Fungal Functional Level to Describe Soil Fungal Composition at Mediterranean Vineyards
Abstract
1. Introduction
2. Materials and Methods
2.1. Location of the Study Sites and Soil Sampling
2.2. Soil DNA Extraction, Fungal Community Sequencing and Taxonomic Identification
2.3. Data Analyses
3. Results
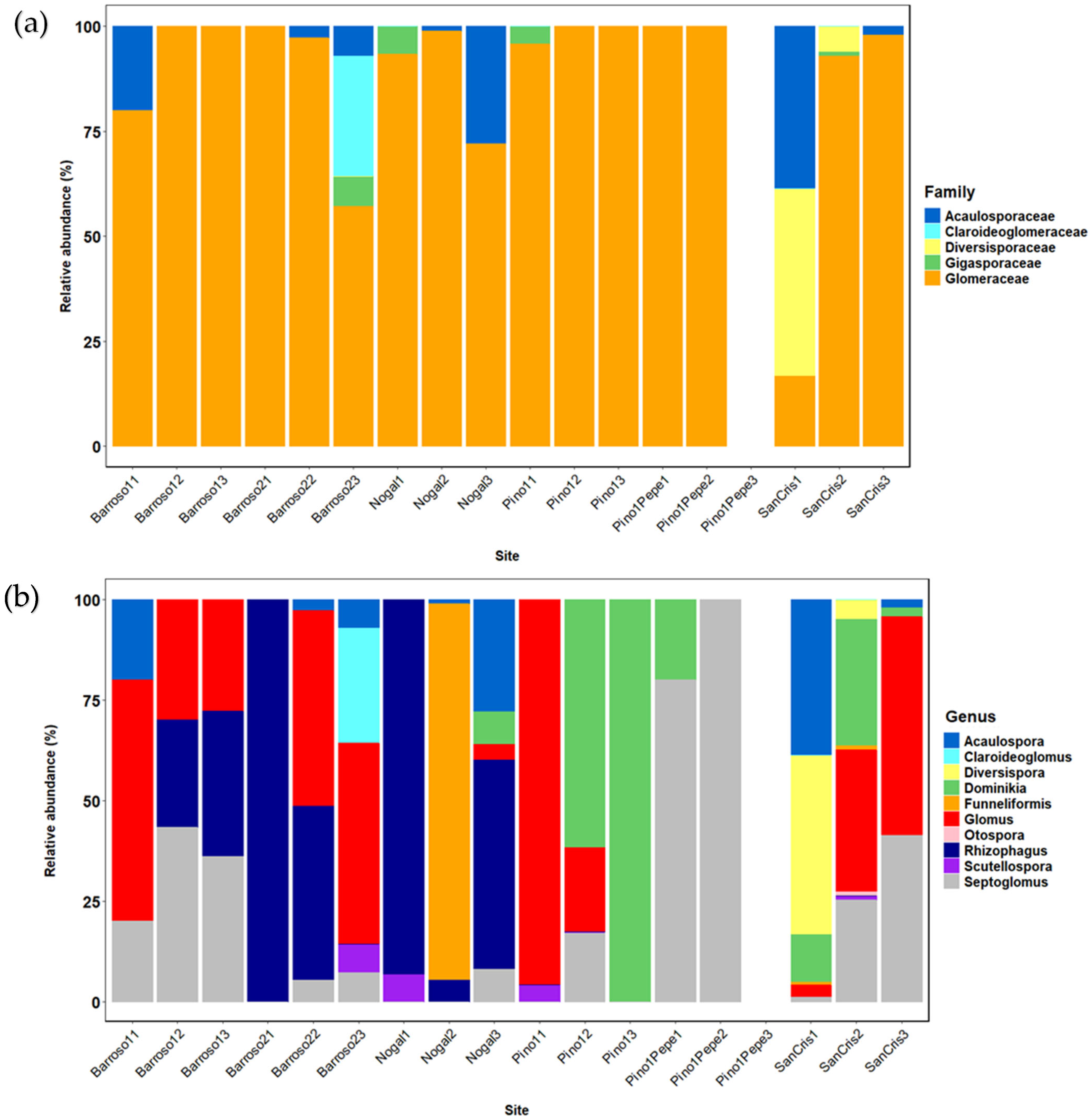
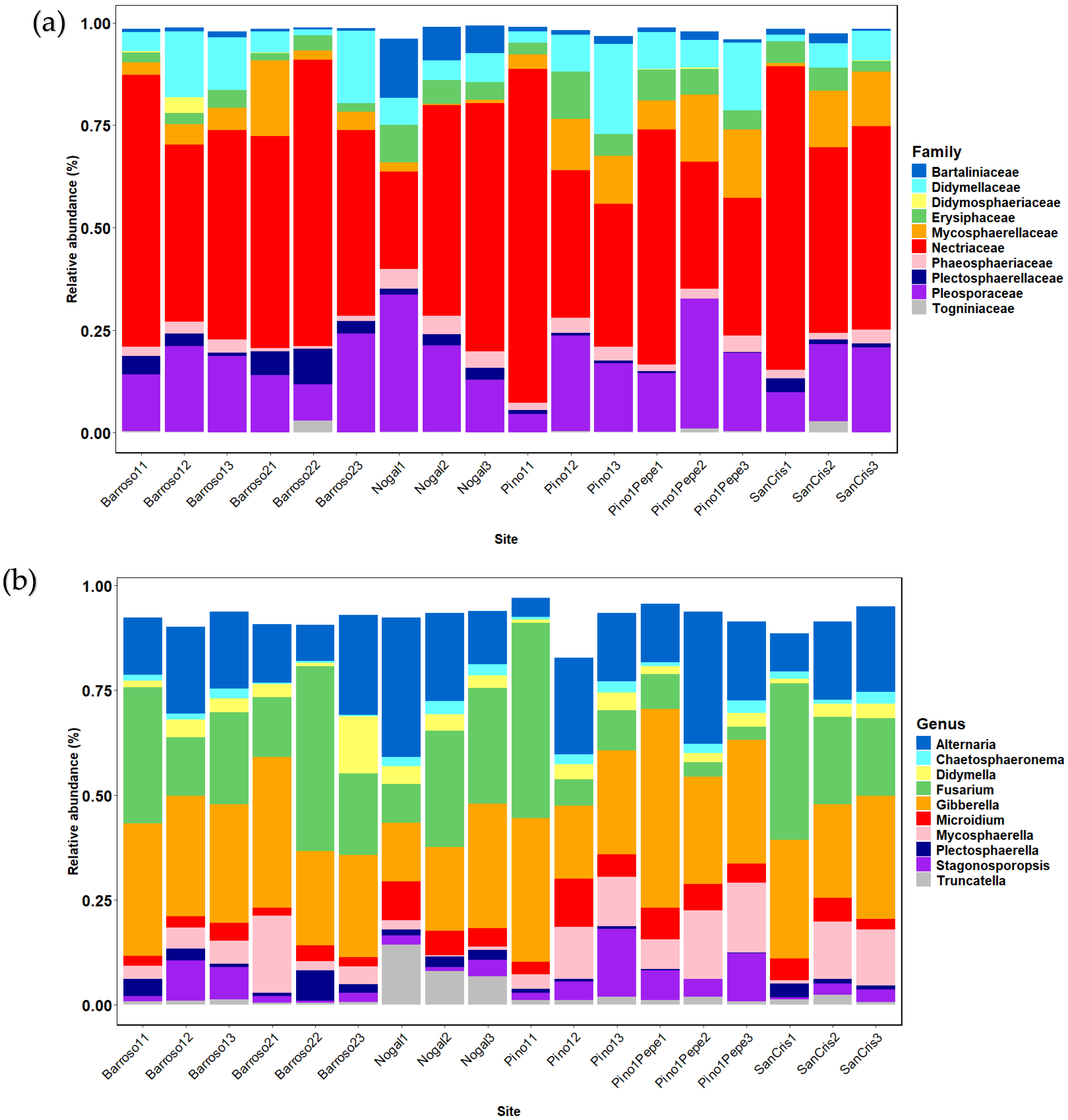
3.1. Community Raw Results
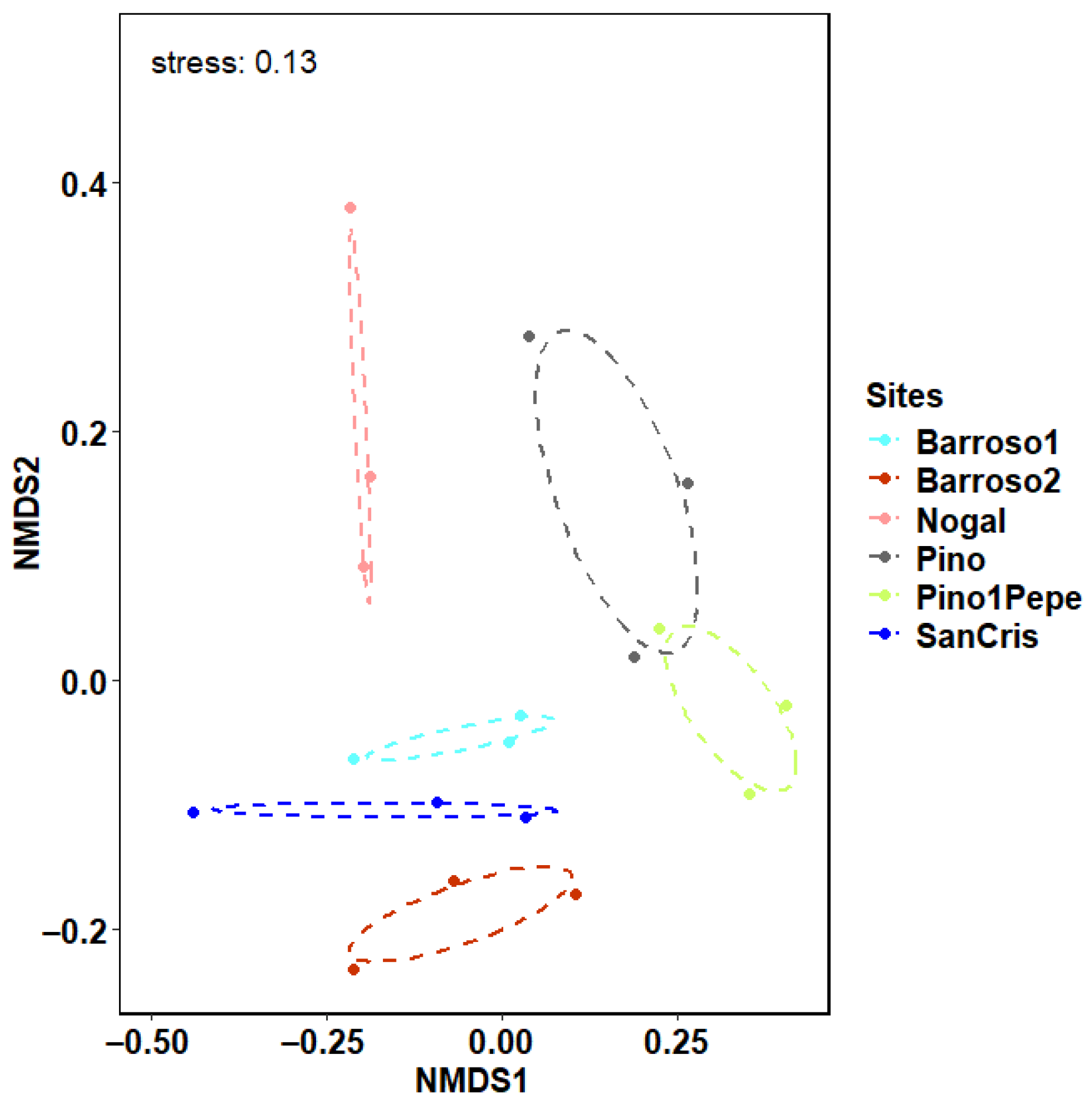
3.2. Community Analyses
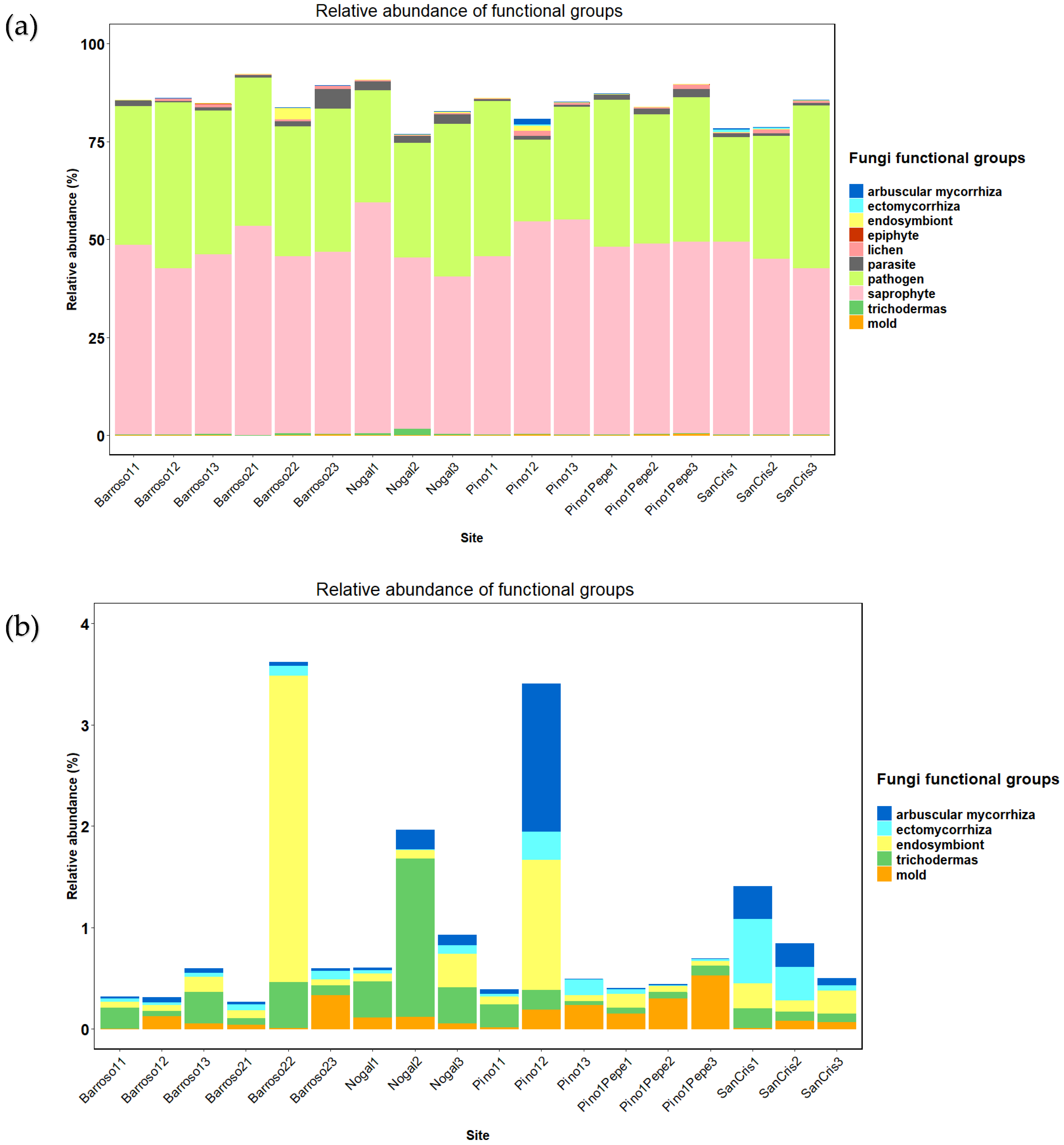
3.3. Functional Guilds Analysis
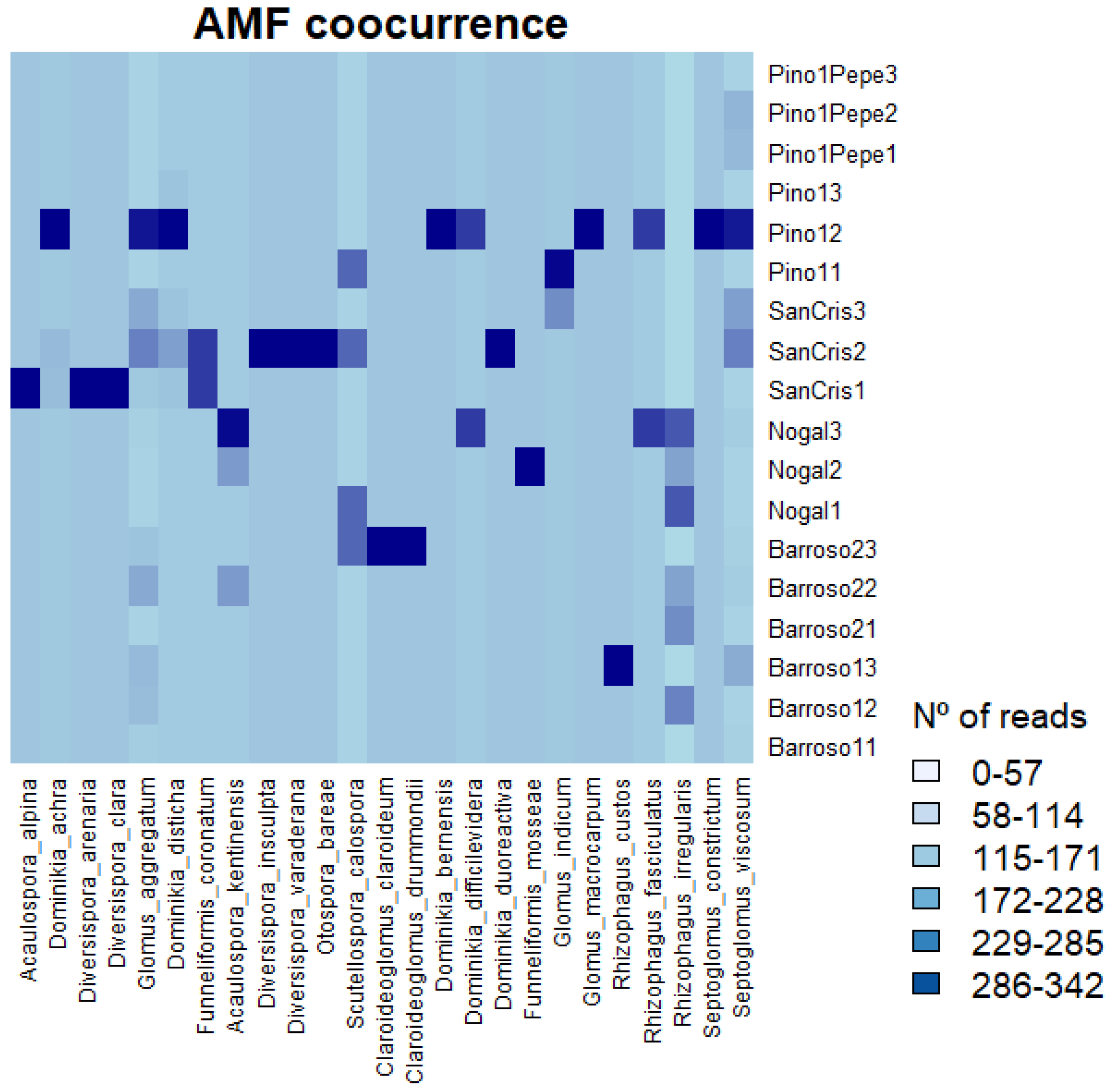
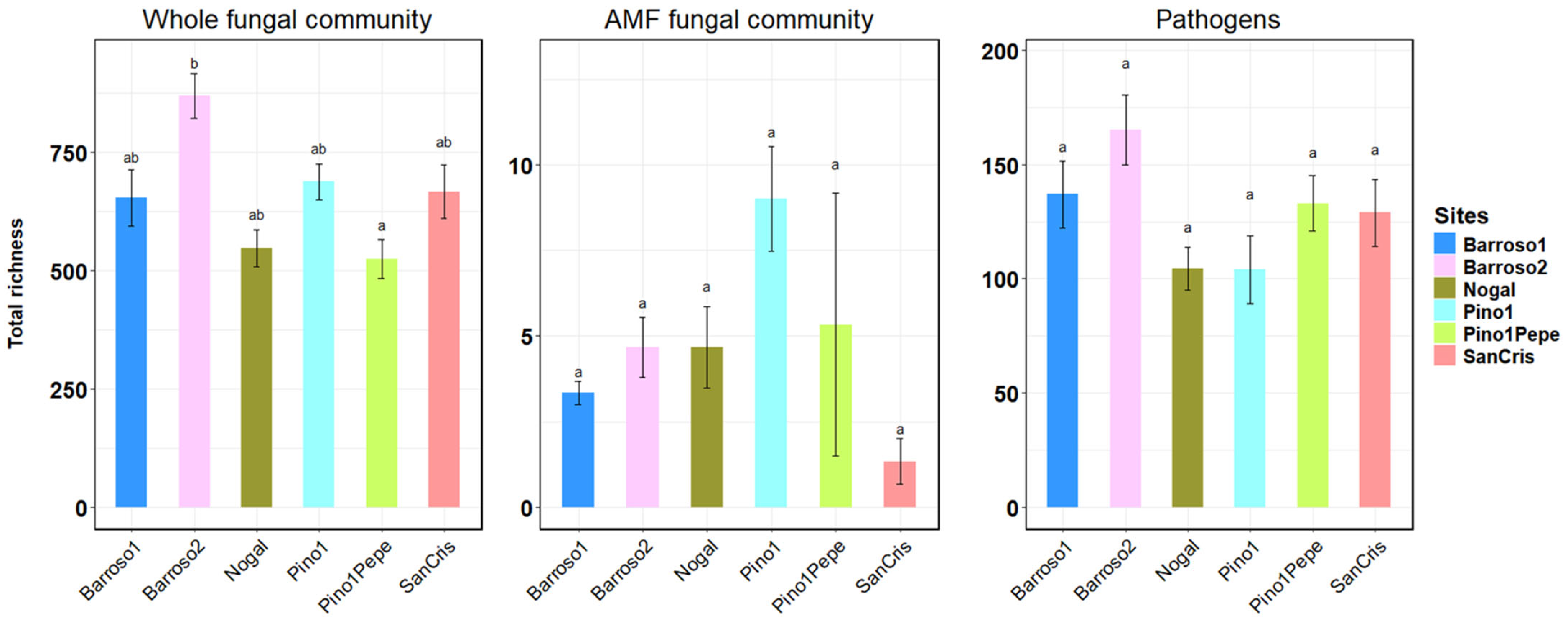
3.4. Most Abundant Species and Fungal Diversity Analyses
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 234, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Mezzatesta, D.; Aguilar, M.O.; Gobbi, A.; Pistorio, M.; Hansen, L.H.; Buscema, F.; Piccoli, P.; Berli, F. Soil-associated fungal and prokaryotic diversity influenced by stoniness, depth and vintage in a high-altitude vineyard. OENO One 2024, 58, 1–12. [Google Scholar] [CrossRef]
- OIV-International Organization of Vine and Wine. International Code of Enological Practices (II, 4 1–4); OIV-International Organization of Vine and Wine: Paris, France, 2020. [Google Scholar]
- Johnston-Monje, D.; Vergara, L.I.; Lopez-Mejia, J.; White, J.F. Plant microbiomes as contributors to agricultural terroir. Front. Agron. 2023, 5, 1216520. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From vineyard soil to wine fermentation: Microbiome approximations to explain the “terroir” concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef]
- Cao, Y.; Fanning, S.; Proos, S.; Jordan, K.; Srikumar, S. A Review on the Applications of Next Generation Sequencing Technologies as Applied to Food-Related Microbiome Studies. Front. Microbiol. 2017, 8, 1829. [Google Scholar] [CrossRef]
- Paolinelli, M.; Martinez, L.E.; Garcia Lampasona, S.C.; Diaz Quirós, C.; Belmonte, M.; Ahumada, G.; Pirrone, M.Á.; Mercado, L.A. Microbiome in soils of Mendoza: Microbial resources for the development of agroecological management in viticulture. OENO One 2023, 57, 191–205. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis; Academic Press: Cambridge, MA, USA, 2010. [Google Scholar]
