Bioremediation of Oil-Contaminated Soils Using Biosurfactants Produced by Bacteria of the Genus Nocardiopsis sp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Biosurfactant Production and Extraction
2.2. Characteristics of Biosurfactant Properties
2.3. Experimental Design
2.4. Determination of Petroleum Hydrocarbon Content
2.5. Evaluation of Microorganism Activity
2.6. Estimation of Bacterial Abundance by qPCR
2.7. Statistical Analysis
3. Results and Discussion
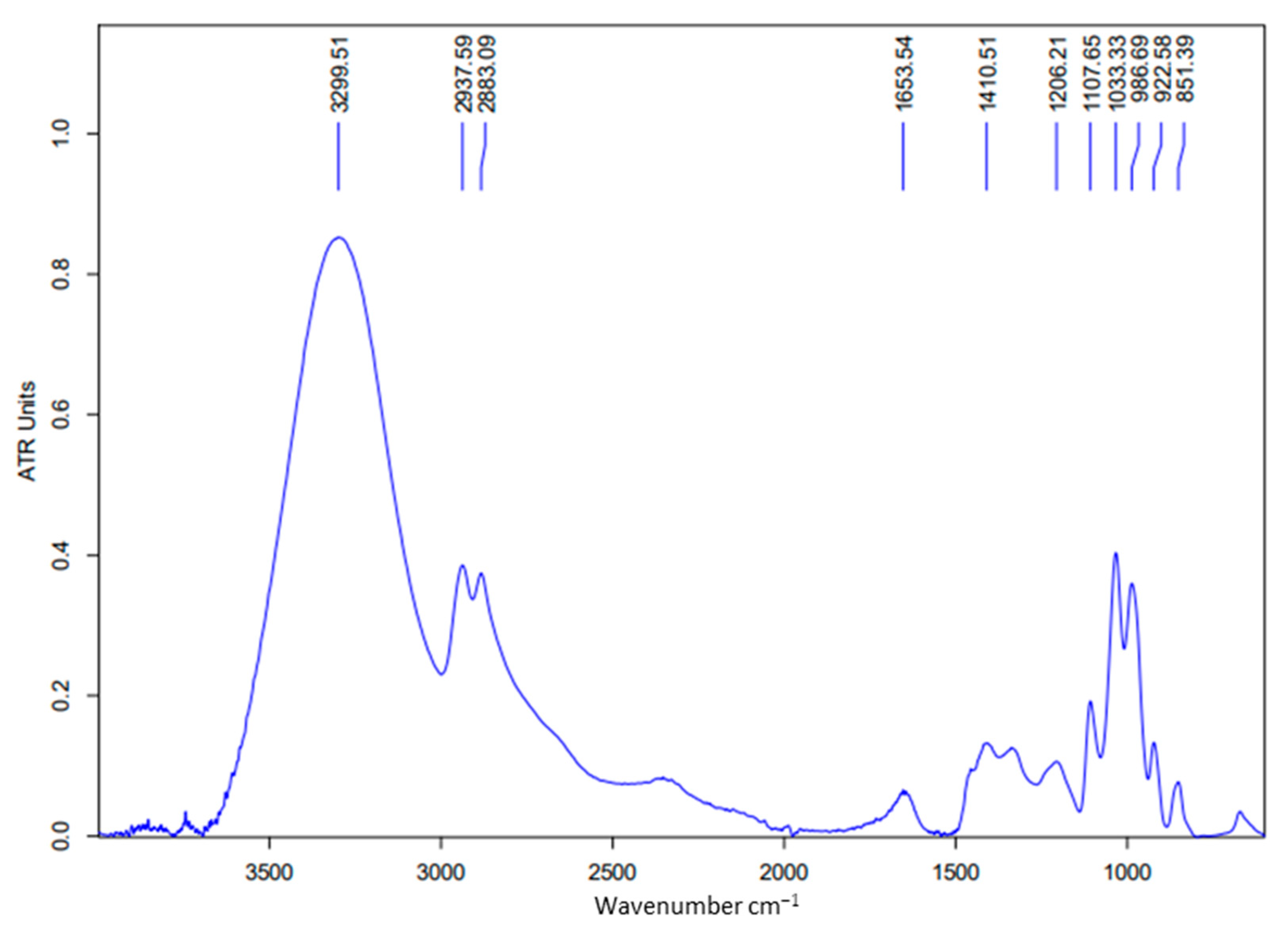
3.1. Evaluation of Biosurfactants Produced by Nocardiopsis sp.
3.2. Laboratory Modeling of the Bioremediation Process
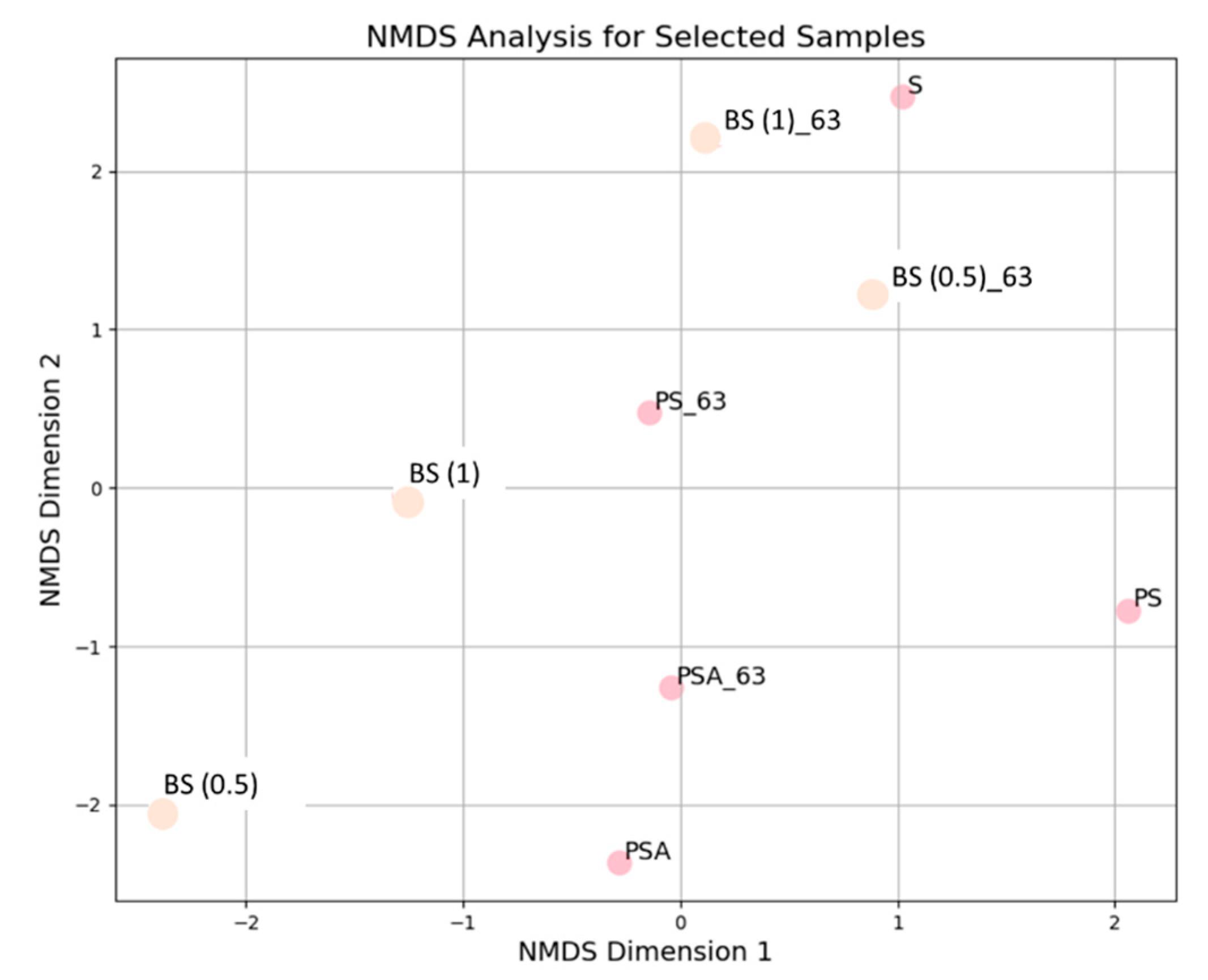
3.3. Evaluation of Metabolic Activity of Microorganisms in the Process of Bioremediation
| Authors | Year | Biosurfactant-Producing Microorganism | Biosurfactant Type | Percentage of Biosurfactant Applied | Object | Percentage of Hydrocarbon Pollution/Type of Pollution | Bioremediation Efficiency |
|---|---|---|---|---|---|---|---|
| Feng et al. [54] | 2021 | sophorolipid | Soil + consortium + 1.5 g kg−1 SL | soil | 0.5 g L−1/diesel oil | 44.5% and 57.7%—isolated consortium and isolated consortium and 1.5 g sophorolipid (SL)kg−1 | |
| Li et al. [44] | 2024 | Raoultella planticola | lipopeptides | 1 g kg−1 (0.1%) (R. planticola together with 1.0 g intracellular lipopeptide) | soil | 7 g kg−1 (0.7%) (dichloromethane to n-hexadecane 20: 1) | 59.0% (R. planticola together with 1.0 g intracellular lipopeptide) |
| Ambust et al. [47] | 2021 | Pseudomonas sp. SA3 | rhamnolipids | 1 L (Pseudomonas sp. SA3 + 300 mL crude biosurfactant) | soil | 1, 3, 5%/crude oil | 10–15% enhancement compared to negative control |
| Das and Kumar [50] | 2018 | Bacillus licheniformis | lipopeptides | B. licheniformis +lipopeptides | soil | 5%/petroleum | reduction in soil toxicity by 40% |
| Bezza and Chirwa [58] | 2015 | B. subtilis | lipopeptides | Bacterial cells (2% v/v) + biosurfactant 0.15% (w/v) | sand | 3% (v/v)/motor oil | 85% |
| Silva et al. [55] | 2018 | Pseudomonas aeruginosa UCP 0992 | rhamnolipids | biosurfactant (0.6 g L−1) and (1.2 g L−1) and 15% of Pseudomonas aeruginosa (107 CFUs/mL) | sand | 10%/motor oil | 90% for 1.2 g L−1 concentration of biosurfactants and Pseudomonas aeruginosa |
| Rahman et al. [52] | 2002 | Pseudomonas sp. DS10-129 | (i) mixed bacterial consortium (MC), (ii) poultry litter (PL), (iii) coir pith (CP), and (iv) rhamnolipid biosurfactant (BS) | 0.1% and 1% | soil | gasoline | 67% and 78% amended with RS + GS + MC + PL + CP + BS at 0.1% and 1% |
| Rahman et al. [53] | 2007 | Pseudomonas aeruginosa | rhamnolipids with bacterial cell | 0.4 mg kg−1 | soil | 10%, 20%/tank bottom sludge | 100% (nC8–nC11), 83–98% (nC12–nC21), 80–85% (nC22–nC31), and 57–73% (nC32–nC40) |
| Patowary et al. [59] | 2018 | Pseudomonas aeruginosa SR17 | rhamnolipids | 1.5 g L−1 | soil | 6800 ppm, 8500 ppm/TPH | 86.1% and 80.5% in two soil samples containing 6800 ppm and 8500 ppm TPH |
| Cubitto et al. [56] | 2004 | Bacillus subtilis O9 | lipopeptides | 1.9, 19.5, 39 mg kg−1 | sandy loam soil | 5%/crude oil | 1.95 and 19.5 mg Bs kg−1 soil—58% and 40% in the RAH concentration |
