathog. 2019, 15, e1007592. [Google Scholar] [CrossRef]
- Roizman, B. Multiplication. In Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; Chapter 42. Available online: https://www.ncbi.nlm.nih.gov/books/NBK8181/ (accessed on 4 May 2025).
- Faisst, S. Propagation of viruses|Animal. In Encyclopedia of Virology, 2nd ed.; Granoff, A., Webster, R.G., Eds.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 1408–1413. Available online: https://www.sciencedirect.com/science/article/pii/B0122270304002363 (accessed on 4 May 2025).
- Sai, L.; Sun, J.; Shao, L.; Chen, S.; Liu, H.; Ma, L. Epidemiology and clinical features of rotavirus and norovirus infection among children in Ji’nan, China. Virol. J. 2013, 10, 302. [Google Scholar] [CrossRef]
- Rackoff, L.A.; Bok, K.; Green, K.Y.; Kapikian, A.Z. Epidemiology and evolution of rotaviruses and noroviruses from an archival WHO Global Study in Children (1976–79) with implications for vaccine design. PLoS ONE 2013, 8, e59394. [Google Scholar] [CrossRef]
- Nirwati, H.; Donato, C.M.; Mawarti, Y.; Mulyani, N.S.; Ikram, A.; Aman, A.T.; Peppelenbosch, M.P.; Soenarto, Y.; Pan, Q.; Hakim, M.S. Norovirus and rotavirus infections in children less than five years of age hospitalized with acute gastroenteritis in Indonesia. Arch Virol 2019, 164, 1515–1525. [Google Scholar] [CrossRef] [PubMed]
- Amimo, J.O.; Raev, S.A.; Chepngeno, J.; Mainga, A.O.; Guo, Y.; Saif, L.; Vlasova, A.N. Rotavirus Interactions With Host Intestinal Epithelial Cells. Front. Immunol. 2021, 12, 793841. [Google Scholar] [CrossRef] [PubMed]