- Aguilera, P.; Ortiz, N.; Becerra, N.; Turrini, A.; Gaínza-Cortés, F.; Silva-Flores, P.; Aguilar-Paredes, A.; Borie, F. Application of arbuscular mycorrhizal fungi in vineyards: Water and biotic stress under a climate change scenario: New challenge for Chilean grapevine crop. Front. Microbiol. 2022, 13, 826571. [Google Scholar] [CrossRef]
- Holland, T.; Vukicevich, E.; Thomsen, C.; Pogiatzis, A.; Hart, M.; Bowen, P. Arbuscular mycorrhizal fungi in viticulture: Should we use biofertilizers? Am. J. Enol. Vitic. 2018, 2 (Suppl. S2), 59–63. [Google Scholar] [CrossRef]
- Scandellari, F. Arbuscular mycorrhizal contribution to nitrogen uptake of grapevines. Vitis 2017, 56, 147–154. [Google Scholar] [CrossRef]
- Yu, K.; Pieterse, C.M.; Bakker, P.A.; Berendsen, R.L. Beneficial microbes going underground of root immunity. Plant Cell Environ. 2019, 42, 2860–2870. [Google Scholar] [CrossRef]
- Kocsis, I.; Petróczy, M.; Markó, G. Bridging boundaries: Exploring vineyard, management and variety characteristics influencing long-term infection of grapevine pathogens. OENO One 2024, 58. [Google Scholar] [CrossRef]
- Altieri, V.; Rossi, V.; Fedele, G. Integration of mathematical modeling and target-based application of biocontrol agents for the control of Botrytis cinerea in vineyards. Pest Manag. Sci. 2024, 80, 4352–4360. [Google Scholar] [CrossRef] [PubMed]
- Gramaje, D.; Úrbez-Torres, J.R.; Sosnowski, M.R. Managing grapevine trunk diseases with respect to etiology and epidemiology: Current strategies and future prospects. Plant Dis. 2018, 102, 12–39. [Google Scholar] [CrossRef] [PubMed]
- Franco, G.C.; Leiva, J.; Nand, S.; Lee, D.M.; Hajkowski, M.; Dick, K.; Withers, B.; Soto, L.; Mingoa, B.-R.; Acholonu, M.; et al. Soil microbial communities and wine terroir: Research gaps and data needs. Foods 2024, 13, 2475. [Google Scholar] [CrossRef] [PubMed]
- Igual, J.M.; Andrades, M.S.; Frade, C.; Carpio, M.J.; Marín-Benito, J.M.; Rodríguez-Cruz, M.S.; Valverde, A. Impacts of spent mushroom substrate amendment and season on microbial communities in a semiarid vineyard soil. Appl. Soil Ecol. 2024, 203, 105689. [Google Scholar] [CrossRef]