| Lai et al. [57] | 2008 | rhamnolipids, surfactin | 0.2% | soils | 3000, 9000 mg kg−1/THP | rhamnolipids, surfactin—23% and 14% (3000 mg kg−1), 63% and 62% (9000 mg kg−1) | |
| Szulc et al. [60] | 2014 | rhamnolipids | 150 mg kg−1 | soils | 1%/crude oil | 52 and 53% for non-bioaugmented plots as well as 88 and 89% for bioaugmented plots | |
| Joe et al. [63] | 2019 | Shewanella sp. BS4 | rhamnolipids | 2% | soil | 10%/crude oil | 60% |
| Jorfi et al. [61] | 2014 | Pseudomonas aeruginosa | rhamnolipid | 240 m L−1 | soil | 500 mg kg−1/pyrene | biosurfactant, without biosurfactant, and controls—86.4%, 59.8%, and 14% |
| Bezza et al. [58] | 2017 | Bacillus cereus SPL-4 | lipopeptide | 0.2 and 0.6% (w/w) | soil | 6745.5 mg kg−1/PAHs | 0.2 and 0.6% (w/w)—34.2 and 63% |
| Millioli et al. [62] | 2009 | rhamnolipid | 1–15 mg g−1 | soil | 50 mg g−1/crude oil | 60% | |
| Guadarrama et al. [46] | 2024 | Pseudomonas sp. | rhamnolipids | 0.1% | soil | 28 g kg−1 (2.8%)/crude oil | 42.7% vs. chemical surfactants—32.3% |
| de França et al. [64] | 2015 | B. subtilis ICA56 | lipopeptides | 40 mL crude biosurfactant | sand | 10%/crude oil or motor oil | 76 and 88% for crude oil and motor oil |
| Nikolopoulou et al. [65] | 2013 | rhamnolipids | 0.175 g kg−1 | sand | 0.5%/crude oil | 97% (n-alkane), 95% (PAH) | |
| Wang et al. [71] | 2014 | rhamnolipids | 0.2 g·kg−1 (0.02%) | soil | PAH | 71.5% | |
| Whang et al. [51] | 2008 | Bacillus subtilis ATCC 21332, Pseudomonas aeruginosa J4 | surfactin, rhamnolipid | 0.05 g kg−1 of rhamnolipids and 0.04 g kg−1 surfactin | sandy loam soil | 7 g kg−1 (0.7%)/diesel | 97% by rhamnolipid |
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Michel, J.; Fingas, M. Oil Spills: Causes, Consequences, Prevention, and Countermeasures. In Fossil Fuels: Current Status and Future Directions; World Scientific Publishing Company: Singapore, 2016; pp. 159–201. [Google Scholar] [CrossRef]
- Kamble, N.S.; Bera, S.; Bhedase, S.A.; Gaur, V.; Chowdhury, D. Review on Applied Applications of Microbiome on Human Lives. Bacteria 2024, 3, 141–159. [Google Scholar] [CrossRef]
- Safiyanu, I.; Abdulwahid Isah, A.; Abubakar, U.S.; Rita Singh, M. Review on Comparative Study on Bioremediation for Oil Spills Using Microbes. Res. J. Pharm. Biol. Chem. Sci. 2015, 6, 783–790. [Google Scholar]
- Varjani, S.J. Remediation Processes for Petroleum Oil Polluted Soil. Indian J. Biotechnol. 2017, 16, 157–163. [Google Scholar]
- Sayed, K.; Baloo, L.; Sharma, N.K. Bioremediation of Total Petroleum Hydrocarbons (TPH) by Bioaugmentation and Biostimulation in Water with Floating Oil Spill Containment Booms as Bioreactor Basin. Int. J. Environ. Res. Public Health 2021, 18, 2226. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.P.; Van Der Gast, C.J.; Ciric, L.; Singer, A.C. Bioaugmentation for Bioremediation: The Challenge of Strain Selection. Environ. Microbiol. 2005, 7, 909–915. [Google Scholar] [CrossRef]
- Kumari, R.; Singha, L.P.; Shukla, P. Biotechnological Potential of Microbial Biosurfactants, Their Significance, and Diverse Applications. FEMS Microbes 2023, 4, xtad015. [Google Scholar] [CrossRef]