- Oluwatoyin Japhet, M.; Adeyemi Adesina, O.; Famurewa, O.; Svensson, L.; Nordgren, J. Molecular epidemiology of rotavirus and norovirus in Ile-Ife, Nigeria: High prevalence of G12P [8] rotavirus strains and detection of a rare norovirus genotype. J. Med. Virol. 2012, 84, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Cilli, A.; Luchs, A.; Morillo, S.G.; Costa, F.F.; Carmona, R.d.e.C.; Timenetsky, M.d.o.C. Characterization of rotavirus and norovirus strains: A 6-year study (2004–2009). J. Pediatr. 2011, 87, 445–449. [Google Scholar] [CrossRef]
- Mousavi Nasab, S.D.; Sabahi, F.; Makvandi, M.; Mirab Samiee, S.; Nadji, S.A.; Ravanshad, M. Epidemiology of Rotavirus-Norovirus Co-Infection and Determination of Norovirus Genogrouping among Children with Acute Gastroenteritis in Tehran, Iran. Iran. Biomed. J. 2016, 20, 280–286. [Google Scholar] [CrossRef]
- El Qazoui, M.; Oumzil, H.; Baassi, L.; Omari, N.E.; Sadki, K.; Amzazi, S.; Benhafid, M.; Aouad, R.E. Rotavirus and Norovirus infections among acute gastroenteritis children in Morocco. BMC Infect. Dis. 2014, 14, 300. [Google Scholar] [CrossRef]
- Quee, F.A.; de Hoog, M.L.A.; Schuurman, R.; Bruijning-Verhagen, P. Community burden and transmission of acute gastroenteritis caused by norovirus and rotavirus in the Netherlands (RotaFam): A prospective household-based cohort study. Lancet Infect. Dis. 2020, 20, 598–606. [Google Scholar] [CrossRef]
- Rönnelid, Y.; Bonkoungou, I.J.O.; Ouedraogo, N.; Barro, N.; Svensson, L.; Nordgren, J. Norovirus and rotavirus in children hospitalised with diarrhoea after rotavirus vaccine introduction in Burkina Faso. Epidemiol. Infect. 2020, 148, e245. [Google Scholar] [CrossRef]
- Santiso-Bellón, C.; Randazzo, W.; Pérez-Cataluña, A.; Vila-Vicent, S.; Gozalbo-Rovira, R.; Muñoz, C.; Buesa, J.; Sanchez, G.; Rodríguez Díaz, J. Epidemiological Surveillance of Norovirus and Rotavirus in Sewage (2016–2017) in Valencia (Spain). Microorganisms 2020, 8, 458. [Google Scholar] [CrossRef]