- Marín, C.; Dittrich, F.; Vasar, M.; Gaínza-Cortés, F.; Silva-Flores, P.; Aguilera, P. Vineyard maturity increases arbuscular mycorrhizal and decreases plant pathogen fungal relative abundance in bulk soil across a 1000 km Chilean gradient. Plants People Planet 2025, 7-4, 987–997. [Google Scholar] [CrossRef]
- Martínez-Diz, M.; Andrés-Sodupe, M.; Bujanda, R.; Díaz-Losada, E.; Eichmeier, A.; Gramaje, D. Soil-plant compartments affect fungal microbiome diversity and composition in grapevine. Fungal Ecol. 2019, 41, 234–244. [Google Scholar] [CrossRef]
- Rotbart, N.; Doniger, T.; Applebaum, I.; Steinberger, Y. Soil Fungal Communities in the Rhizosphere of Sauvignon Blanc Grapes Subjected to Various Agricultural Management Practices. Land 2025, 14, 667. [Google Scholar] [CrossRef]
- Tanunchai, B.; Ji, L.; Schroeter, S.A.; Wahdan, S.F.M.; Hossen, S.; Delelegn, Y.; Buscot, F.; Lehnert, A.S.; Alves, E.G.; Hilke, I.; et al. FungalTraits vs. FUNGuild: Comparison of Ecological Functional Assignments of Leaf- and Needle-Associated Fungi Across 12 Temperate Tree Species. Microb. Ecol. 2023, 85, 411–428. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wu, X.; Yang, J.; Ruan, H.; Wang, S.; Yang, Y.; Naeem, I.; Liu, L.; Wang, D. The diversity and co-occurrence network of soil bacterial and fungal communities and their implications for a new indicator of grassland degradation. Ecol. Indic. 2021, 129, 107989. [Google Scholar] [CrossRef]
- Chen, J.; Shi, Z.; Liu, S.; Zhang, M.; Cao, X.; Chen, M.; Li, F.; Feng, Q. Altitudinal variation influences soil fungal community composition and diversity in Alpine–Gorge region on the Eastern Qinghai–Tibetan Plateau. J. Fungi 2022, 8, 807. [Google Scholar] [CrossRef]
- Gobbi, A.; Acedo, A.; Imam, N.; Santini, R.G.; Ortiz-Álvarez, R.; Ellegaard-Jensen, L.; Belda, I.; Hansen, L.H. A global microbiome survey of vineyard soils highlights the microbial dimension of viticultural terroirs. Commun. Biol. 2022, 5, 241. [Google Scholar] [CrossRef]
- Knight, S.J.; Karon, O.; Goddard, M.R. Small scale fungal community differentiation in a vineyard system. Food Microbiol. 2020, 87, 103358. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Wang, X.; Yan, J.; Luo, L. Characterizing the intra-vineyard variation of soil bacterial and fungal communities. Front. Microbiol. 2019, 10, 1239. [Google Scholar] [CrossRef] [PubMed]
