Abstract
Antibiotics like colistin can save patients infected with carbapenem-resistant Pseudomonas aeruginosa. However, patients can succumb to such infections even if they undergo colistin therapy. This prompted us to investigate the probable antimicrobial resistance mechanisms and virulence determinants involved in colistin- and carbapenem-resistant P. aeruginosa (CCRPA). Of the 448 P. aeruginosa clinical strains, 19 isolates were resistant to both colistin and carbapenem. Carbapenemases and efflux pump encoding genes were assessed by multiplex PCR and qPCR, respectively. blaVIM was detected among six CCRPA isolates and blaIMP in one strain. The expression levels of pmrA and phoP, as well as pmrB genes and their association with colistin resistance, were assessed by qPCR and semi-quantitate PCR, respectively. pmrA and phoP genes were significantly enhanced in three and nine CCRPA isolates, respectively. We also phenotypically evaluated biofilms, pyocyanin, and alginate production among CCRPA strains. Alginate production was observed in 15 isolates, followed by biofilm (n = 8) and pyocyanin (n = 5). Our results highlighted the coexistence of colistin and carbapenem resistance and biofilm formation among clinical isolates of CCRPA. Further studies are required to trace the source and the origin of colistin and carbapenem resistance in this specific environment.
1. Introduction
Pseudomonas aeruginosa causes severe nosocomial infections in cystic fibrosis (CF) and immunocompromised patients [1,2]. The spread of carbapenem-resistant P. aeruginosa (CARPA) and the infections it causes are a burgeoning problem, particularly for hospitalized patients. The World Health Organization (WHO) considers CARPA to be one of the most high-priority and concerning drug-resistant organisms. Patients infected with CARPA can be saved by antibiotics like polymyxin (colistin or polymyxin B). However, patients succumb to such infections even when receiving polymyxin therapy [3]. The spread of colistin- and carbapenem-resistant P. aeruginosa (CCRPA) isolates has become a severe problem associated with high morbidity and mortality rates worldwide. The recurrent usage of polymyxin against CARPA has resulted in the emergence of colistin resistance in P. aeruginosa worldwide [4,5].
Colistin is considered to be a last-line treatment against P. aeruginosa, showing resistance to multiple classes of antimicrobial agents, including polymyxin B, aminoglycosides, carbapenems, cephalosporin, and fluoroquinolones. Colistin is a cationic polypeptide that binds at the anionic lipopolysaccharide (LPS) region and causes cell death by displacing the Mg2+ and Ca2+ ions [6]. Bacteria can encounter these interactions by expressing and adding an enormous amount of phosphoethanolamine and/or 4-amino4-deoxy-l-arabinose to the anionic region of LPSs. In colistin-resistant isolates, the electrostatic interactions between colistin and LPSs might be reduced due to the masking of anionic regions of LPSs with phosphoethanolamine and/or 4-amino4-deoxy-l-arabinose. The activated expression of two-component regulatory systems (pmrAB and phoPQ) mediates colistin resistance by adding an enormous amount of 4-amino-4-deoxy-L-arabinose (L-Ara4N) and/or phosphoethanolamine (PEtn) to the lipid A component of the outer membrane of lipopolysaccharides (LPSs). This interaction blocks the binding of cationic colistin with anionic LPSs and allows bacteria to survive in the presence of colistin [4,5,6]. However, plasmid-mediated colistin resistance has been found worldwide, and in India, it is called MCR (mobile colistin resistance) [7,8,9]. MCR is a phosphoethanolamine transferase that catalyzes the addition of a phosphoethanolamine group to lipid A, leading to a decreased affinity of colistin for the lipopolysaccharide. MCR-1 is located on plasmid pHNSHP45 and can be easily transferable within E. coli or other species. This plasmid is highly prone to transfer MCR-1 to K. pneumonia and P. aeruginosa via transformations [10,11]. This indicates the plasmid’s ability to confer colistin resistance in non-fermentative bacilli. A study by Liu et al. found MCR-1-harboring K. pneumonia isolates in patients admitted to a hospital [8]. From that study onwards, several studies have confirmed MCR-1-harboring Gram-negative isolates in animals, humans, and environments [12,13].
The production of carbapenemases and their occurrence and colistin resistance mechanisms represent a challenge for clinical management. Carbapenemases are β-lactamase enzymes that play an essential role in the hydrolysis of carbapenems, penicillins, and cephalosporins. According to their Ambler classification, carbapenemases are differentiated into classes A, B, and D based on the hydrolytic mechanisms at their active sites. Class A and D carbapenemases have serine amino acids at the active site (serine-dependent); hence, they are called serine carbapenemases [14]. Class B carbapenemases are called metallo-β-lactamases (MBLs) because they have zinc (zinc-dependent). In the active site of MBLs, the zinc-binding motif histidine-X-aspartic acid synchronizes the arrangement of water molecules, which play an important role in hydrolysis. Class A carbapenemases encoding genes are plasmid-mediated or chromosomal. Class A carbapenemases include blaKPC, blaIMI, blaSME, blaGES, and blaNMC families, and they are more frequently identified in Klebsiella pneumonia. MBL encoding genes are plasmid-mediated or chromosomal in some cases. The most common MBL enzymes are blaNDM (New Delhi metallo-β-lactamases), blaIMP (imipenemase), blaVIM (Verona imipenemase), blaSPM (São Paolo metallo-β-lactamase), and blaGIM (German imipenemase) [14,15]. The class D carbapenemases are referred to as OXA-type carbapenemases, and they are subdivided into blaOXA-23, blaOXA-24/40, blaOXA-48, and blaOXA-58 [16]. OXA-type carbapenemases are oxacillinases, which hydrolyze isoxazolyl penicillin oxacillin much more commonly and faster than classical penicillins. Penicillin is the preferred substrate for blaOXA [16,17]. The activated expression of efflux pump systems contributes to multidrug resistance, which expels various antimicrobial agents and chemicals, including metabolic products, organic solvents, dyes, detergents, and biocides [18]. In P. aeruginosa, efflux pump transporters belong to the RND (resistance–nodulation–cell division) family, which consists of three parts: a transporter, linker, and outer membrane pore. The RND-type efflux pumps encoded by mexAB-oprM, mexCD oprJ, mexEF-oprN, and mexXY(oprA) genes contribute to dangerous multi-drug resistance phenotypes. Meropenem was sensitive to the overexpression of MexAB-OprM, MexCD-OprJ, or MexXY-OprM, whereas imipenem was not significantly affected. It was evident from several studies that the overexpression of mexAB plays a vital role in the extrusion of meropenem, which acts as a last-line treatment against multi-drug-resistant P. aeruginosa [19,20,21]. In addition to mexAB, mexCD also shows an important role in effluxing meropenem antibiotics. Hence, targeting the expression of both genes in pan-drug-resistant P. aeruginosa isolates is crucial.
