Copper Tolerance of Trichoderma koningii Tk10
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain
2.2. Isolation and Screening of Copper-Resistant Trichoderma Strains
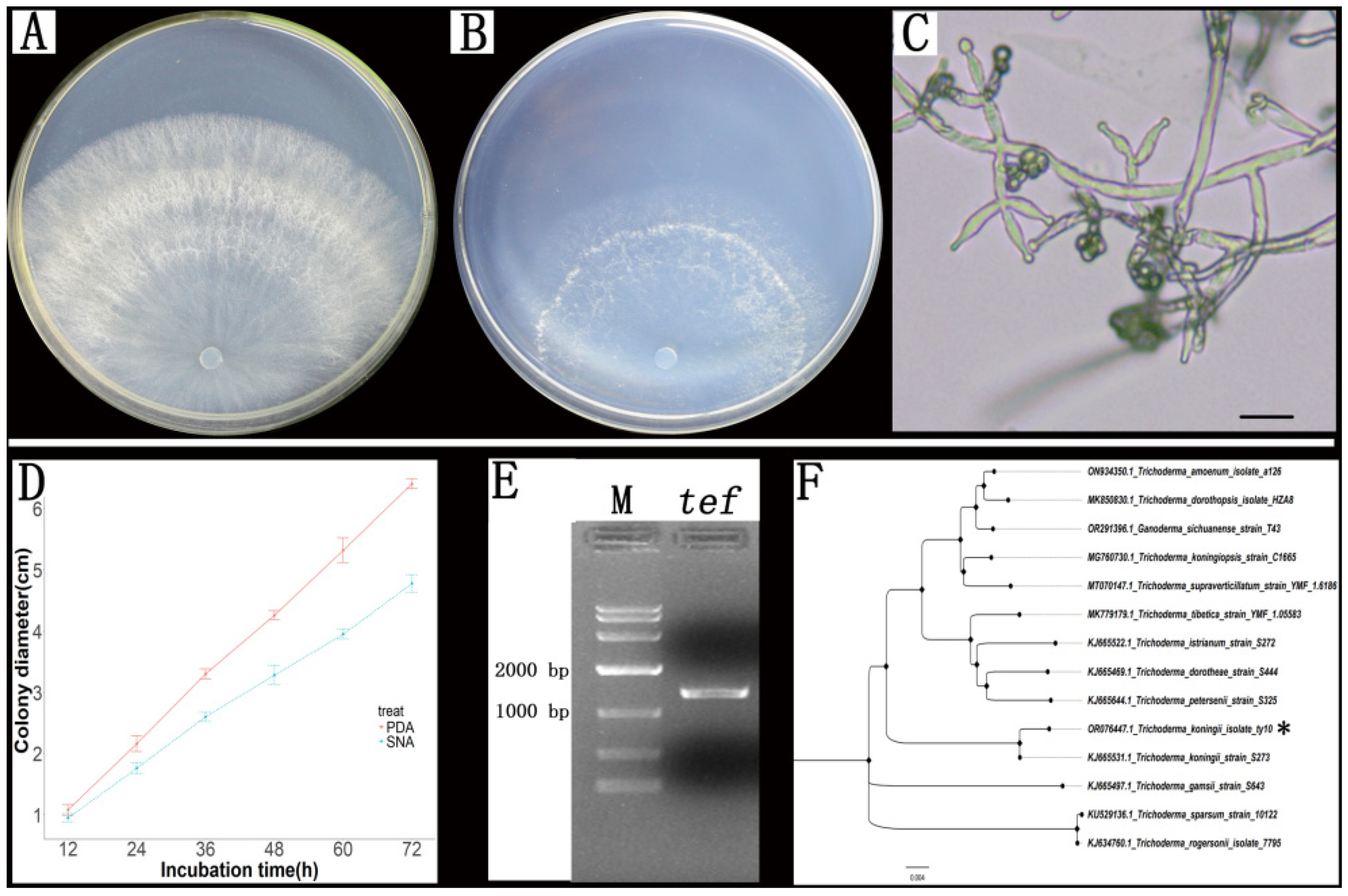
2.3. Morphological and Molecular Analysis
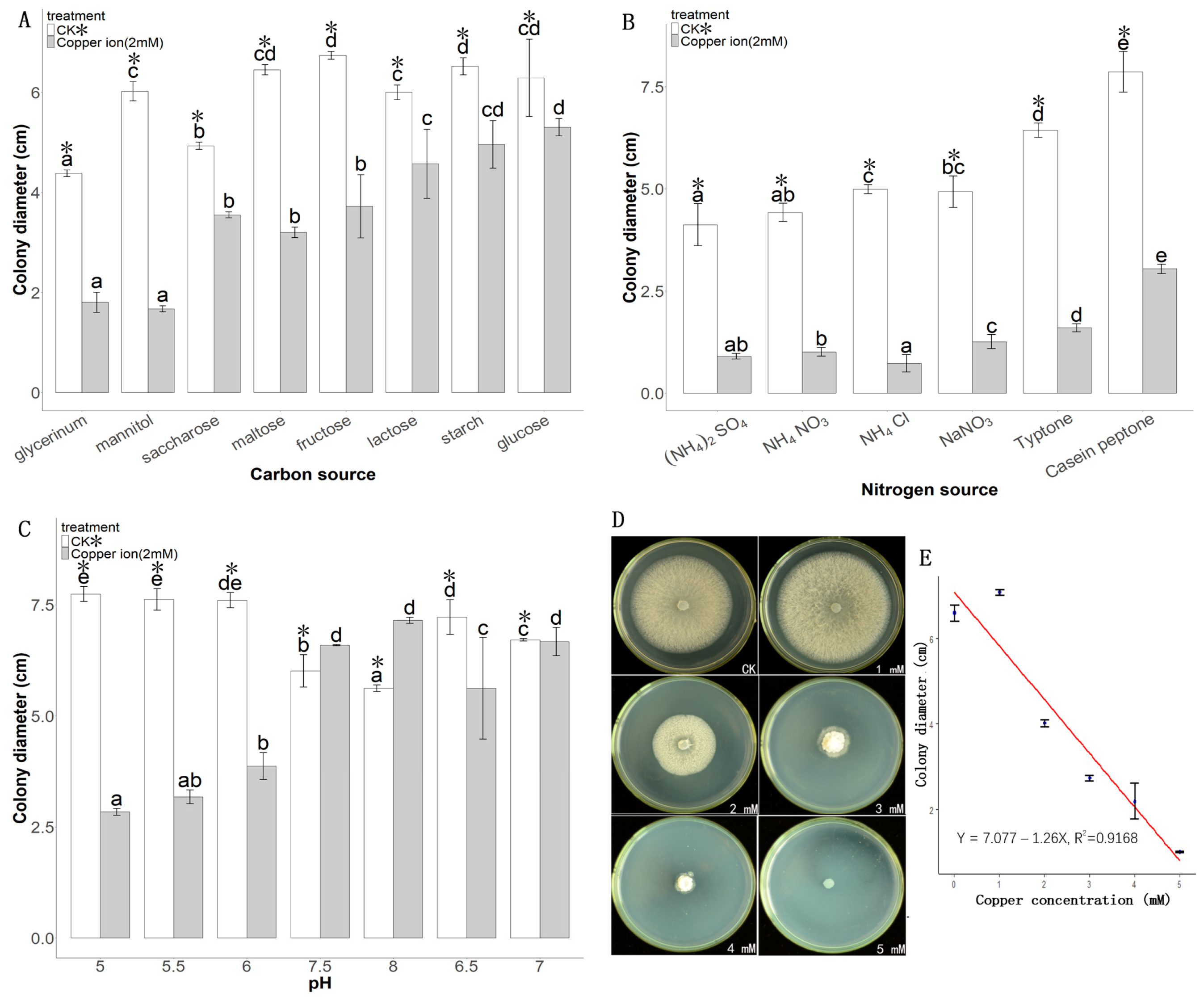
2.4. Single Factor Optimization of Culture Conditions
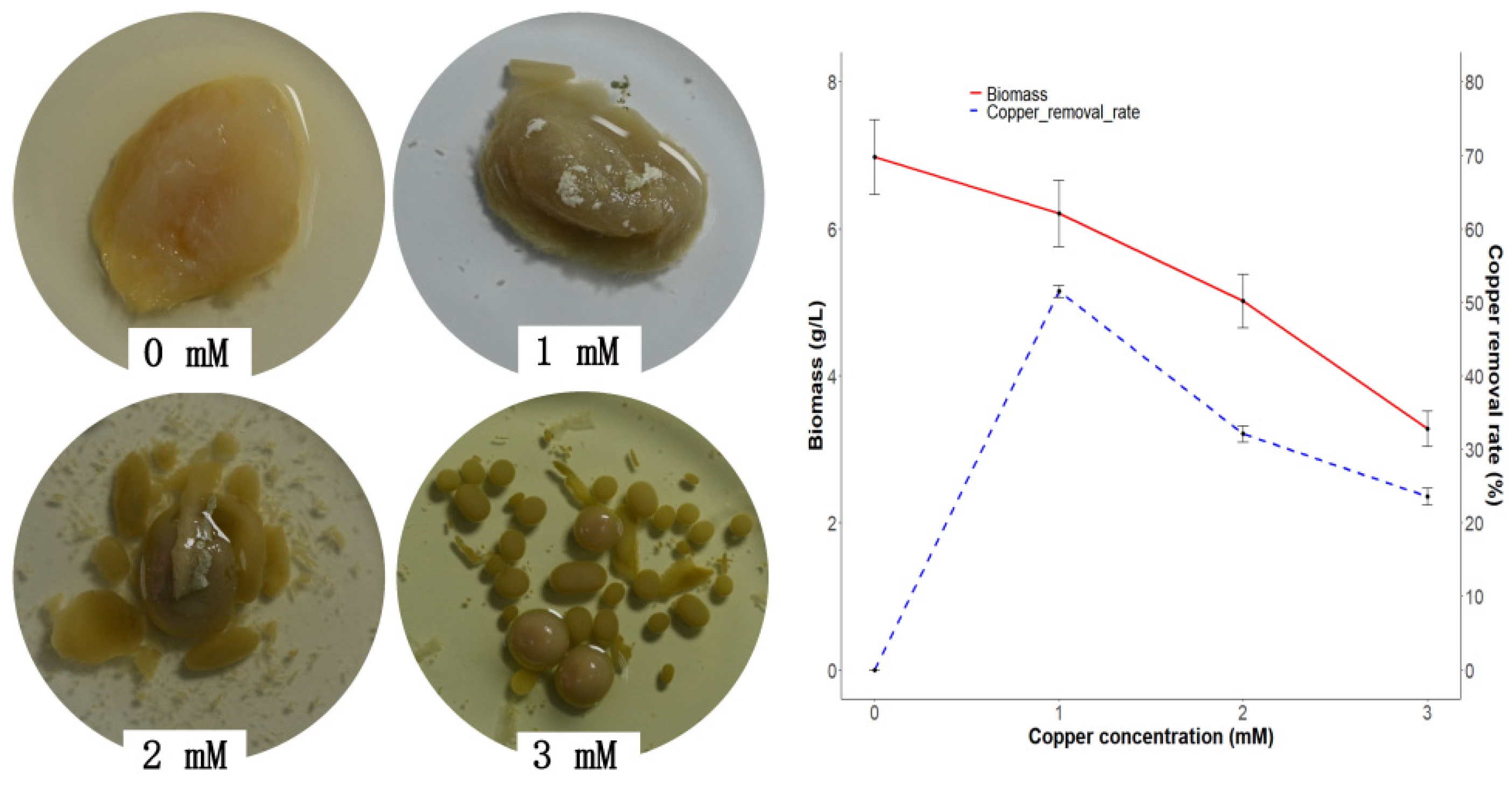
2.5. Copper Bioremediation Properties
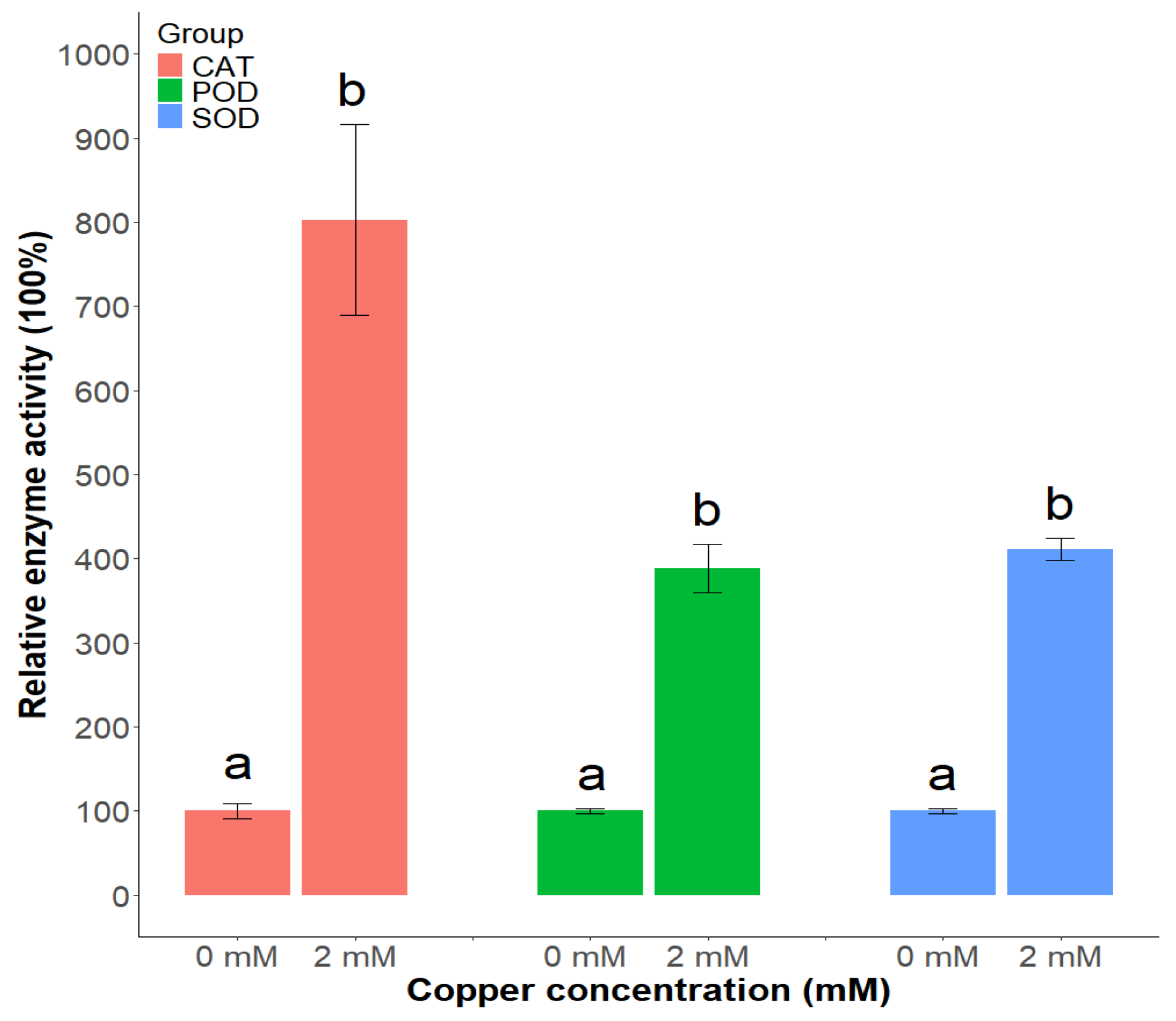
2.6. Measured Enzyme Activities
2.7. Whole-Genome Sequencing
2.8. Comparison and Analysis of the Proteins
2.9. Quantitative RT-PCR
2.10. Statistical Analysis
3. Results
3.1. Strain Isolation and Identification
3.2. Optimization of Growth Conditions of Tk10 to Increase Copper Tolerance
3.3. Copper Bioaccumulation