- Adamo, I.; Piñuela, Y.; Bonet, J.A.; Castaño, C.; de Aragón, J.M.; Parladé, J.; Pera, J.; Alday, J.G. Sampling forest soils to describe fungal diversity and composition. Which is the optimal sampling size in Mediterranean pure and mixed pine oak forests? Fungal Biol. 2021, 125, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Nilsson, R.H.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. vegan: Community Ecology Package; R package version 2.5-7; 2020; https://CRAN.R-project.org/package=vegan (accessed on 3 March 2025).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Chambers, J.M.; Freeny, A.; Heiberger, R.M. Analysis of Variance; Designed Experiments; Chapter 5 of Statistical Models in S; Routledge: New York, NY, USA, 2017; pp. 145–193. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Oliveira, M.C.O.; Alves, A.; Fidalgo, C.; de Freitas, J.G.; Pinheiro de Carvalho, M.A. Variations in the structure and function of the soil fungal communities in the traditional cropping systems from Madeira Island. Front. Microbiol. 2024, 15, 1426957. [Google Scholar] [CrossRef]
- Aguilar-Paredes, A.; Turrini, A.; Avio, L.; Stuardo, C.; Velásquez, A.; Becerra, J.; Giovannetti, M.; Seeger, M. Agricultural managements influence the diversity of arbuscular mycorrhizal fungi in vineyards from Chilean Mediterranean climate ecosystems. J. Soil Sci. Plant Nutr. 2024, 24, 6099–6112. [Google Scholar] [CrossRef]
- Betancur-Agudelo, M.; Meyer, E.; Lovato, P.E. Arbuscular mycorrhizal fungus richness in the soil and root colonization in vineyards of different ages. Rhizosphere 2021, 17, 100307. [Google Scholar] [CrossRef]
- Ortiz-Vidal, N.; Borie, F.; Castillo, C.; Sieverding, E.; Aguilera, P. Mycorrhizal Symbiosis in Southern Chile: Exploring root-fungus Associations and Fungal Species Identification in La Araucanía Region Vineyards. J. Soil Sci. Plant Nutr. 2025, 25, 5875–5883. [Google Scholar] [CrossRef]
- Schreiner, R.P. Depth structures the community of arbuscular mycorrhizal fungi amplified from grapevine (Vitis vinifera L.) roots. Mycorrhiza 2020, 30, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Lv, B.; He, Z.; Li, H.; Wang, H. Rhizosphere metabolite dynamics in continuous cropping of vineyards: Impact on microflora diversity and co-occurrence networks. Microbiol. Res. 2025, 296, 128134. [Google Scholar] [CrossRef]