- Mishra, S.; Lin, Z.; Pang, S.; Zhang, Y.; Bhatt, P.; Chen, S. Biosurfactant Is a Powerful Tool for the Bioremediation of Heavy Metals from Contaminated Soils. J. Hazard. Mater. 2021, 418, 126253. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. A New Look on Factors Affecting Microbial Degradation of Petroleum Hydrocarbon Pollutants. Int. Biodeterior. Biodegrad. 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 11. [Google Scholar] [CrossRef]
- Chakraborty, J.; Das, S. Biosurfactant-Based Bioremediation of Toxic Metals. In Microbial Biodegradation and Bioremediation; Das, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 167–201. [Google Scholar] [CrossRef]
- Payal Beura, P.; Kumar Raul, S. Biosurfactants: New Insights in Bioengineering and Bioremediation of Crude Oil Contamination. In Biosurfactants: A Boon to Healthcare, Agriculture & Environmental Sustainability; Bentham Science Publishers: Sharjah, United Arab Emirates, 2024; pp. 136–158. [Google Scholar] [CrossRef]
- Bjerk, T.R.; Severino, P.; Jain, S.; Marques, C.; Silva, A.M.; Pashirova, T.; Souto, E.B. Biosurfactants: Properties and Applications in Drug Delivery, Biotechnology and Ecotoxicology. Bioengineering 2021, 8, 115. [Google Scholar] [CrossRef]
- Sowbaranika, U.; Ashok Kumar, K.; Jayanthi, M.; Abirami, G.; Suganthi, M.; Wong, L.S.; Durgadevi, R.; Prakash, B.; Suresh, D. Bioremediation of Hydrocarbon-Contaminated Environments: Harnessing the Potential of Biosurfactants—A Review. J. Adv. Zool. 2023, 44, 522–526. [Google Scholar] [CrossRef]
- Ibrahim, A.H.; Desoukey, S.Y.; Fouad, M.A.; Kamel, M.S.; Gulder, T.A.M.; Abdelmohsen, U.R. Natural Product Potential of the Genus Nocardiopsis. Mar. Drugs 2018, 16, 147. [Google Scholar] [CrossRef] [PubMed]
- Poomalai, P.; Krishnan, J.; Ravichandran, A.; Sureshkumar, R. Biosurfactants: Sustainable Alternative to Synthetic Surfactants and Their Applications. Int. J. Appl. Pharm. 2024, 16, 34–43. [Google Scholar] [CrossRef]
- Usman, S. Bioremediation Techniques for the Management of Agricultural Soils Contamination by Oil Spilling. Bulg. J. Soil. Sci. Agrochem. Ecol. 2024, 58, 25–36. [Google Scholar] [CrossRef]
- Ahmad, Z.; Zhang, X.; Imran, M.; Zhong, H.; Andleeb, S.; Zulekha, R.; Liu, G.; Ahmad, I.; Coulon, F. Production, Functional Stability, and Effect of Rhamnolipid Biosurfactant from Klebsiella sp. on Phenanthrene Degradation in Various Medium Systems. Ecotoxicol. Environ. Saf. 2021, 207, 111514. [Google Scholar] [CrossRef]
- Jahan, R.; Bodratti, A.M.; Tsianou, M.; Alexandridis, P. Biosurfactants, Natural Alternatives to Synthetic Surfactants: Physicochemical Properties and Applications. Adv. Colloid Interface Sci. 2020, 275, 102061. [Google Scholar] [CrossRef]
- Mnif, I.; Sahnoun, R.; Ellouze-Chaabouni, S.; Ghribi, D. Evaluation of B. Subtilis SPB1 Biosurfactants’ Potency for Diesel-Contaminated Soil Washing: Optimization of Oil Desorption Using Taguchi Design. Environ. Sci. Pollut. Res. 2014, 21, 851–861. [Google Scholar] [CrossRef]
- Busi, S.; Rajkumari, J. Biosurfactant: A Promising Approach toward the Remediation of Xenobiotics, a Way to Rejuvenate the Marine Ecosystem. In Marine Pollution and Microbial Remediation; Naik, M., Dubey, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 87–104. [Google Scholar] [CrossRef]