3.4. Enzyme Activity Analysis of Catalase (CAT), Peroxidase (POD), and Superoxide Dismutase (SOD) Under Copper Stress
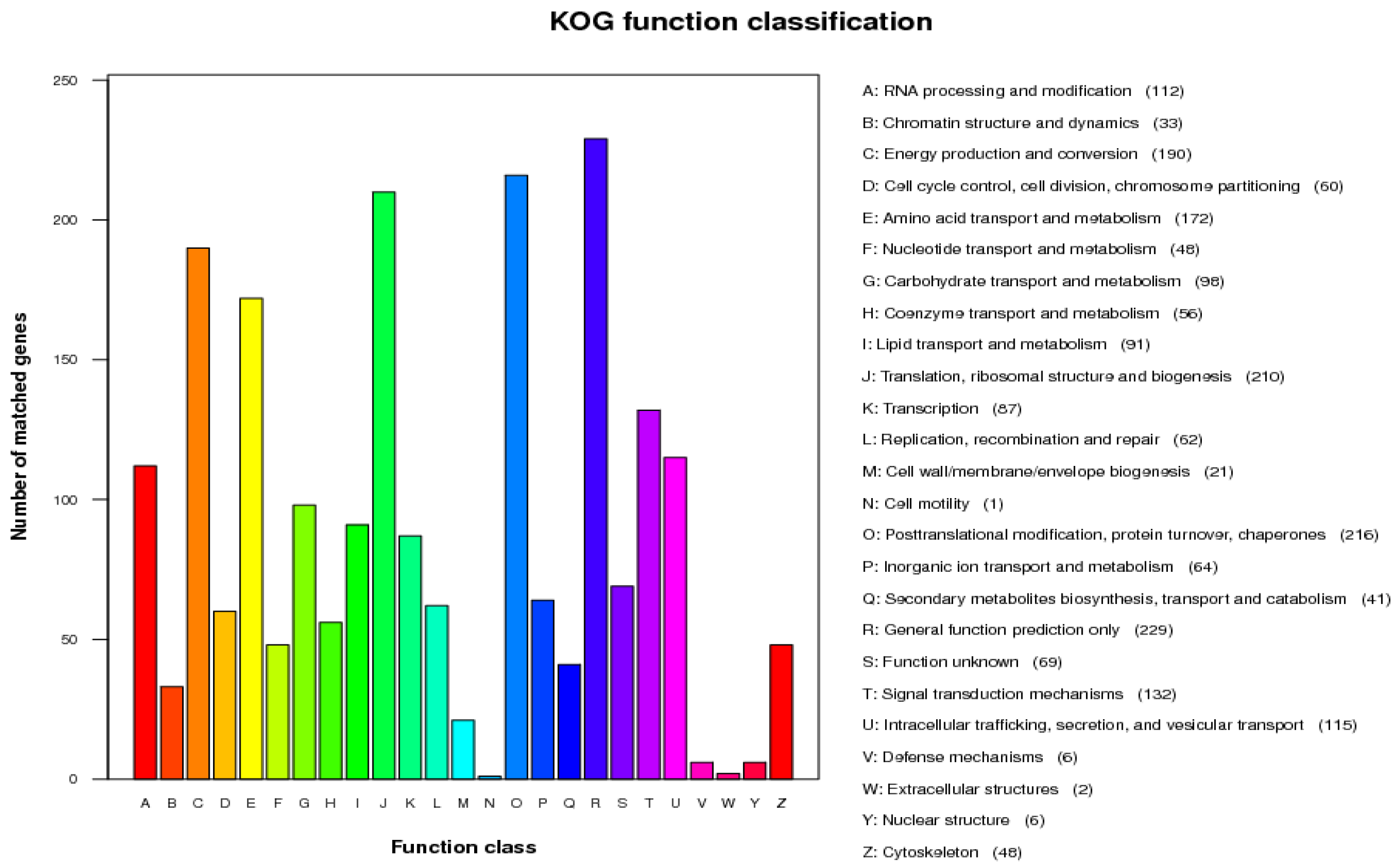
3.5. Genome Sequencing
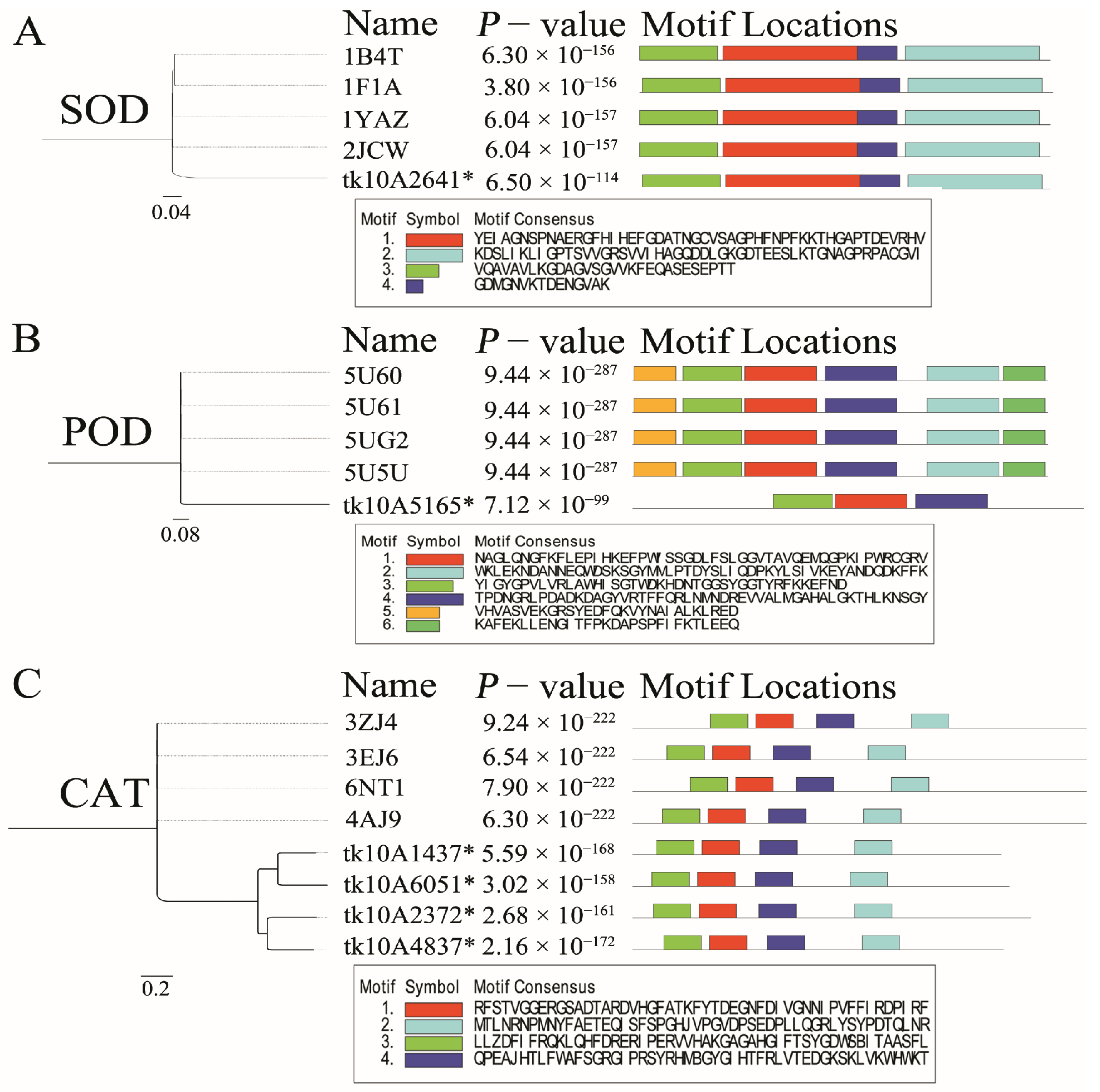
3.6. Copper-Related Genes Prediction
3.7. Protein Structural Analysis
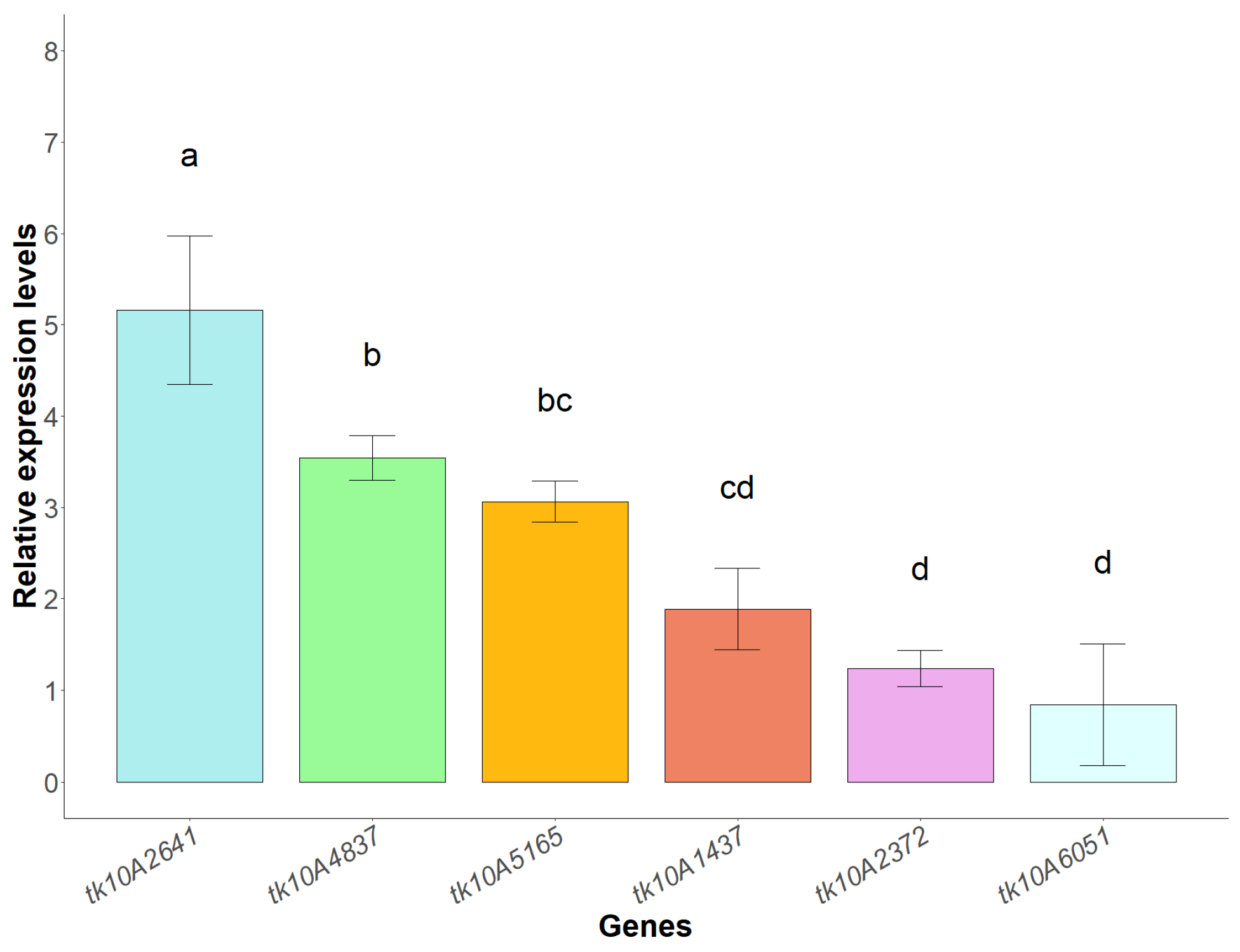
3.8. Expression Profiles of SOD, CAT and POD Genes Under Copper Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ruiz, L.M.; Libedinsky, A.; Elorza, A.A. Role of copper on mitochondrial function and metabolism. Front. Mol. Biosci. 2021, 8, 711227. [Google Scholar] [CrossRef] [PubMed]
- Salah, I.; Parkin, I.P.; Allan, E. Copper as an antimicrobial agent: Recent advances. RSC Adv. 2021, 11, 18179–18186. [Google Scholar] [CrossRef] [PubMed]