- del Pilar Martínez-Diz, M.; Díaz-Losada, E.; Díaz-Fernández, Á.; Bouzas-Cid, Y.; Gramaje, D. Protection of grapevine pruning wounds against Phaeomoniella chlamydospora and Diplodia seriata by commercial biological and chemical methods. Crop Prot. 2021, 143, 105465. [Google Scholar] [CrossRef]
- Díaz, G.A.; Reveglia, P.; Tomoiagă, L.L.; Chedea, V.S. Fungal pathogens causing the grapevine trunk diseases-biology and identification. Front. Fungal Biol. 2023, 4, 1186166. [Google Scholar] [CrossRef]
- Bai, C.; Yao, Y.; Wang, H.; Li, H.; Wei, R. The Fungal Microbiome in the Vineyard Ecosystem Plays a Key Role in Shaping the Regional Characteristics of Wine. Foods 2025, 14, 1211. [Google Scholar] [CrossRef]
- Castañeda, L.E.; Barbosa, O. Metagenomic analysis exploring taxonomic and functional diversity of soil microbial communities in Chilean vineyards and surrounding native forests. PeerJ 2017, 5, e3098. [Google Scholar] [CrossRef]
- Prendes, L.P.; Merín, M.G.; Zachetti, V.G.L.; Pereyra, A.; Ramirez, M.L.; Morata de Ambrosini, V.I. Impact of antagonistic yeasts from wine grapes on growth and mycotoxin production by Alternaria alternata. J. Appl. Microbiol. 2021, 131, 833–843. [Google Scholar] [CrossRef]
- Torres-Palazzolo, C.; Ferreyra, S.; Iribas, F.; Chimeno, V.; Rojo, M.C.; Casalongue, C.; Combina, M.; Ponsone, M.L. Biocontrol of Alternaria alternata in cold-stored table grapes using psychrotrophic yeasts and bioactive compounds of natural sources. Int. J. Food Microbiol. 2024, 415, 110640. [Google Scholar] [CrossRef]
- Patanita, M.; Albuquerque, A.; Campos, M.D.; Materatski, P.; Varanda, C.M.; Ribeiro, J.A.; Felix, M.D.R. Metagenomic assessment unravels fungal microbiota associated to grapevine trunk diseases. Horticulturae 2022, 8, 288. [Google Scholar] [CrossRef]
- Lehmann, A.; Rillig, M.C. Understanding mechanisms of soil biota involvement in soil aggregation: A way forward with saprobic fungi? Soil Biol. Biochem. 2015, 88, 298–302. [Google Scholar] [CrossRef]