To invade and enhance its pathogenicity against host defense mechanisms, P. aeruginosa uses several virulence mechanisms, including biofilm formation, alginate, and pyocyanin production [22]. The coexistence of multiple virulence factors (biofilm, alginate, and pyocyanin) and colistin and carbapenem resistance mechanisms in the same strain is a cause for concern. Biofilms are organized communities of bacteria enclosed in a matrix of self-secreted extracellular polymeric substances (EPSs) [23]. Compared to planktonic bacteria, the formation of mature biofilms mediates resistance to several antimicrobial agents and host immune defense. P. aeruginosa forms biofilms more effectively on the surface of medical devices. Biofilm formation is clinically important in CF patients. Notably, the biofilm-forming isolates harboring multi-drug resistance mechanisms and the production of exopolysaccharide alginate are causes of concern. Alginate is responsible for the growth and maturity of the biofilm and related protective mechanisms against environmental stresses [23]. Alginate is a polysaccharide composed of polyuronic acid, and in capsule form, alginate is exposed outside of the cell. Studies have reported that polymorphonuclear neutrophils produce hydrogen peroxide, which converts the non-mucoid phenotype to the mucoid phenotype [24]. Alginate production inhibits antibiotic penetration and allows the bacteria to survive persistently in the respiratory tract [25]. The activated production of alginate helps P. aeruginosa to survive successfully in cystic fibrosis patients. Pyocyanin is the only pigment synthesized by P. aeruginosa. The antibiotic property of pyocyanin allows P. aeruginosa to eradicate competing bacteria. Pyocyanin is toxic to cells and plays an important role in inhibiting cellular respiration and lymphocyte proliferation [26].
Based on this information and background, we aimed to address the colistin and carbapenem resistance mechanisms together with the production of various virulence factors, including alginate, biofilms, and pyocyanin production in CCRPA isolates collected from patients hospitalized in a South Indian tertiary care hospital.
2. Materials and Methods
2.1. Pseudomonas aeruginosa Isolates
A total of 448 P. aeruginosa isolates were collected from clinical specimens of patients admitted in different wards and ICUs of a tertiary care hospital in South India during the study period (January 2014 to February 2016). Ethical approval for this study was received from the Institutional Ethics Committee (project no. JIP/IEC/2014/5/325). The samples were collected randomly from different clinical specimens. Antimicrobial susceptibility test (AST) was performed with a battery of antimicrobial agents, such as ceftazidime (30 μg), amikacin (30 μg), gentamycin (10 μg), ciprofloxacin (5 μg), piperacillin/tazobactam (PTZ), imipenem (10 μg), meropenem (10 μg), colistin (10 μg), and polymyxin B (10 μg) using disk diffusion method. AST tests were carried out according to the guidelines of CLSI 2014 [27]. The P. aeruginosa isolates were considered as colistin- and carbapenem- resistant if the MIC of colistin (Sigma) and meropenem (Orchid, Chennai) was ≥4 µg/mL and >32 μg/mL, respectively, using the broth microdilution method. Colistin- and polymyxin-B-resistant patterns were re-confirmed with colistin and polymixin B E-Strips (bioMérieux). Of 448 P. aeruginosa isolates, 19 strains were resistant to all tested antimicrobial agents, including colistin and carbapenem. P. aeruginosa ATCC 27853 was employed as a reference strain.
2.2. Transcriptional Analysis of pmrA, phoP, mexA, and mexC
Total RNA was extracted from the mid-log phase of bacterial culture (optical density at 600 nm of approximately 0.5) of 19 CCRPA and 5 CCSPA (colistin- and carbapenem- sensitive P. aeruginosa) isolates using Roche total RNA extraction kit. Total RNA was quantified using NanoDrop spectrophotometer (Thermo Scientific). First-strand cDNA was synthesized using reverse transcription kit (Roche). According to the manufacturer’s protocol (Roche), reverse transcription reactions were carried out. Quantitative Real-Time PCR (qRT-PCR) was optimized to evaluate the expression levels of pmrA and phoP (two-component regulatory systems) and mexA and mexC (efflux pump) genes using ABI StepOne Real-Time PCR machine. TaqMan Master Mix was used during qRT-PCR experiments. The expression of 30S ribosomal gene, rpsL, was evaluated in parallel to normalize the transcriptional levels of pmrA, phoP, mexA, and mexC. The data obtained were compared with the rpsL gene using the threshold cycle (∆∆CT) method (relative). The obtained values were then normalized against the values of CCSPA isolates. The qRT-PCR experiments were conducted in triplicate. The primers and probes are listed in Table 1.