- Raffa, N.; Osherov, N.; Keller, N.P. Copper Utilization, Regulation, and Acquisition by Aspergillus fumigatus. Int. J. Mol. Sci. 2019, 20, 1980. [Google Scholar] [CrossRef] [PubMed]
- Dusengemungu, L.; Kasali, G.; Gwanama, C.; Ouma, K.O. Recent Advances in Biosorption of Copper and Cobalt by Filamentous Fungi. Front. Microbiol. 2020, 11, 582016. [Google Scholar] [CrossRef] [PubMed]
- Leong, Y.K.; Chang, J.-S. Bioremediation of heavy metals using microalgae: Recent advances and mechanisms. Bioresour. Technol. 2020, 303, 122886. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.R.; Isikhuemhen, O.S.; Anike, F.N. Fungal-Metal Interactions: A Review of Toxicity and Homeostasis. J. Fungi 2021, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Racić, G.; Vukelić, I.; Kordić, B.; Radić, D.; Lazović, M.; Nešić, L.; Panković, D. Screening of Native Trichoderma Species for Nickel and Copper Bioremediation Potential Determined by FTIR and XRF. Microorganisms 2023, 11, 815. [Google Scholar] [CrossRef] [PubMed]
- Vaksmaa, A.; Guerrero-Cruz, S.; Ghosh, P.; Zeghal, E.; Hernando-Morales, V.; Niemann, H. Role of fungi in bioremediation of emerging pollutants. Front. Mar. Sci. 2023, 10, 1070905. [Google Scholar] [CrossRef]
- Smith, A.D.; Logeman, B.L.; Thiele, D.J. Copper Acquisition and Utilization in Fungi. Annu. Rev. Microbiol. 2017, 71, 597–623. [Google Scholar] [CrossRef] [PubMed]
- Palanivel, T.M.; Pracejus, B.; Novo, L.A.B. Bioremediation of copper using indigenous fungi Aspergillus species isolated from an abandoned copper mine soil. Chemosphere 2023, 314, 137688. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Jiang, Y.; Yang, Y.; Peng, Y.; Li, C. Copper metabolism in Saccharomyces cerevisiae: An update. BioMetals 2020, 34, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef] [PubMed]