- Ortiz-Álvarez, R.; Ortega-Arranz, H.; Ontiveros, V.J.; de Celis, M.; Ravarani, C.; Acedo, A.; Belda, I. Network properties of local fungal communities reveal the anthropogenic disturbance consequences of farming practices in vineyard soils. Msystems 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Barceló, M.; van Bodegom, P.M.; Tedersoo, L.; den Haan, N.; Veen, G.F.; Ostonen, I.; Trimbos, K.; Soudzilovskaia, N.A. The abundance of arbuscular mycorrhiza in soils is linked to the total length of roots colonized at ecosystem level. PLoS ONE 2020, 15, e0237256. [Google Scholar] [CrossRef]
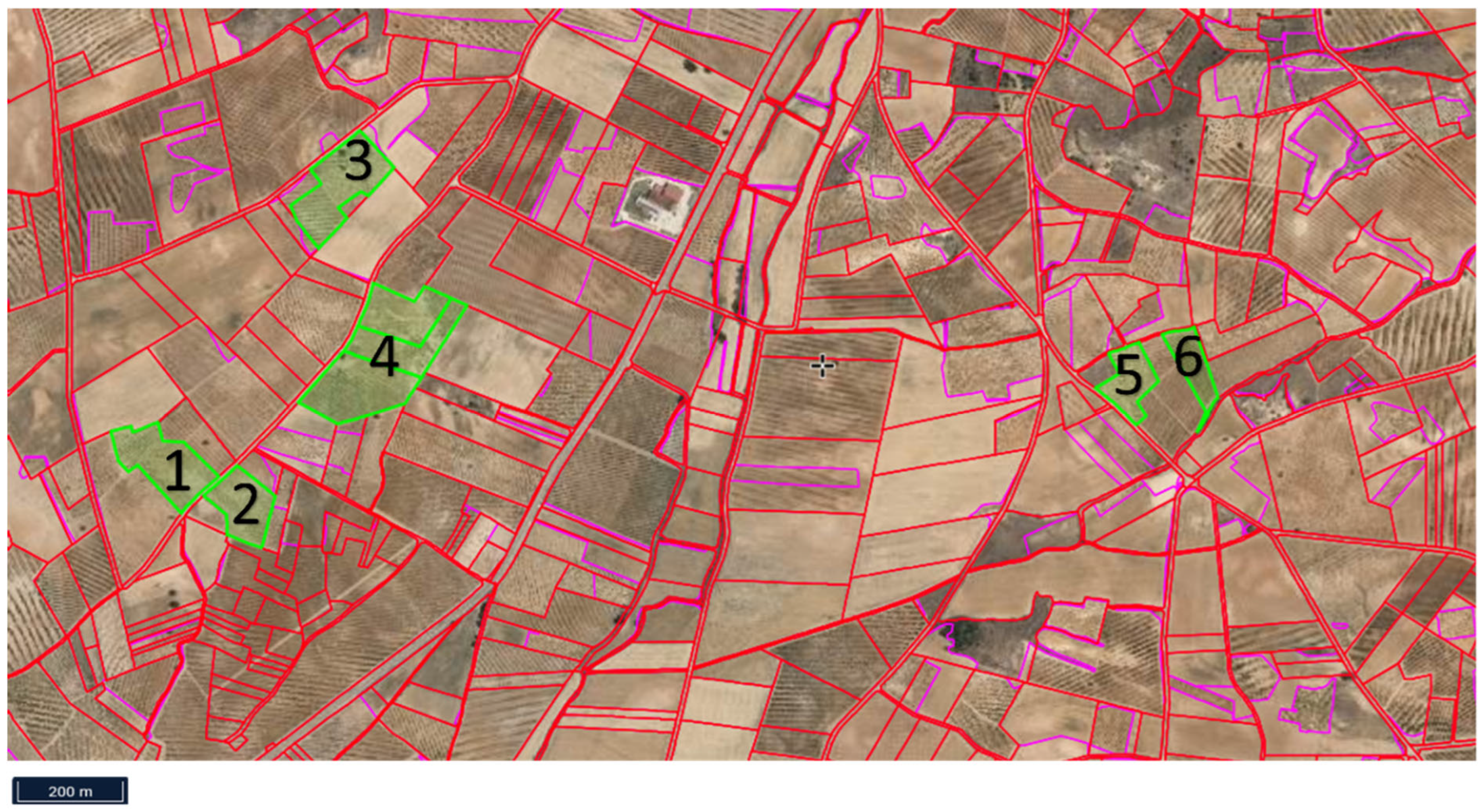
| Vineyard/Site | Density (Vine/ha) | Grape Variety | Management | Pesticide and Fertilizer Applications | Plant Cover |
|---|---|---|---|---|---|
| Barroso1 | 2022 | Tempranillo | Biodynamic | Cu and S* Biodynamic compost amendment | Yes |
| Barroso2 | 2000 | Tempranillo | Biodynamic | Cu and S* Biodynamic compost amendment | Yes |
| Nogal | 1736 | Tempranillo | Biodynamic | Cu and S* Biodynamic compost amendment | Yes |
| Pino1 | 4000 | Tempranillo | Biodynamic | Cu and S* Biodynamic compost amendment | Yes |
| Pino1Pepe | 2222 | Tempranillo | Biodynamic | Cu and S* Biodynamic compost amendment | Yes |
| SanCris | 2645 | Tempranillo | Ecologic | Cu and S* No amendment | No |
| Sites | ||||||
|---|---|---|---|---|---|---|
| Soil properties | Barroso1 | Barroso2 | San Cris | Nogal | Pino1 | Pino1Pepe |
| pH (1:2,5) | 8.68 | 8.39 | 8.85 | 8.59 | 8.75 | 8.49 |
| Electrical conductivity (dS m-1) | 0.07 | 0.06 | 0.05 | 0.07 | 0.07 | 0.11 |
| Sand (%) | 76.72 | 82.72 | 71.72 | 82.72 | 77.72 | 58.44 |
| Silt (%) | 6.28 | 6.28 | 7.28 | 5.28 | 7.28 | 15.28 |
| Clay (%) | 17 | 11 | 21 | 12 | 15 | 26.28 |
| Texture | Loamy | Loamy | Loamy | Loamy | Loamy | Clay |
| Organic matter (%) | 0.9 | 0.56 | 0.94 | 0.64 | 0.48 | 0.68 |
| CaCo3 (%) | 6 | 6 | 6 | No | 6 | 8.6 |
| Available P (mg/Kg) | 4 | 10.4 | 6.5 | 6.8 | 9.2 | 8.6 |
| Available K (mg/Kg) | 279 | 179 | 289 | 141 | 146 | 177 |
| Ca (mmolc L-1) | 31.2 | 6 | 30 | 20.5 | 30.6 | 39.1 |
| Mg (mmolc L-1) | 0.9 | 0.42 | 1.17 | 0.43 | 0.79 | 1.69 |
| Na (mmolc L-1) | 0.04 | 0.04 | 0.09 | 0.05 | 0.07 | 0.08 |
| Ntotal (%) | 0.08 | 0.05 | 0.06 | 0.06 | 0.04 | 0.07 |
| Site | Barroso1 | Barroso2 | Nogal | Pino1 | Pino1 Pepe | SanCris | p-Value |
|---|---|---|---|---|---|---|---|