2.3. Semi-Quantitative PCR of pmrB
We evaluated the expression of pmrB using semi-quantitative PCR. The first-strand cDNA of CCRPA isolates was used as a template for pmrB amplification. PCR cycle parameters for pmrB amplification were as follows: initial denaturation at 95 °C for 10 min, followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 81 °C for 30 s, and extension at 72 °C for 2 min, with a final extension at 72 °C for 10 min. The amplified cDNA of pmrB and rpsL was purified using gel purification kit (Nucleospin extract II, Macherey-Nagel, Dueren, Germany) and confirmed by sequencing (ABI sequencer). The rpsL gene was used as an internal control. The expressions of pmrB and rpsL genes were visualized by 1.5% agarose gel electrophoresis.
2.4. DNA Extraction and 16SrRNA PCR
DNA extraction from 19 CCRPA cultures was performed by using boiling lysis method. The extracted DNA was subjected to PCR as recommended by Igbinosa et al. [28]. The 16SrRNA was amplified and confirmed by sequencing by using Pseudomonas species primers (Table 2).



Table 1.
Primers and probes used for qPCR assays.
Table 1.
Primers and probes used for qPCR assays.
| Gene | Left Primer (5′ to 3′) | Probe | Right Primer (5′ to 3′) | Reference |
|---|---|---|---|---|
| rpsL | CTTCCGGGTGTGCGTTAC | CTGGACAC | CCCTGCTTACGGTCTTTGAC | [29] |
| pmrA | GTCGAGAGCAACGCCATC | CCACCACC | CAACTGGTTGCCGAGCTT | This study |
| phoP | TGCTGGTAGTGGAAGACGAG | CCACCACC | GTTCACCCAGGCGGGTAT | This study |
| mexA | CTGGAGGACGGTAGCCAATA | GCTGGAAG | GACGGAAACCTCGGAGAAT | [29] |
| mexC | AGCCAGCAGGACTTCGATAC | CCTGGAGA | CAGTGACCGAGGCGTAGC | [29] |

Table 2.
List of primers used for conventional and semi-quantitative PCR.
Table 2.
List of primers used for conventional and semi-quantitative PCR.
| Gene | Sequence (5′ → 3″) | Length (bp) | References |
|---|---|---|---|
| rpsL-F | GTGGTGAAGGTCACAACCTG | 135 | [4] |
| rpsL-R | CCTGCTTACGGTCTTTGACA | ||
| pmrB-F | GGATCCATGTCCCGTGCCGCCGTCCC | 1400 | This study |
| pmrB-R | AAGCTTGCCGGCGGCCGCGTCGCGT | ||
| 16SrRNA-F | GACGGGTGAGTAATGCCTA | 618 | [28] |
| 16SrRNA-R | CACTGGTGTTCCTTCCTATA |
2.5. Plasmid Extraction and MCR-1 PCR
All 19 CCRPA isolates were subjected to plasmid extraction as recommended by the manufacturer (NucleoSpin Plasmid, Macherey-Nagel). The extracted plasmid from the study isolates was subjected to MCR-1 PCR as described by Elnahriry et al. [13]. The primer pairs used for the MCR-1 gene amplification are F: 5′CTCATGATGCAGCATACTTC3′ and 5′CGAATGGAGTGTGCGGTG3′.
2.6. Detection of Carbapenemases’ Genes
Multiplex PCR was optimized to detect carbapenemases’ (blaNDM-1, blaVIM, blaIMP, blaKPC, and blaOXA48) genes in the study isolates. The primers used to detect carbapenemases’ encoding genes were selected from our previous study [29]. The parameters of multiplex PCR were initial denaturation for 10 min at 95 °C, followed by 30 cycles of denaturation for 1 min at 94 °C, annealing for 30 s at 59 °C, and extension for 2 min at 72 °C, with a final extension at 72 °C for 10 min [29].
2.7. Virulence Assay
2.7.1. Quantitative Biofilm Assay
Mueller–Hinton Broth (MHB) supplemented with glucose (0.5%) was used for biofilm formation analysis. The overnight cultures were inoculated in MHB and left in a microtiter plate for two days at 37 °C. After two days, the plate was washed rapidly with tap water, dried, and stained with 0.2% crystal violet for 10 min. After staining, the plates were washed, air-dried, and treated with ethanol (95%). After 15 min, the absorbance was measured spectrophotometrically at 570 nm. The experiments were conducted in triplicate. Both positive (P. aeruginosa ATCC27853) and negative (uninoculated sterile broth) controls were run concurrently. An optical density (OD) value > 0.2 was considered to indicate biofilm formation [30,31].
2.7.2. Alginate Production Assay
Carbazole assay was carried out to evaluate alginate production in the study isolates [32,33]. The cultures grown in Luria–Bertani (LB) broth were centrifuged (14,000× g, 30 min), and the supernatant was treated with ethanol (95%). After 15 min of centrifugation, the alginate solution was treated with borate-sulphuric acid and carbazole reagent (0.1%) (Himedia, Mumbai, India). The reaction mixture was heated at 55 °C for 30 min, and absorbance was measured spectrophotometrically at 500 nm. Alginate (sigma) was used as standard, and uninoculated sterile broth was used as negative control.
2.7.3. Pyocyanin Assay
The cultures grown in LB broth were centrifuged (10,000 rpm, 10 min), and the supernatant was filtered (2 μm filter, Millipore). The filtrate treated with chloroform was centrifuged at 10,000 rpm for 10 min. The chloroform phase was collected separately and mixed with hydrochloric acid (0.2 M). The total reaction mixture was centrifuged, and the samples were measured spectrophotometrically at 530 nm. The above experiments were carried out in triplicate [34,35]. Both positive (P. aeruginosa strain blue-green colonies) and negative (uninoculated sterile broth) controls were run concurrently.
2.8. Statistical Analysis
The demographic profiles, carbapenemases’ gene positivity, and virulence factors (biofilms, alginate, and pyocyanin) are expressed as percentages. The differential expression of mexA, mexC, pmrA, and phoP genes among CCRPA and CCSPA strains were analyzed using Student’s t-test with the normal significance level (p ≤ 0.05). IBM SPSS Statistics v.20.0 (IBM Corp., Armonk, NY, USA) was used for statistical analyses.