- Sood, M.; Kapoor, D.; Kumar, V.; Sheteiwy, M.S.; Ramakrishnan, M.; Landi, M.; Araniti, F.; Sharma, A. Trichoderma: The “Secrets” of a Multitalented Biocontrol Agent. Plants 2020, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Ladi, E.; Shukla, N.; Bohra, Y.; Tiwari, A.K.; Kumar, J. Copper tolerant Trichoderma asperellum increases bio-efficacy of copper against Phytophthora infestans in dual combination. Phytoparasitica 2020, 48, 357–370. [Google Scholar] [CrossRef]
- Babu, A.G.; Shim, J.; Bang, K.-S.; Shea, P.J.; Oh, B.-T. Trichoderma virens PDR-28: A heavy metal-tolerant and plant growth-promoting fungus for remediation and bioenergy crop production on mine tailing soil. J. Environ. Manag. 2014, 132, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Ernesto Juniors, P.-T.; Valeria, C.-L.; Santiago, P.-O.; Mario, R.-M.; Gabriela, S.-J. Tolerance to oxidative stress caused by copper (Cu) in Trichoderma asperellum To. Biocatal. Agric. Biotechnol. 2020, 29, 101783. [Google Scholar] [CrossRef]
- Saed-Moucheshi, A.; Sohrabi, F.; Fasihfar, E.; Baniasadi, F.; Riasat, M.; Mozafari, A.A. Superoxide dismutase (SOD) as a selection criterion for triticale grain yield under drought stress: A comprehensive study on genomics and expression profiling, bioinformatics, heritability, and phenotypic variability. BMC Plant Biol. 2021, 21, 148. [Google Scholar] [CrossRef] [PubMed]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2019, 54, 287–293. [Google Scholar] [CrossRef]