- Piedade, J.; Nordgren, J.; Esteves, F.; Esteves, A.; Teodósio, R.; Svensson, L.; Istrate, C. Molecular epidemiology and host genetics of norovirus and rotavirus infections in Portuguese elderly living in aged care homes. J. Med. Virol. 2019, 91, 1014–1021. [Google Scholar] [CrossRef]
| Name | mC | mH | mO | mN | mP | mS | Mr (kDa) |
|---|---|---|---|---|---|---|---|
| Norovirus particle | 548,504 | 817,571 | 193,862 | 157,282 | 7525 | 3060 | 13,048 |
| Norovirus RNA | 71,684 | 88,571 | 52,202 | 28,762 | 7525 | 0 | 2421.4 |
| Norovirus VP1 | 2649 | 4050 | 787 | 714 | 0 | 17 | 59.035 |
| Name | Content (%-Mass) |
|---|---|
| RNA | 18.6 |
| Proteins | 81.4 |
| Name | nH | nO | nN | nP | nS | Mr (g/C-mol) |
|---|---|---|---|---|---|---|
| Norovirus particle | 1.4905 | 0.3534 | 0.2867 | 0.013719 | 0.005579 | 23.79 |
| Norovirus RNA | 1.2356 | 0.7282 | 0.4012 | 0.104975 | 0.000000 | 33.78 |
| Norovirus VP1 | 1.5289 | 0.2971 | 0.2695 | 0.000000 | 0.006418 | 22.29 |
| Name | ΔfH0 (kJ/C-mol) | Sm0 (J/C-mol K) | ΔfG0 (kJ/C-mol) |
|---|---|---|---|
| Norovirus particle | −76.06 | 31.30 | −35.49 |
| Norovirus RNA | −170.69 | 38.09 | −121.32 |
| Norovirus VP1 | −61.83 | 30.28 | −22.58 |
| Name | Reactants | → | Products | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Amino acid | O2 | HPO42− | HCO3− | Bio | SO42− | H2O | HCO3− | H2CO3 | ||
| Norovirus particle | 1.2760 | 0.3628 | 0.0137 | 0.0188 | → | 1 | 0.0231 | 0.1232 | 0.0000 | 0.2948 |
| Norovirus RNA | 1.7855 | 1.1408 | 0.1050 | 0.0000 | → | 1 | 0.0401 | 0.3190 | 0.1297 | 0.6558 |
| Norovirus VP1 | 1.1994 | 0.2459 | 0.0000 | 0.0411 | → | 1 | 0.0205 | 0.0937 | 0.0000 | 0.2405 |
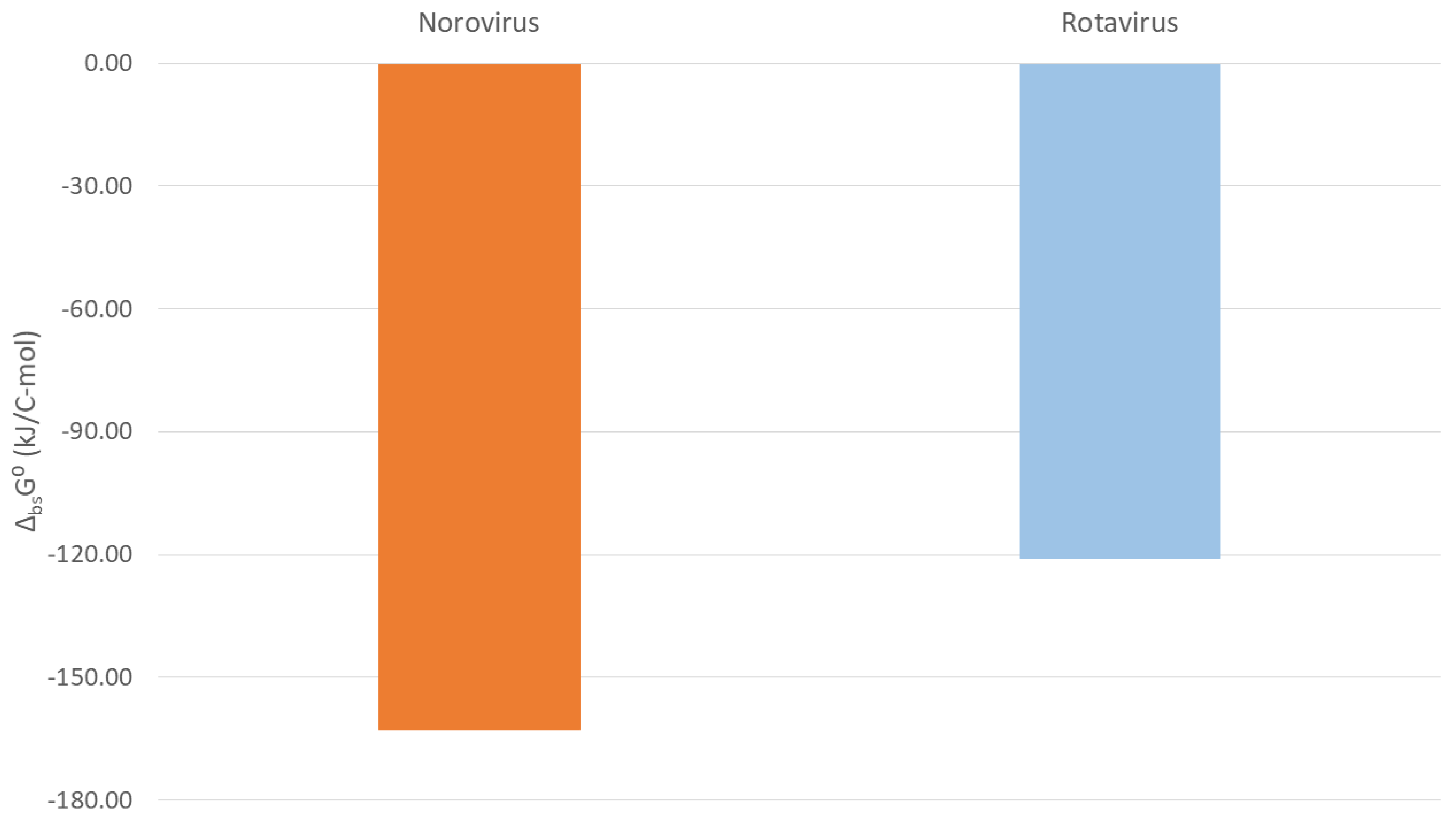