3. Results
During the study period, most of the strains were collected from wound swabs (30%, n = 136), followed by blood (21%, n = 92), urine (15%, n = 65), pus (10%, n = 46), sputum (6%, n = 27), tracheal aspirate (6%, n = 26), tissue (5%, n = 22), peritoneal fluid (4%, n = 16), CSF (2%, n = 10), and bronchial wash (2%, n = 8) (Figure 1).
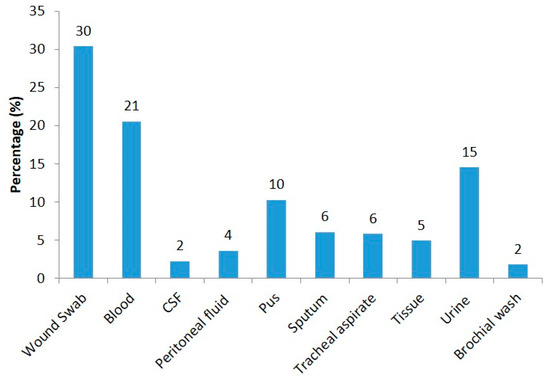
Figure 1.
Distribution of P. aeruginosa isolates collected from different clinical samples.
The actual sites of origin for the wound swabs were a carbuncle, pemphigus vulgaris, dermatitis, non-healing ulcer, wound gap, bullous pemphigoid, and umbilical swab. Those for pus were cervical lymph nodes, cellulitis, a paraspinal abscess, a gluteal abscess, an appendicular perforation, a hydatid cyst, pyoderma, perforation peritonitis, an abscess in the back, and vasculitis. And those for tissue were a diabetic foot, necrotizing fasciitis, Fournier’s gangrene, PVD, cellulitis, an infected stump, and necrotizing pancreatitis. Of the total of 448 P. aeruginosa isolates, 89% of the strains were found to be ceftazidime-resistant, followed by gentamycin (60%), ciprofloxacin (59%), PTZ (56%), amikacin (50%), imipenem (37%), meropenem (35%), polymyxin B (6%), and colistin (4%) (Figure 2).
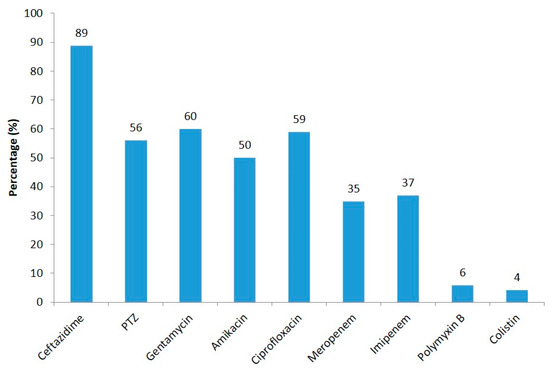
Figure 2.
Resistance pattern of P. aeruginosa isolates against different antibiotics.
About 19 (4%) of the CCRPA strains were found to be resistant to all tested antibiotics, including colistin and carbapenem. Colistin and Polymyxin B resistance was confirmed using E-Strips (Table 3 and Figure 3).

Table 3.
Specimen, resistance, and virulence profiles of colistin-resistant P. aeruginosa isolates collected during the study period from 2014 to 2016.

Figure 3.
E-Strips showing colistin and Polymyxin B resistance in P. aeruginosa isolates.
All 19 CCRPA isolates were confirmed as P. aeruginosa by 16SrRNA gene sequencing (Table 2). Most of the CCRPA strains were collected from wound swabs (n = 15), followed by blood (n = 2), pus (n = 1), and urine (n = 1). In this study, ten CCRPA strains were collected from male patients and nine from female patients (Table 3). Overall, the expression levels of the pmrA and phoP genes in the CCRPA isolates were significantly enhanced in three and nine CCRPA isolates, respectively, compared to the CCSPA strains (Figure 4). Figure 5 depicts that the transcript level of the pmrB gene in a CCRPA isolate (MIC, 8 µg/mL) was enhanced promisingly as compared to colistin-sensitive strains in a semi-quantitative PCR analysis. The rpsL gene was used as the control gene. In this study, we observed that six strains were found to be have an enhanced expression of the pmrB gene. The 16SrRNA sequences obtained from these six strains were subjected to a phylogenetic tree analysis by the maximum-likelihood algorithm MEGA-X (Figure 6). The phylogenetic tree demonstrated two main branches from the root. One branch includes PA456, PA330, PA399, and PA282 isolates, and the other consists of two strains—PA306 and PA461.
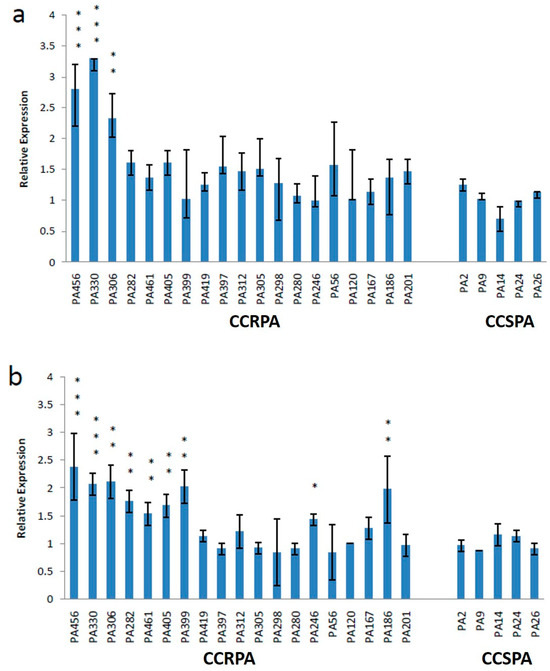
Figure 4.