- Coelho, E.; Reis, T.A.; Cotrim, M.; Rizzutto, M.; Corrêa, B. Bioremediation of water contaminated with uranium using Penicillium piscarium. Biotechnol. Prog. 2020, 36, e30322. [Google Scholar] [CrossRef] [PubMed]
| Feature | Value |
|---|---|
| Raw Data (Mb) | 7503 |
| Filtered Reads (%) | 11.67 |
| Clean Data (Mb) | 6628 |
| Genome size (Mb) | 37.2 |
| GC content (%) | 48.75 |
| Predicted genes | 6992 |
| Gene ID | Function | Subject ID | Gene Length (bp) | Intron | Protein (aa) |
|---|---|---|---|---|---|
| tk10A1437 | Catalase | gi|358392193|gb|EHK41597.1 | 2267 | 1 | 737 |
| tk10A6051 | gi|358396285|gb|EHK45666.1 | 2463 | 10 | 493 | |
| tk10A2372 | gi|358395560|gb|EHK44947.1 | 1863 | 5 | 504 | |
| tk10A4837 | gi|358392347|gb|EHK41751.1 | 1864 | 3 | 533 | |
| tk10A5165 | Peroxidase | gi|358395813|gb|EHK45200.1 | 1122 | 1 | 350 |
| tk10A2641 | Superoxide dismutase | gi|358398082|gb|EHK47440.1 | 1010 | 4 | 154 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, K.; Fan, L.; Ji, J.; Qiu, X. Copper Tolerance of Trichoderma koningii Tk10. Microbiol. Res. 2025, 16, 18. https://doi.org/10.3390/microbiolres16010018
Fu K, Fan L, Ji J, Qiu X. Copper Tolerance of Trichoderma koningii Tk10. Microbiology Research. 2025; 16(1):18. https://doi.org/10.3390/microbiolres16010018
Chicago/Turabian StyleFu, Kehe, Lili Fan, Jiaming Ji, and Xiayu Qiu. 2025. "Copper Tolerance of Trichoderma koningii Tk10" Microbiology Research 16, no. 1: 18. https://doi.org/10.3390/microbiolres16010018
APA StyleFu, K., Fan, L., Ji, J., & Qiu, X. (2025). Copper Tolerance of Trichoderma koningii Tk10. Microbiology Research, 16(1), 18. https://doi.org/10.3390/microbiolres16010018