- Sharma, N.; Lavania, M.; Lal, B. Biosurfactant: A Next-Generation Tool for Sustainable Remediation of Organic Pollutants. Front. Microbiol. 2022, 12, 821531. [Google Scholar] [CrossRef]
- Khopade, A.; Biao, R.; Liu, X.; Mahadik, K.; Zhang, L.; Kokare, C. Production and Stability Studies of the Biosurfactant Isolated from Marine Nocardiopsis sp. B4. Desalination 2012, 285, 198–204. [Google Scholar] [CrossRef]
- Gandhimathi, R.; Seghal Kiran, G.; Hema, T.A.; Selvin, J.; Rajeetha Raviji, T.; Shanmughapriya, S. Production and Characterization of Lipopeptide Biosurfactant by a Sponge-Associated Marine Actinomycetes Nocardiopsis alba MSA10. Bioprocess Biosyst. Eng. 2009, 32, 825–835. [Google Scholar] [CrossRef]
- El-Gendy, M.M.A.A.; El-Bondkly, A.M.A. Evaluation and Enhancement of Heavy Metals Bioremediation in Aqueous Solutions by Nocardiopsis sp. MORSY1948, and Nocardia sp. MORSY2014. Braz. J. Microbiol. 2016, 47, 571–586. [Google Scholar] [CrossRef] [PubMed]
- Sivaperumal, P.; Kamala, K.; Rajaram, R. Adsorption of Cesium Ion by Marine Actinobacterium Nocardiopsis sp. 13H and Their Extracellular Polymeric Substances (EPS) Role in Bioremediation. Environ. Sci. Pollut. Res. 2018, 25, 4254–4267. [Google Scholar] [CrossRef] [PubMed]
- Behera, S.; Das, S. Potential and Prospects of Actinobacteria in the Bioremediation of Environmental Pollutants: Cellular Mechanisms and Genetic Regulations. Microbiol. Res. 2023, 273, 127399. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Verma, A.; Gangola, S.; Bhandari, G.; Chen, S. Microbial Glycoconjugates in Organic Pollutant Bioremediation: Recent Advances and Applications. Microb. Cell Factories 2021, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Adlin Jenifer, J.S.C.; Michaelbabu, M.; Eswaramoorthy Thirumalaikumar, C.L.; Jeraldin Nisha, S.R.; Uma, G.; Citarasu, T. Antimicrobial Potential of Haloalkaliphilic Nocardiopsis sp. AJ1 Isolated From Solar Salterns In India. J. Basic Microbiol. 2019, 59, 288–301. [Google Scholar] [CrossRef]
- Roy, S.; Hens, D.; Biswas, D.; Biswas, D.; Kumar, R. Survey of Petroleum-Degrading Bacteria in Coastal Waters of Sunderban Biosphere Reserve. World J. Microbiol. Biotechnol. 2002, 18, 575–581. [Google Scholar] [CrossRef]
- Biktasheva, L.; Gordeev, A.; Kirichenko, A.; Kuryntseva, P.; Selivanovskaya, S. Screening of Microorganisms from Wastes and Identification of the Optimal Substrate for Biosurfactant Production. Microbiol. Res. 2024, 15, 152–163. [Google Scholar] [CrossRef]
- Mishra, S.; Jyot, J.; Kuhad, R.C.; Lal, B. In Situ Bioremediation Potential of an Oily Sludge-Degrading Bacterial Consortium. Curr. Microbiol. 2001, 43, 328–335. [Google Scholar] [CrossRef]
- ISO 16072:2002(En); Soil Quality—Laboratory Methods for Determination of Microbial Soil Respiration. International Organization for Standardization: Geneva, Switzerland, 2002.
- García, C.; Hernandez, T.; Costa, F. Potential Use of Dehydrogenase Activity as an Index of Microbial Activity in Degraded Soils. Commun. Soil Sci. Plant Anal. 1997, 28, 123–134. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Papiernik, S.K.; Gan, J.; Yates, S.R.; Yang, C.H.; Crowley, D.E. Impact of Fumigants on Soil Microbial Communities. Appl. Environ. Microbiol. 2001, 67, 3245–3257. [Google Scholar] [CrossRef]