Expression levels of pmrA (a) and phoP (b) genes in 19 CCRPA and 5 CCSPA isolates. Mean values of pmrA and phoP expression levels of CCSPA isolates were 1.02 and 1.01, respectively. The error bar represents the standard deviation of three repeats. Asterisks indicate significant difference (compared with the control: *** p = 0.001; ** p = 0.01; and * p = 0.05).
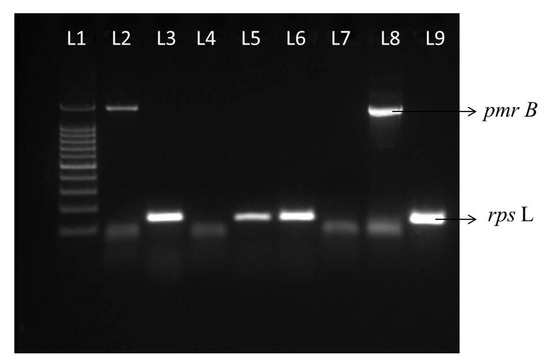
Figure 5.
Transcript levels of pmrB and rpsl genes. Lane 1, 100 bp ladder. L2 and L3 show the transcript levels of pmrB and rpsl genes from a CSPA (colistin-susceptible P. aeruginosa) isolate. L4, L7, L5, and L6 show the absence of pmrB and presence of rpsl expression in two CSPA strains, respectively. L8 and L9 show transcript levels of pmrB and rpsl genes from a CCRPA (PA456), respectively.
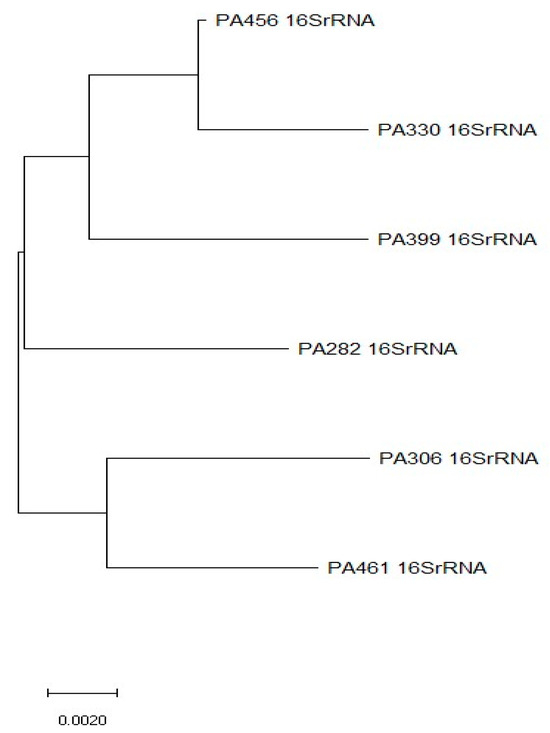
Figure 6.
Rooted neighbor-joining (N-J) phylogenetic tree was constructed from 16SrRNA gene sequences in MEGA X program. This N-J tree shows the distribution and phylogenetic relationships between 6 CCRPA strains with enhanced pmrB expressions.
In addition, a PCR assay was carried out to detect the presence of MCR-1-mediated colistin resistance in CCRPA isolates (Table 2). No MCR-1 gene was detected in this study. In the multiplex PCR test, six CCRPA isolates were found to have blaVIM, and one isolate possessed blaIMP (Table 3). None of the study isolates showed an activated expression of mexA and mexC genes. Of the 19 CCRPA isolates, alginate, biofilm, and pyocyanin were produced in fifteen, eight, and five strains, respectively. Both biofilm formation and alginate production were observed in six CCRPA strains (Table 3). A CCRPA isolate (PA330, Colistin MIC (8 µg/mL)) with blaIMP was found to be have an enhanced expression of both pmrA and phoP genes together with the production of alginate and biofilms (Table 3).
4. Discussion
The spread of carbapenem-resistant Pseudomonas aeruginosa causes severe infection in hospitalized patients. Antibiotics like colistin can save these patients. However, the emergence of colistin resistance among such infections has become an alarming threat. In the present study, we have characterized two-component regulatory systems (pmrAB and phoP) and their association with colistin resistance; carbapenemases’ and RND efflux pumps’ (mexA and mexC) activities in mediating carbapenem resistance; and the co-occurrence of virulence factors (biofilms, alginate, and pyocyanin) among CCRPA clinical strains isolated from a tertiary care hospital in South India. In this study, we observed that the expression levels of pmrA and phoP genes were significantly enhanced in three and nine CCRPA isolates, respectively, compared to colistin-sensitive strains. Similar to our study, Lee et al. highlighted that the enhanced expression of pmrA and phoP may affect colistin resistance in P. aeruginosa [4]. They observed that seven and eight colistin-resistant P. aeruginosa isolates displayed an increased expression of pmrA and phoP, respectively. Our study also highlighted that phoP was enhanced in more isolates (n = 9) than pmrA (n = 3). Similar to our research, Ramedani et al. reported that phoP expression was upregulated in 15 colistin non-susceptible isolates compared to pmrA [36]. In addition, our semi-quantitative PCR testing highlighted that the expression level of the pmrB gene in colistin-resistant P. aeruginosa (MIC, 8 µg/mL) was enhanced promisingly compared to colistin-sensitive strains (Figure 5).