| AMF | 0.04 ± 0.01 a | 0.03 ± 0.01 a | 0.11 ± 0.05 a | 0.50 ± 0.48 a | 0.01 ± 0.00 a | 0.21 ± 0.07 a | 0.51 |
| ECM | 0.03 ± 0.01 a | 0.08 ± 0.01 a | 0.04 ± 0.02 a | 0.15 ± 0.07 a | 0.02 ± 0.01 a | 0.34 ± 0.17 a | 0.09 |
| endo | 0.09 ± 0.03 a | 1.05 ± 0.98 a | 0.16 ± 0.08 a | 0.47 ± 0.40 a | 0.08 ± 0.03 a | 0.19 ± 0.04 a | 0.6 |
| epi | 0.02 ± 0.01 a | 0.02 ± 0.00 a | 0.03 ± 0.03 a | 0.01 ± 0.00 a | 0.04 ± 0.01 a | 0.02 ± 0.01 a | 0.78 |
| lichen | 0.51 ± 0.25 a | 0.50 ± 0.17 a | 0.27 ± 0.04 a | 0.66 ± 0.36 a | 0.50 ± 0.34 a | 0.56 ± 0.23 a | 0.93 |
| mold | 0.06 ± 0.04 a | 0.13 ± 0.10 a | 0.10 ± 0.02 a | 0.15 ± 0.07 a | 0.33 ± 0.11 a | 0.05 ± 0.02 a | 0.13 |
| paras | 0.88 ± 0.30 a | 2.29 ± 1.36 a | 2.13 ± 0.19 a | 0.71 ± 0.16 a | 1.68 ± 0.25 a | 0.69 ± 0.09 a | 0.25 |
| path | 38.18 ± 2.12 a | 35.87 ± 1.33 a | 32.27 ± 3.35 a | 29.76 ± 5.44 a | 35.77 ± 1.41 a | 33.23 ± 4.37 a | 0.57 |
| sapro | 45.49 ± 1.73 a | 48.28 ± 2.56 a | 47.58 ± 5.79 a | 51.42 ± 3.02 a | 48.39 ± 0.29 a | 45.49 ± 1.96 a | 0.76 |
| Trich | 0.19 ± 0.08 a | 0.21 ± 0.13 a | 0.76 ± 0.40 a | 0.15 ± 0.06 a | 0.07 ± 0.01 a | 0.12 ± 0.03 a | 0.15 |
| Unknown | 14.51 ± 0.36 a | 11.55 ± 2.52 a | 16.55 ± 4.05 a | 16.01 ± 1.63 a | 13.11 ± 1.70 a | 19.09 ± 2.35 a | 0.34 |
| Phylum | Barroso11 | Barroso12 | Barroso13 | Barroso21 | Barroso22 | Barroso23 | Nogal1 | Nogal2 | Nogal3 | SanCris1 | SanCris2 | SanCris3 | Pino11 | Pino12 | Pino13 | Pino1Pepe1 | Pino1Pepe2 | Pino1Pepe3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ascomycota | 79.5 | 75.9 | 75.0 | 69.9 | 76.3 | 75.5 | 59.8 | 71.5 | 74.4 | 79.8 | 73.9 | 79.6 | 70.6 | 73.5 | 66.8 | 76.2 | 75.7 | 83.7 |
| Basidiomycota | 14.2 | 17.8 | 18.4 | 24.8 | 16.5 | 19.5 | 33.8 | 18.6 | 14.9 | 11.0 | 16.1 | 14.6 | 21.2 | 14.0 | 25.1 | 14.4 | 20.2 | 10.2 |
| Mortierellomycota | 2.8 | 3.6 | 3.7 | 3.3 | 5.0 | 1.9 | 2.6 | 5.0 | 5.1 | 5.5 | 4.1 | 2.7 | 4.7 | 6.3 | 3.4 | 3.2 | 2.2 | 2.6 |
| Chytridiomycota | 1.5 | 1.6 | 1.2 | 0.8 | 1.1 | 1.8 | 2.7 | 1.6 | 2.3 | 1.7 | 2.2 | 2.0 | 1.4 | 1.8 | 2.0 | 2.8 | 0.9 | 2.4 |
| Mucoromycota | 0.6 | 0.1 | 0.1 | 0.1 | 0.2 | 0.1 | 0.1 | 1.0 | 0.2 | 0.4 | 0.3 | 0.1 | 0.3 | 0.0 | 0.1 | 0.9 | 0.0 | 0.0 |
| Glomeromycota | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | 0.1 | 0.4 | 0.2 | 0.1 | 0.1 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 |