- Khaziev, F.K. Methods of Soil Enzymology, 10th ed.; Nauka: Moscow, Russia, 2005; p. 254. (In Russian) [Google Scholar]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of Complex Microbial Populations by Denaturing Gradient Gel Electrophoresis Analysis of Polymerase Chain Reaction-Amplified Genes Coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Faith, D.P.; Minchin, P.R.; Belbin, L. Compositional Dissimilarity as a Robust Measure of Ecological Distance. Vegetatio 1987, 69, 57–68. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-Parametric Multivariate Analyses of Changes in Community Structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Patel, P.; Patel, R.; Mukherjee, A.; Munshi, N.S. Microbial Biosurfactants for Green Agricultural Technology. In Sustainable Agriculture Reviews 60; Singh, N., Chattopadhyay, A., Lichtfouse, E., Eds.; Springer: Cham, Switzerland, 2023; Volume 60, pp. 389–413. [Google Scholar] [CrossRef]
- Chakraborty, S.; Ghosh, M.; Chakraborti, S.; Jana, S.; Sen, K.K.; Kokare, C.; Zhang, L. Biosurfactant Produced from Actinomycetes Nocardiopsis A17: Characterization and Its Biological Evaluation. Int. J. Biol. Macromol. 2015, 79, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Cao, W.; Jiang, M.; Saren, G.; Liu, J.; Cao, J.; Ali, I.; Yu, X.; Peng, C.; Naz, I. Isolation and Characterization of Biosurfactant-Producing and Diesel Oil Degrading Pseudomonas sp. CQ2 from Changqing Oil Field, China. RSC Adv. 2018, 8, 39710–39720. [Google Scholar] [CrossRef]
- Dardouri, M.; Mendes, R.M.; Frenzel, J.; Costa, J.; Ribeiro, I.A.C. Seeking Faster, Alternative Methods for Glycolipid Biosurfactant Characterization and Purification. Anal. Bioanal. Chem. 2021, 413, 4311–4320. [Google Scholar] [CrossRef]
- Li, S.; Jiang, X.; Zhao, C.; Ren, Y.; Luo, L. Evaluation of Lipopeptides Biosurfactant from Raoultella planticola for Bioremediation in N-Hexadecane-Contaminated Soil. J. Environ. Chem. Eng. 2024, 12, 112622. [Google Scholar] [CrossRef]
- Edgcomb, V.P.; Teske, A.P.; Mara, P. Microbial Hydrocarbon Degradation in Guaymas Basin—Exploring the Roles and Potential Interactions of Fungi and Sulfate-Reducing Bacteria. Front. Microbiol. 2022, 13, 831828. [Google Scholar] [CrossRef]
- Guadarrama, M.A.M.; Mier, M.V.; Callejas, R.L.; Ramos, A.L.; Valencia, M.N.R. Remediation of Soils Contaminated with Heavy Fraction Hydrocarbons Using Biosurfactant Produced by Pseudomonas sp. Int. J. Adv. Res. 2024, 12, 573–582. [Google Scholar] [CrossRef]
- Ambust, S.; Das, A.J.; Kumar, R. Bioremediation of Petroleum Contaminated Soil through Biosurfactant and Pseudomonas sp. SA3 Amended Design Treatments. Curr. Res. Microb. Sci. 2021, 2, 100031. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. Production and Applications of Lipopeptide Biosurfactant for Bioremediation and Oil Recovery by Bacillus subtilis CN2. Biochem. Eng. J. 2015, 101, 168–178. [Google Scholar] [CrossRef]