In our study, we observed that the association of pmrA and phoP among the 10 CCRPA strains was not related to colistin’s MICs, suggesting the involvement of additional mechanisms contributing to colistin resistance in P. aeruginosa. Studies have reported that other two-component regulatory systems, like ParPS and CprRS, mediate polymyxin resistance in P. aeruginosa [4]. However, it has been suggested that the enhanced expression of pmrA and phoP plays a significant role in mediating colistin resistance compared to ParPS and CprRS systems [37]. Although several studies have reported colistin-resistant P. aeruginosa worldwide, it is pertinent to point out that similar studies and data are lacking from this country. Very few studies from this geographical area have reported colistin resistance and its mechanisms in Gram-negative pathogens [38,39,40]. An investigation from India identified MCR-1 harboring E. coli for the first time [9]. Worryingly, MCR-1 has been found in CTX-M and carbapenemases producing Enterobacteriaceae [41]. Distribution studies of MCR-1 have reported that MCR-1 possessing plasmids is present in different incompatibility groups such as IncI2, X4, HI2, P, and F [42]. Animals are a reservoir of MCR-1, which is evidenced by the insertion sequence (ISApl1) originating from Pasteurella multocida [42,43]. Our present study did not observe the MCR-1 gene among the study CCRPA isolates.
The co-occurrence of colistin resistance in carbapenem-resistant isolates and its ability to produce multiple virulence factors cause severe life-threatening infections in hospitalized patients. Hence, efforts were made to analyze carbapenemases, RND efflux pump expression, and virulence factor production among clinical P. aeruginosa isolates found to be resistant to all tested antimicrobial agents, including colistin and carbapenem. Here, we have also highlighted the coexistence of metallo-β-lactamases (MBLs) among clinical P. aeruginosa strains that are resistant to colistin. Six CCRPA strains were harbored in blaVIM, and one isolate in blaIMP. A study from North India revealed the coexistence of blaNDM among colistin-resistant P. aeruginosa harboring the MCR-1 gene [9]. A study from Turkey by Vatansever et al. highlighted the coexistence of OXA-48 and NDM-1 in CCRPA isolates and suggested the importance of the continuous surveillance of high-risk clones of colistin-resistant P. aeruginosa strains harboring carbapenemases [44]. Additionally, qPCR assays from our study revealed that the expression levels of the mexA and mexC genes (efflux pumps) remained steady among the CCRPA strains.
P. aeruginosa biofilms cause chronic lung infections in CF patients, and treating such biofilm-associated infections remains challenging, especially for the infections induced by MDR bacteria. Studies have shown that biofilm production may enhance colistin resistance through modifications in LPSs via the activation of TCSs like PmrAB and PhoPQ [45]. A recent study by Ramedani et al. revealed that all 18 colistin-resistant P. aeruginosa strains were biofilm producers [36]. They also observed alginates encoding gene- algD expression in all biofilm-producing colistin-resistant isolates. Another recent study from Bangladesh by Kabir et al. reported that biofilm-forming P. aeruginosa strains were significantly resistant to colistin [46]. In our study, we observed that among the 19 CCRPA strains, eight strains were found to be forming biofilms. Additionally, we observed that fifteen and five CCRPA isolates produce alginate and pyocyanin, respectively. Worryingly, we found that six CCRPA strains were found to be producing both biofilms and alginate. The ability of colistin-resistant P. aeruginosa to produce biofilms and alginate poses a significant challenge when choosing a treatment against such infections. A study from Azimi et al. reported that 52% of colistin-sensitive P. aeruginosa isolates were found to be potent producers of biofilm; however, none of the colistin-resistant P. aeruginosa strains were strong producers [47]. Heidari et al. observed that 95% of Carbapenem-resistant P. aeruginosa isolates were found to be producing biofilms, of which 23.3% were strong biofilm producers [48]. A study from Iran by Davarzani et al. reported a significant relationship between the amount of alginate production and biofilm formation levels [49]. Hence, our study may help clinicians look into the further dissemination of such isolates in this geographical area. Moreover, the current study warrants the inclusion of more colistin-resistant P. aeruginosa strains and the characterization of other two-component regulatory systems that might play roles in colistin resistance in P. aeruginosa.
5. Conclusions
Our results shed light on the spread of the coexistence of MBLs among clinical isolates of colistin-resistant P. aeruginosa together with biofilm and alginate production in a South Indian population. We strongly recommend that further attention is required to screen the high-risk clones of colistin- and carbapenem-resistant P. aeruginosa strains, as well as their probable dissemination and resistance mechanisms, in this geographical area. Future studies highlighting the mutations in PmrAB and PhoPQ genes will advance our understanding of colistin resistance mechanisms in P. aeruginosa and improve our clinical understanding of and the rational use of polymyxin.
Author Contributions
Conceptualization, E.K. and H.B.N.; methodology, E.K.; software, E.K., A.A. and H.B.N.; validation, E.K., A.A. and H.B.N.; formal analysis, E.K.; investigation, E.K.; resources, E.K.; data curation, E.K.; writing—original draft preparation, E.K. and A.A.; writing—review and editing, E.K., A.A., H.B.N. and R.S.; visualization, E.K., A.A., H.B.N. and R.S.; supervision, E.K. and H.B.N.; project administration, E.K.; funding acquisition, E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported financially by Jawaharlal Institute of Post Graduate Medical Education and Research (JIPMER), Puducherry, India.
Institutional Review Board Statement
Ethical approval for this study was received from the Institutional Ethics Committee (project no. JIP/IEC/2014/5/325).