| Name | ΔbsH0 (kJ/C-mol) | ΔbsS0 (J/C-mol K) | ΔbsG0 (kJ/C-mol) |
|---|---|---|---|
| Norovirus particle | −170.79 | −27.12 | −162.85 |
| Norovirus RNA | −519.57 | −104.02 | −489.79 |
| Norovirus VP1 | −118.35 | −15.56 | −113.70 |
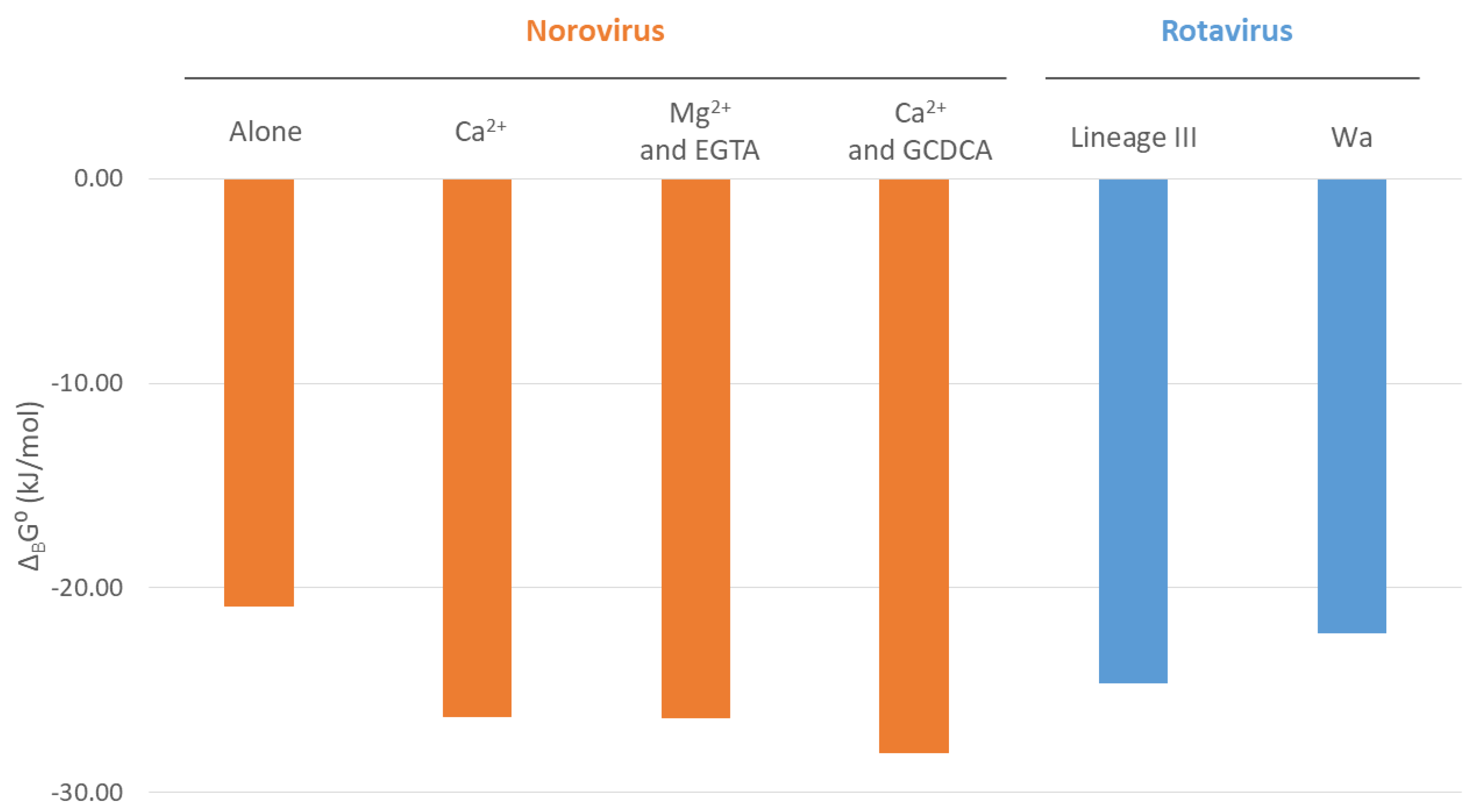
| Interaction | Conditions | Kd (M) | KB (M−1) | ΔBG0 (kJ/mol) |
|---|---|---|---|---|
| VP1 protruding (P) domain with CD300lf receptor | Alone | 2.19 × 10−4 | 4.57 × 103 | −20.89 |
| Ca2+ | 2.45 × 10−5 | 4.08 × 104 | −26.32 | |
| Mg2+ and EGTA | 2.43 × 10−5 | 4.12 × 104 | −26.34 | |
| Ca2+ and GCDCA | 1.20 × 10−5 | 8.31 × 104 | −28.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Popović, M.E.; Tadić, V.; Pantović Pavlović, M. Biothermodynamic Analysis of Norovirus: Mechanistic Model of Virus–Host Interactions and Virus–Virus Competition Based on Gibbs Energy. Microbiol. Res. 2025, 16, 112. https://doi.org/10.3390/microbiolres16060112
Popović ME, Tadić V, Pantović Pavlović M. Biothermodynamic Analysis of Norovirus: Mechanistic Model of Virus–Host Interactions and Virus–Virus Competition Based on Gibbs Energy. Microbiology Research. 2025; 16(6):112. https://doi.org/10.3390/microbiolres16060112
Chicago/Turabian StylePopović, Marko E., Vojin Tadić, and Marijana Pantović Pavlović. 2025. "Biothermodynamic Analysis of Norovirus: Mechanistic Model of Virus–Host Interactions and Virus–Virus Competition Based on Gibbs Energy" Microbiology Research 16, no. 6: 112. https://doi.org/10.3390/microbiolres16060112
APA StylePopović, M. E., Tadić, V., & Pantović Pavlović, M. (2025). Biothermodynamic Analysis of Norovirus: Mechanistic Model of Virus–Host Interactions and Virus–Virus Competition Based on Gibbs Energy. Microbiology Research, 16(6), 112. https://doi.org/10.3390/microbiolres16060112