| Kickxellomycota | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.2 | 0.4 | 0.3 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | 0.0 | 0.0 |
| Monoblepharomycota | 0.2 | 0.1 | 0.0 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 | 0.1 | 0.0 | 0.1 |
| Olpidiomycota | 0.2 | 0.1 | 0.1 | 0.0 | 0.1 | 0.1 | 0.0 | 0.1 | 0.2 | 0.0 | 0.2 | 0.1 | 0.1 | 0.3 | 0.1 | 0.0 | 0.0 | 0.0 |
| Rozellomycota | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.2 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 |
| Entomophthoromycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 |
| Aphelidiomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Blastocladiomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Entorrhizomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Zoopagomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Calcarisporiellomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Neocallimastigomycota | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Unidentified | 0.7 | 0.6 | 1.3 | 0.9 | 0.6 | 0.8 | 0.5 | 1.4 | 1.7 | 0.7 | 2.6 | 0.7 | 1.2 | 2.1 | 2.4 | 2.3 | 0.8 | 0.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piñuela, Y.; Hernández, M.; Escudero, I.; Sisseck, P.; Olaizola, J. Fungal Functional Level to Describe Soil Fungal Composition at Mediterranean Vineyards. Microbiol. Res. 2025, 16, 217. https://doi.org/10.3390/microbiolres16100217
Piñuela Y, Hernández M, Escudero I, Sisseck P, Olaizola J. Fungal Functional Level to Describe Soil Fungal Composition at Mediterranean Vineyards. Microbiology Research. 2025; 16(10):217. https://doi.org/10.3390/microbiolres16100217
Chicago/Turabian StylePiñuela, Yasmin, María Hernández, Iván Escudero, Peter Sisseck, and Jaime Olaizola. 2025. "Fungal Functional Level to Describe Soil Fungal Composition at Mediterranean Vineyards" Microbiology Research 16, no. 10: 217. https://doi.org/10.3390/microbiolres16100217
APA StylePiñuela, Y., Hernández, M., Escudero, I., Sisseck, P., & Olaizola, J. (2025). Fungal Functional Level to Describe Soil Fungal Composition at Mediterranean Vineyards. Microbiology Research, 16(10), 217. https://doi.org/10.3390/microbiolres16100217