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to express our sincere thanks to Glenmark Pharmaceuticals Ltd., India, for providing the colistin and polymyxin B E-strips for this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Gellatly, S.L.; Hancock, R.E.W. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Fernández, L.; Álvarez-Ortega, C.; Wiegand, I.; Olivares, J.; Kocíncová, D.; Lam, J.S.; Martínez, J.L.; Hancock, R.E. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2013, 57, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Howard-Anderson, J.; Davis, M.; Page, A.M.; Bower, C.W.; Smith, G.; Jacob, J.T.; Andersson, D.I.; Weiss, D.S.; Satola, S.W. Prevalence of colistin heteroresistance in carbapenem-resistant Pseudomonas aeruginosa and association with clinical outcomes in patients: An observational study. J. Antimicrob. Chemother. 2022, 77, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Chung, E.S.; Na, I.Y.; Kim, H.; Shin, D.; Ko, K.S. Development of colistin resistance in pmrA-, phoP-, parR- and cprR- inactivated mutants of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014, 69, 2966–2971. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Ko, K.S. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 2014, 78, 271–276. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.D.; Brinkman, F.S.; Hancock, R.E.W. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory system to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995–4006. [Google Scholar] [CrossRef]
- Stojanoski, V.; Sankaran, B.; Prasad, B.V.; Poirel, L.; Nordmann, P.; Palzkill, T. Structure of the catalytic domain of the colistin resistance enzyme MCR-1. BMC Biol. 2016, 14, 81. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Wang, Y.; Walsh, T.R.; Yi, L.X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis. 2016, 16, 161–168. [Google Scholar] [CrossRef]
- Kumar, M.; Saha, S.; Subudhi, E. More Furious than Ever: E. coli acquired Co-Resistance towards Colistin and Carbapenems. Clin. Infect. Dis. 2016, 63, 1267–1268. [Google Scholar]
- Karaiskos, I.; Souli, M.; Galani, I.; Giamarellou, H. Colistin: Still a lifesaver for the 21st century? Expert Opin. Drug Metab. Toxicol. 2017, 13, 59–71. [Google Scholar] [CrossRef]
- Stoesser, N.; Mathers, A.J.; Moore, C.E.; Day, N.P.; Crook, D.W. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 2016, 16, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Amira, E.; Gamal, E.S. Screening of mcr-1 among Gram-Negative Bacteria from Different Clinical Samples from ICU Patients in Alexandria, Egypt: One-Year Study. Pol. J. Microbiol. 2022, 71, 83–90. [Google Scholar]
- Elnahriry, S.S.; Khalifa, H.O.; Soliman, A.M.; Ahmed, A.M.; Hussein, A.M.; Shimamoto, T.; Shimamoto, T. Emergence of plasmid-mediated colistin resistance gene mcr-1 in a clinical Escherichia coli isolate from Egypt. Antimicrob. Agents Chemother. 2016, 60, 3249. [Google Scholar] [CrossRef] [PubMed]
- Queenan, A.M.; Bush, K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007, 20, 440–458. [Google Scholar] [CrossRef] [PubMed]
- Sacha, P.; Wieczorek, P.; Hauschild, T.; Zórawski, M.; Olszańska, D.; Tryniszewska, E. Metallo-beta-lactamases of Pseudomonas aeruginosa-a novel mechanism resistance to beta-lactam antibiotics. Folia Histochem. Cytobiol. 2008, 46, 137–142. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Høiby, N. OXA-type carbapenemases. J. Antimicrob. Chemother. 2006, 57, 373–383. [Google Scholar] [CrossRef]
- Turton, J.F.; Woodford, N.; Glover, J.; Yarde, S.; Kaufmann, M.E.; Pitt, T.L. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J. Clin. Microbiol. 2006, 44, 2974–2976. [Google Scholar] [CrossRef]
- Fernández, L.; Hancock, R.E. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2013, 26, 163. [Google Scholar] [CrossRef][Green Version]
- Poole, K. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 2004, 10, 12–26. [Google Scholar] [CrossRef]
- Köhler, T.; Michea-Hamzehpour, M.; Plesiat, P.; Kahr, A.L.; Pechere, J.C. Differential selection of multidrug efflux systems by quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1997, 41, 2540–2543. [Google Scholar] [CrossRef]
- Köhler, T.; Michéa-Hamzehpour, M.; Henze, U.; Gotoh, N.; Kocjancic Curty, L.; Pechère, J.C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 1997, 23, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Huang, X.; Wang, Q.; Yao, D.; Lu, W. Virulence factors of Pseudomonas aeruginosa and antivirulence strategies to combat its drug resistance. Front. Cell. Infect. Microbiol. 2022, 12, 926758. [Google Scholar] [CrossRef] [PubMed]
- Shree, P.; Singh, C.K.; Sodhi, K.K.; Surya, J.N.; Singh, D.K. Biofilms: Understanding the structure and contribution towards bacterial resistance in antibiotics. Med. Microecol. 2023, 30, 100084. [Google Scholar] [CrossRef]
- Tan, Q.; Qing, A.; Qi, X.; Fang, L.; Jialin, Y. Polymorphonuclear leukocytes or hydrogen peroxide enhance biofilm development of mucoid Pseudomonas aeruginosa. Mediat. Inflam. 2018, 2018, 8151362. [Google Scholar] [CrossRef]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef]
- Mudaliar, S.B.; Bharath Prasad, A.S. A biomedical perspective of pyocyanin from Pseudomonas aeruginosa: Its applications and challenges. World J. Microbiol. Biotechnol. 2024, 40, 90. [Google Scholar] [CrossRef]
- Wayne, P.A.; Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 24th Informational Supplement; CLSI Document M100-S24; Clinical and Laboratory Standards Institute (CLSI): Berwyn, PA, USA, 2014. [Google Scholar]