- Silva, E.J.; Correa, P.F.; Almeida, D.G.; Luna, J.M.; Rufino, R.D.; Sarubbo, L.A. Recovery of Contaminated Marine Environments by Biosurfactant-Enhanced Bioremediation. Colloids Surf. B Biointerfaces 2018, 172, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Das, A.J.; Kumar, R. Bioslurry Phase Remediation of Petroleum-Contaminated Soil Using Potato Peels Powder through Biosurfactant Producing Bacillus Licheniformis J1. Int. J. Environ. Sci. Technol. 2018, 15, 525–532. [Google Scholar] [CrossRef]
- Whang, L.M.; Liu, P.W.G.; Ma, C.C.; Cheng, S.S. Application of Biosurfactants, Rhamnolipid, and Surfactin, for Enhanced Biodegradation of Diesel-Contaminated Water and Soil. J. Hazard. Mater. 2008, 151, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.S.M.; Banat, I.M.; Thahira, J.; Thayumanavan, T.; Lakshmanaperumalsamy, P. Bioremediation of gasoline contaminated soil by a bacterial consortium amended with poultry litter, coir pith and rhamnolipid biosurfactant. Bioresour. Technol. 2002, 81, 25–32. [Google Scholar] [CrossRef]
- Rahman, K.; Rahman, T.; Banat, I.; Lord, R.; Street, G. Bioremediation of Petroleum Sludge using Bacterial Consortium with Biosurfactant. In Environmental Bioremediation Technologies; Singh, S.N., Tripathi, R.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 391–408. [Google Scholar] [CrossRef]
- Feng, L.; Jiang, X.; Huang, Y.; Wen, D.; Fu, T.; Fu, R. Petroleum hydrocarbon-contaminated soil bioremediation assisted by isolated bacterial consortium and sophorolipid. Environ. Pollut. 2021, 273, 116476. [Google Scholar] [CrossRef]
- Silva, M.d.G.C.d.; da Silva, M.E.P.; de Medeiros, A.O.; Meira, H.M.; Sarubbo, L.A. Surface Properties and Biological Activities on Bacteria Cells by Biobased Surfactants for Antifouling Applications. Surfaces 2022, 5, 383–394. [Google Scholar] [CrossRef]
- Cubitto, M.A.; Morán, A.C.; Commendatore, M.; Chiarello, M.N.; Baldini, M.D.; Siñeriz, F. Effects of Bacillus subtilis O9 biosurfactant on the bioremediation of crude oil-polluted soils. Biodegradation 2004, 15, 281–287. [Google Scholar] [CrossRef]
- Lai, C.-C.; Huang, Y.-C.; Wei, Y.-H.; Chang, J.-S. Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J. Hazard. Mater. 2009, 167, 609–614. [Google Scholar] [CrossRef]
- Bezza, F.A.; Chirwa, E.M.N. The role of lipopeptide biosurfactant on microbial remediation of aged polycyclic aromatic hydrocarbons (PAHs)-contaminated soil. Chem. Eng. J. 2017, 309, 563–576. [Google Scholar] [CrossRef]
- Patowary, R.; Patowary, K.; Kalita, M.C.; Deka, S. Application of biosurfactant for enhancement of bioremediation process of crude oil contaminated soil. Int. Biodeterior. Biodegrad. 2018, 129, 50–60. [Google Scholar] [CrossRef]
- Szulc, A.; Ambrożewicz, D.; Sydow, M.; Ławniczak, L.; Piotrowska-Cyplik, A.; Marecik, R.; Chrzanowski, L. The influence of bioaugmentation and biosurfactant addition on bioremediation efficiency of diesel-oil contaminated soil: Feasibility during field studies. J. Environ. Manag. 2014, 132, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Jorfi, S.; Rezaee, A.; Jaafarzadeh, N.A.; Esrafili, A.; Akbari, H. Bioremediation of Pyrene-Contaminated Soils Using Biosurfactant. Jentashapir J. Cell Mol. Biol. 2014, 5, e23228. [Google Scholar] [CrossRef]