- Igbinosa, I.H.; Nwodo, U.U.; Sosa, A.; Tom, M.; Okoh, A.I. Commensal Pseudomonas species isolated from wastewater and freshwater milieus in the Eastern Cape Province, South Africa as reservoir of antibiotic resistant determinants. Int. J. Environ. Res. Public Health 2012, 9, 2537–2549. [Google Scholar] [CrossRef] [PubMed]
- Ellappan, K.; Narasimha, H.B.; Kumar, S. Coexistence of multidrug resistance mechanisms and virulence genes in carbapenem-resistant Pseudomonas aeruginosa strains from a tertiary care hospital in South India. J. Glob. Antimicrob. Resist. 2018, 12, 37–43. [Google Scholar] [CrossRef]
- Ghadaksaz, A.; Fooladi, A.A.I.; Hosseini, H.M.; Amin, M. The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J. Appl. Biomed. 2015, 13, 61–68. [Google Scholar] [CrossRef]
- Kalaiarasan, E.; Thirumalaswamy, K.; Harish, B.N.; Gnanasambandam, V.; Sali, V.K.; John, J. Inhibition of quorum sensing-controlled biofilm formation in Pseudomonas aeruginosa by quorum-sensing inhibitors. Microb. Pathog. 2017, 111, 99–107. [Google Scholar] [CrossRef]
- Mathee, K.; Ciofu, O.; Sternberg, C.; Lindum, P.W.; Campbell, J.I.; Jensen, P.; Johnsen, A.H.; Givskov, M.; Ohman, D.E.; Søren, M.; et al. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: A mechanism for virulence activation in the cystic fibrosis lung. Microbiology 1999, 145, 1349–1357. [Google Scholar] [CrossRef]
- May, T.B.; Chakrabarty, A.M. Isolation and assay of Pseudomonas aeruginosa alginate. Methods Enzymol. 1994, 235, 295–304. [Google Scholar] [PubMed]
- Wagner, T.; Soong, G.; Sokol, S.; Saiman, L.; Prince, A. Effects of azithromycin on clinical isolates of Pseudomonas aeruginosa from cystic fibrosis patients. Chest 2005, 128, 912–919. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Carlsson, M.; Shukla, S.; Petersson, A.C.; Segelmark, M.; Hellmark, T. Pseudomonas aeruginosa in cystic fibrosis: Pyocyanin negative strains are associated with BPI-ANCA and progressive lung disease. J. Cyst. Fibros. 2011, 10, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Jafari-Ramedani, S.; Nazari, M.; Arzanlou, M.; Peeri-Dogaheh, H.; Sahebkar, A.; Khademi, F. Prevalence and molecular characterization of colistin resistance in Pseudomonas aeruginosa isolates: Insights from a study in Ardabil hospitals. BMC Microbiol. 2024, 24, 152. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.S.; Lee, J.Y.; Rhee, J.Y.; Ko, K.S. Colistin resistance in Pseudomonas aeruginosa that is not linked to arnB. J. Med. Microbiol. 2017, 66, 833–841. [Google Scholar] [CrossRef]
- Mohit Kumar, M.K. Colistin and Tigecycline Resistance in Carbapenem-Resistant Enterobacteriaceae: Checkmate to Our Last Line Of Defense. Infect. Control Hosp. Epidemiol. 2016, 37, 624–625. [Google Scholar] [CrossRef][Green Version]
- Kumar, M.; Gupta, A.; Sahoo, R.K.; Jena, J.; Debata, N.K.; Subudhi, E. Functional Genome Screening to Elucidate the Colistin Resistance Mechanism. Sci. Rep. 2016, 6, 23156. [Google Scholar] [CrossRef][Green Version]
- Ghafur, A.; Lakshmi, V.; Kannain, P.; Murali, A.; Ma, T. Emergence of Pan-drug resistance amongst gram negative bacteria! The First case series from India. J. Microbiol. Infect. Dis. 2014, 4, 86–91. [Google Scholar] [CrossRef]
- McGann, P.; Snesrud, E.; Maybank, R.; Corey, B.; Ong, A.C.; Clifford, R.; Hinkle, M.; Whitman, T.; Lesho, E.; Schaecher, K.E. Escherichia coli Harboring mcr-1 and blaCTX-M on a Novel IncF Plasmid: First report of mcr-1 in the USA. Antimicrob. Agents Chemother. 2016, 60, 4420–4421. [Google Scholar] [CrossRef]
- Zelendova, M.; Papagiannitsis, C.C.; Valcek, A.; Medvecky, M.; Bitar, I.; Hrabak, J.; Gelbicova, T.; Barakova, A.; Kutilova, I.; Karpiskova, R.; et al. Characterization of the complete nucleotide sequences of mcr-1-encoding plasmids from Enterobacterales isolates in retailed raw meat products from the Czech Republic. Front. Microbiol. 2021, 11, 604067. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Plasmid-mediated colistin resistance: An additional antibiotic resistance menace. Clin. Microbiol. Infect. 2016, 22, 398–400. [Google Scholar] [CrossRef]
- Vatansever, C.; Menekse, S.; Dogan, O.; Gucer, L.S.; Ozer, B.; Ergonul, O.; Can, F. Co-existence of OXA-48 and NDM-1 in colistin resistant Pseudomonas aeruginosa ST235. Emerg. Microbes Infect. 2020, 9, 152–154. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008, 4, e1000213. [Google Scholar] [CrossRef] [PubMed]
- Kabir, R.B.; Ahsan, T.; Rahman, M.F.; Jobayer, M.; Shamsuzzaman, S.M. Biofilm-Producing and Specific Antibiotic Resistance Genes in Pseudomonas aeruginosa Isolated from Patients Admitted to a Tertiary Care Hospital, Bangladesh. Bangladesh J. Microbiol. 2023, 40, 60–65. [Google Scholar] [CrossRef]
- Azimi, L.; Lari, A.R. Colistin-resistant Pseudomonas aeruginosa clinical strains with defective biofilm formation. GMS Hyg. Infect. Control 2019, 14, Doc12. [Google Scholar]
- Heidari, R.; Farajzadeh Sheikh, A.; Hashemzadeh, M.; Farshadzadeh, Z.; Salmanzadeh, S.; Saki, M. Antibiotic resistance, biofilm production ability and genetic diversity of carbapenem-resistant Pseudomonas aeruginosa strains isolated from nosocomial infections in southwestern Iran. Mol. Biol. Rep. 2022, 49, 3811–3822. [Google Scholar] [CrossRef]
- Davarzani, F.; Saidi, N.; Besharati, S.; Saderi, H.; Rasooli, I.; Owlia, P. Evaluation of antibiotic resistance pattern, alginate and biofilm production in clinical isolates of Pseudomonas aeruginosa. Iran. J. Public. Health 2021, 50, 341. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).