- Millioli, V.S.; Servulo, E.-L.C.; Sobral, L.G.S.; De Carvalho, D.D. Bioremediation of Crude Oil-Bearing Soil: Evaluating the Effect of Rhamnolipid Addition to Soil Toxicity and to Crude Oil Biodegradation Efficiency. Glob. NEST J. 2009, 11, 181–188. [Google Scholar]
- Joe, M.M.; Gomathi, R.; Benson, A.; Shalini, D.; Rengasamy, P.; Henry, A.J.; Truu, J.; Truu, M.; Sa, T. Simultaneous Application of Biosurfactant and Bioaugmentation with Rhamnolipid-Producing Shewanella for Enhanced Bioremediation of Oil-Polluted Soil. Appl. Sci. 2019, 9, 3773. [Google Scholar] [CrossRef]
- De Franҫa, Í.W.L.; Lima, A.P.; Lemos, J.A.M.; Lemos, C.G.F.; Melo, V.M.M.; De Sant’ana, H.B.; Gonҫalves, L.R.B. Production of a Biosurfactant by Bacillus subtilis ICA56 Aiming Bioremediation of Impacted Soils. Catal. Today 2015, 255, 10–15. [Google Scholar] [CrossRef]
- Nikolopoulou, M.; Pasadakis, N.; Norf, H.; Kalogerakis, N. Enhanced Ex Situ Bioremediation of Crude Oil Contaminated Beach Sand by Supplementation with Nutrients and Rhamnolipids. Mar. Pollut. Bull. 2013, 77, 37–44. [Google Scholar] [CrossRef]
- Mambu, S.M. Soil Dehydrogenase Activity: A Comparison Between the TTC And INT Method—A review. J. Ilm. SAINS 2014, 14, 87–94. [Google Scholar] [CrossRef]
- Feyzi, H.; Chorom, M.; Bagheri, G. Urease Activity and Microbial Biomass of Carbon in Hydrocarbon Contaminated Soils—A Case Study of Cheshmeh-Khosh Oil Field, Iran. Ecotoxicol. Environ. Saf. 2020, 199, 110664. [Google Scholar] [CrossRef]
- Li, Q.; Huang, Y.; Wen, D.; Fu, R.; Feng, L. Application of Alkyl Polyglycosides for Enhanced Bioremediation of Petroleum Hydrocarbon-Contaminated Soil Using Sphingomonas changbaiensis and Pseudomonas stutzeri. Sci. Total Environ. 2020, 719, 137456. [Google Scholar] [CrossRef]
- Krishnan Venkatesan, S.; Mandava, R.R.D.; Srinivasan, V.R.; Prasad, M.; Kandasamy, R. Strategic Biosurfactant-Advocated Bioremediation Technologies for the Removal of Petroleum Derivatives and Other Hydrophobic Emerging Contaminants. In Industrial Applications of Biosurfactants and Microorganisms; Elsevier: Amsterdam, The Netherlands, 2024; pp. 151–191. ISBN 9780443132889. [Google Scholar]
- Darwesh, O.M.; Mahmoud, M.S.; Barakat, K.M.; Abuellil, A.; Ahmad, M.E. Improving the Bioremediation Technology of Contaminated Wastewater Using Biosurfactants Produced by Novel Bacillus Isolates. Heliyon 2021, 7, e08616. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.P.; Li, J.; Jiang, Y.; Zhang, Z.Y. Enhanced Bioremediation of Field Agricultural Soils Contaminated with PAHs and OCPs. Int. J. Environ. Res. 2014, 8, 1271–1278. [Google Scholar] [CrossRef]
- Bell, T.H.; Camillone, N.; Abram, K.; Bruns, M.A.; Yergeau, E.; St-Arnaud, M. Hydrocarbon Substrate Richness Impacts Microbial Abundance, Microbiome Composition, and Hydrocarbon Loss. Appl. Soil. Ecol. 2021, 165, 104015. [Google Scholar] [CrossRef]
- Smith, R.J.; Jeffries, T.C.; Adetutu, E.M.; Fairweather, P.G.; Mitchell, J.G. Determining the Metabolic Footprints of Hydrocarbon Degradation Using Multivariate Analysis. PLoS ONE 2013, 8, e81910. [Google Scholar] [CrossRef] [PubMed]
- Wijesekara, S.; Seneviratne, M.; Vithanage, M. Role of Biosurfactants on Microbial Degradation of Oil-Contaminated Soils. In Agro-Environmental Sustainability; Singh, J., Seneviratne, G., Eds.; Springer: Cham, Switzerland, 2017; Volume 2, pp. 165–181. [Google Scholar]
- Shahsavari, E.; Aburto-Medina, A.; Taha, M.; Ball, A.S. A Quantitative PCR Approach for Quantification of Functional Genes Involved in the Degradation of Polycyclic Aromatic Hydrocarbons in Contaminated Soils. MethodsX 2016, 9, 205–211. [Google Scholar] [CrossRef]
- Fowler, S.J.; Toth, C.R.A.; Gieg, L.M. Community Structure in Methanogenic Enrichments Provides Insight into Syntrophic Interactions in Hydrocarbon-Impacted Environments. Front. Microbiol. 2016, 7, 562. [Google Scholar] [CrossRef]
- Pal, S.; Roy, A.; Kazy, S.K. Exploring Microbial Diversity and Function in Petroleum Hydrocarbon Associated Environments Through Omics Approaches. In Microbial Diversity in the Genomic Era; Das, S., Dash, H.R., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 171–194. [Google Scholar] [CrossRef]


| Sample | Level of Biodegradation of Hydrocarbons, % | |
|---|---|---|
| Gravimetric Method | IR Spectroscopy | |
| PS | 2.0 ± 0.1 | 65.3 ± 2.0 |
| PSA | 5.0 ± 0.1 | 68.2 ± 2.7 |
| BS(0.5) | 29.4 ± 0.9 | 86.6 ± 3.5 |
| BS(1) | 35.0 ± 1.2 | 83.5 ± 3.3 |
| Sample | AWCD | Shannon Index (H) | ||
|---|---|---|---|---|
| 1 Day | 63 Day | 1 Day | 63 Day | |
| PS | 0.80 | 0.75 | 3.08 | 2.98 |
| PSA | 0.81 | 0.80 | 2.98 | 3.13 |
| BS(0.5) | 0.66 | 0.30 | 2.88 | 2.79 |
| BS(1) | 0.55 | 0.35 | 2.89 | 2.83 |
| S | 0.24 | 2.72 | ||
| Sample | 1 Day | 28 Day | 63 Day |
|---|---|---|---|
| PS | (1.7 ± 0.0) × 106 | (1.5 ± 0.0) × 107 | (1.7 ± 0.0) × 107 |
| PSA | (1.5 ± 0.0) × 106 | (8.1 ± 0.0) × 106 | (2.1 ± 0.0) × 107 |
| BS(0.5) | (8.4 ± 0.0) × 105 | (7.8 ± 0.0) × 106 | (2.8 ± 0.0) × 107 |
| BS(1) | (1.9 ± 0.0) × 106 | (6.6 ± 0.0) × 106 | (3.3 ± 0.0) × 107 |
| S | (1.3 ± 0.0) × 107 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Biktasheva, L.; Gordeev, A.; Usova, A.; Kirichenko, A.; Kuryntseva, P.; Selivanovskaya, S. Bioremediation of Oil-Contaminated Soils Using Biosurfactants Produced by Bacteria of the Genus Nocardiopsis sp. Microbiol. Res. 2024, 15, 2575-2592. https://doi.org/10.3390/microbiolres15040171
Biktasheva L, Gordeev A, Usova A, Kirichenko A, Kuryntseva P, Selivanovskaya S. Bioremediation of Oil-Contaminated Soils Using Biosurfactants Produced by Bacteria of the Genus Nocardiopsis sp. Microbiology Research. 2024; 15(4):2575-2592. https://doi.org/10.3390/microbiolres15040171
Chicago/Turabian StyleBiktasheva, Liliya, Alexander Gordeev, Arina Usova, Anastasia Kirichenko, Polina Kuryntseva, and Svetlana Selivanovskaya. 2024. "Bioremediation of Oil-Contaminated Soils Using Biosurfactants Produced by Bacteria of the Genus Nocardiopsis sp." Microbiology Research 15, no. 4: 2575-2592. https://doi.org/10.3390/microbiolres15040171
APA StyleBiktasheva, L., Gordeev, A., Usova, A., Kirichenko, A., Kuryntseva, P., & Selivanovskaya, S. (2024). Bioremediation of Oil-Contaminated Soils Using Biosurfactants Produced by Bacteria of the Genus Nocardiopsis sp. Microbiology Research, 15(4), 2575-2592. https://doi.org/10.3390/microbiolres15040171






